α-Cyclodextrin/Moringin Induces an Antioxidant Transcriptional Response Activating Nrf2 in Differentiated NSC-34 Motor Neurons
Abstract
1. Introduction
1.1. Cruciferous Vegetables and Their Phytochemicals
1.2. Oxidative Stress and Its Role in Motor Neuron Degeneration
1.3. The Antioxidant Defense System and the Role of MOR
2. Materials and Methods
2.1. Preparation of the α-CD/MOR Complex
2.2. NSC-34 Culture, Differentiation, and Treatment
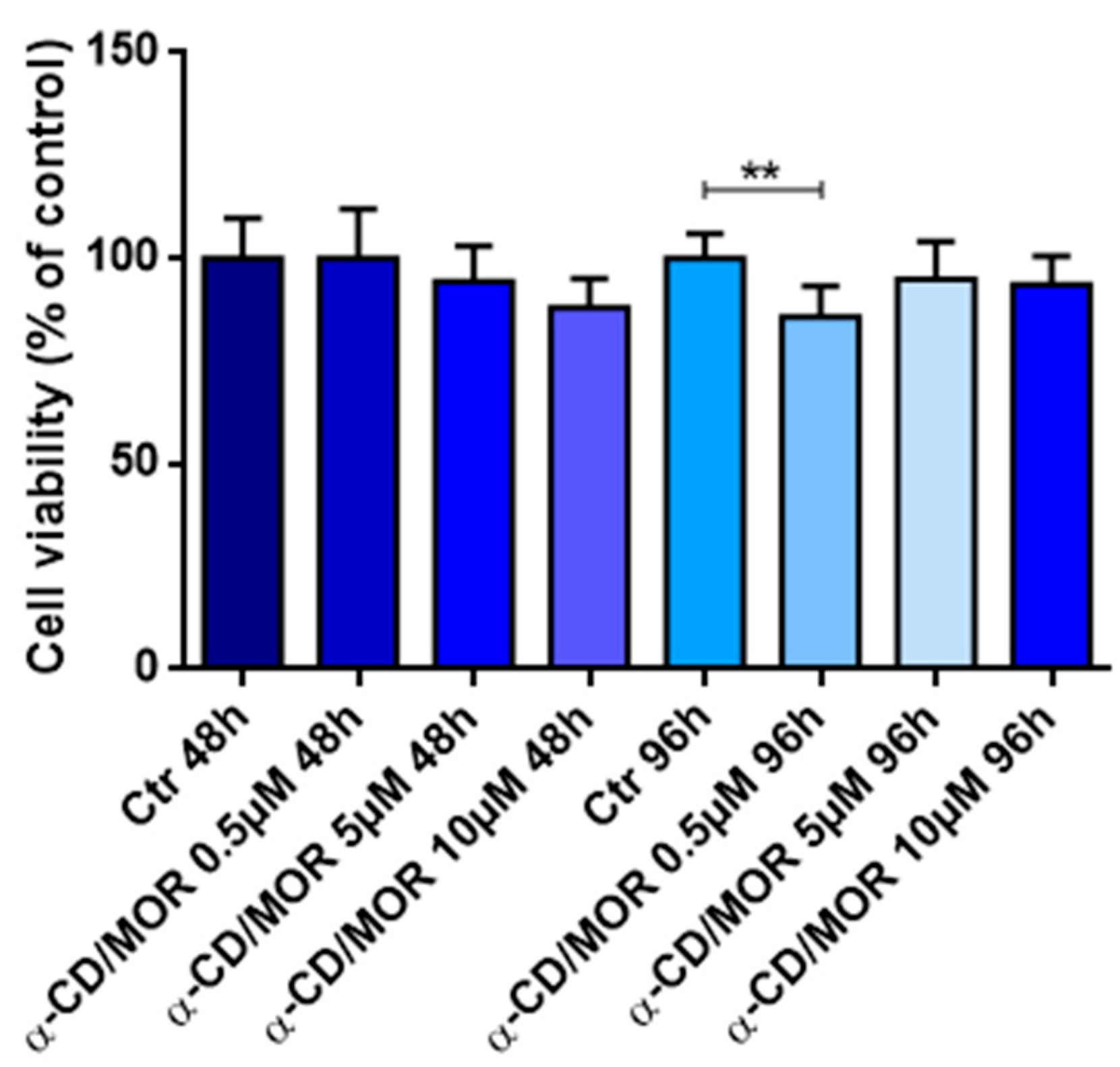
2.3. Cell Viability
2.4. RNA Extraction and Library Preparation
2.5. Comparative Transcriptomic and Linear Model Prediction
2.6. Nrf2 Network and Gene Ontology Analysis
2.7. Protein Extraction and Western Blot
2.8. Statistical Analysis
3. Results
3.1. Cell Viability
3.2. Transcriptomic Comparative Analysis
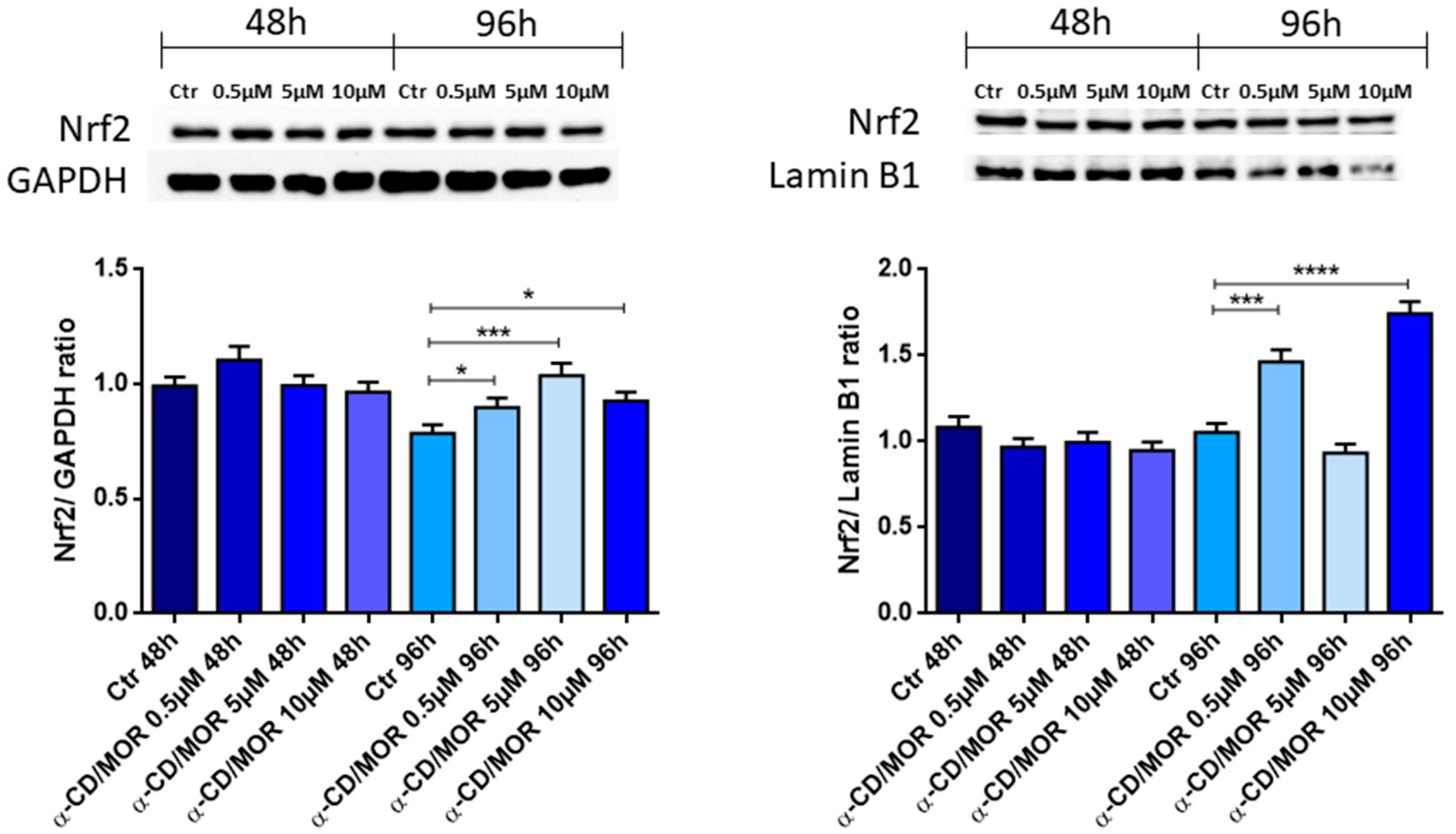
3.3. Nrf2 Gene and Protein Expression Analysis
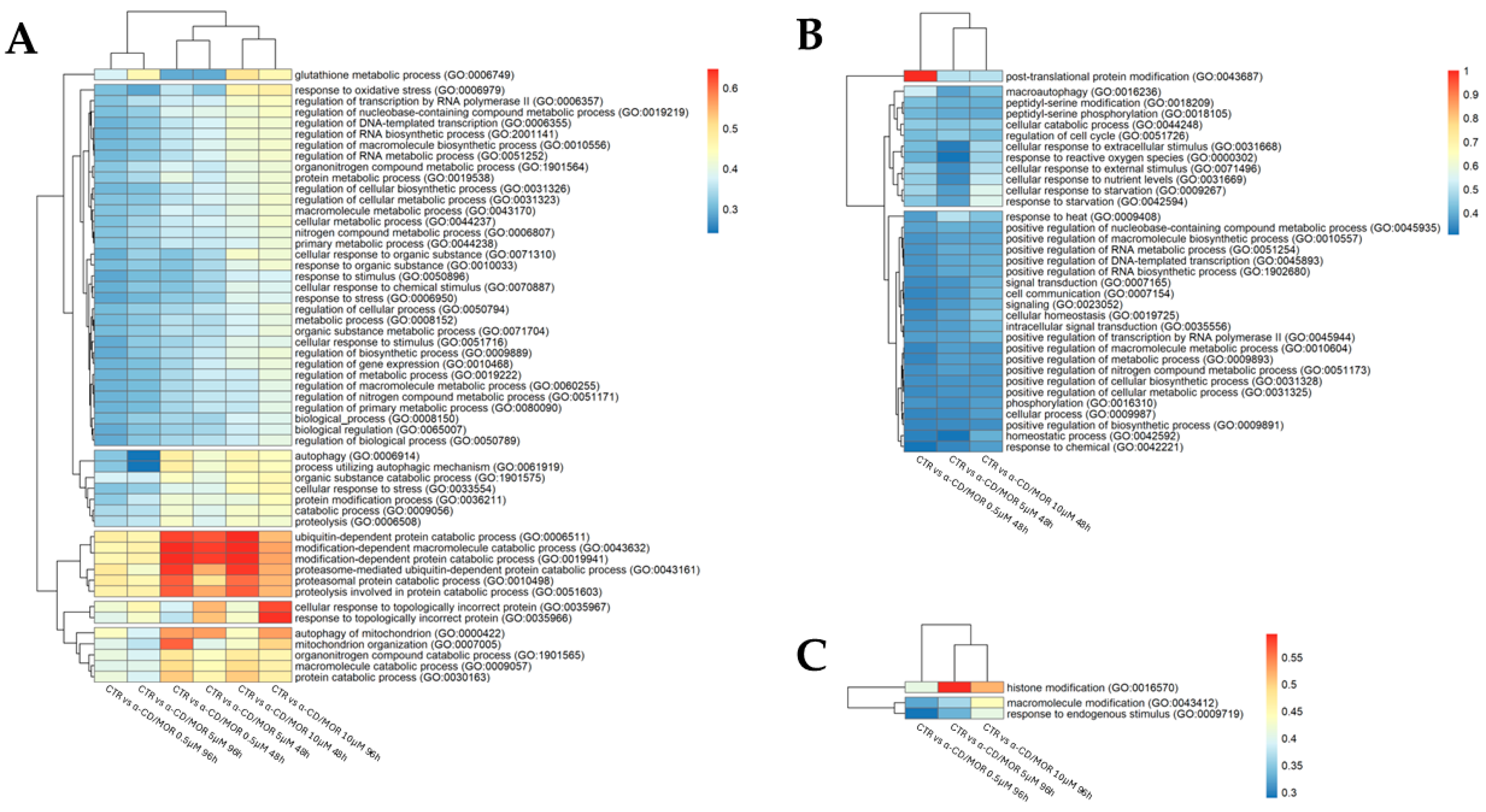
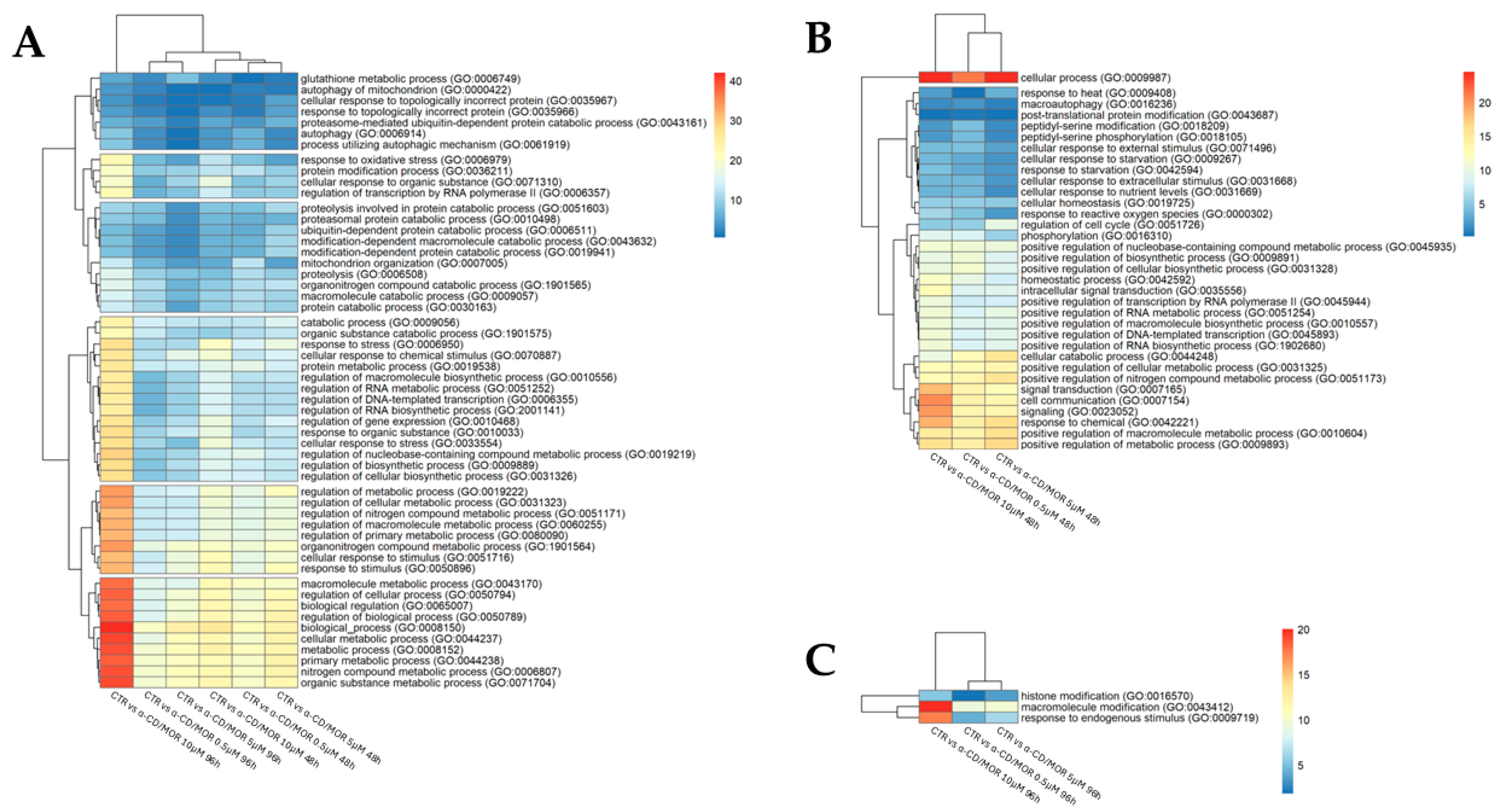
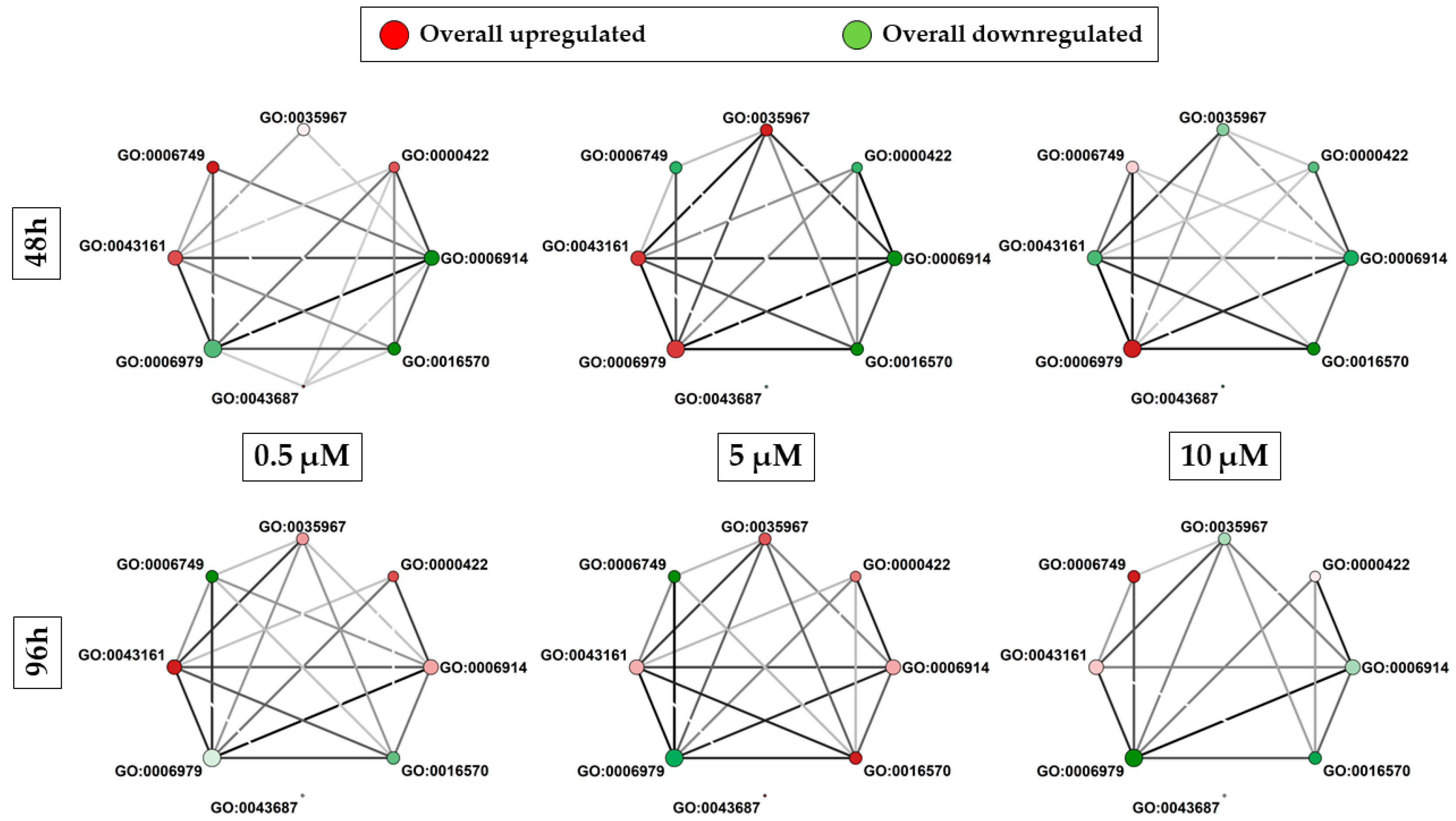
3.4. Nrf2 Interactor Selection and Gene Ontology (GO) Analysis
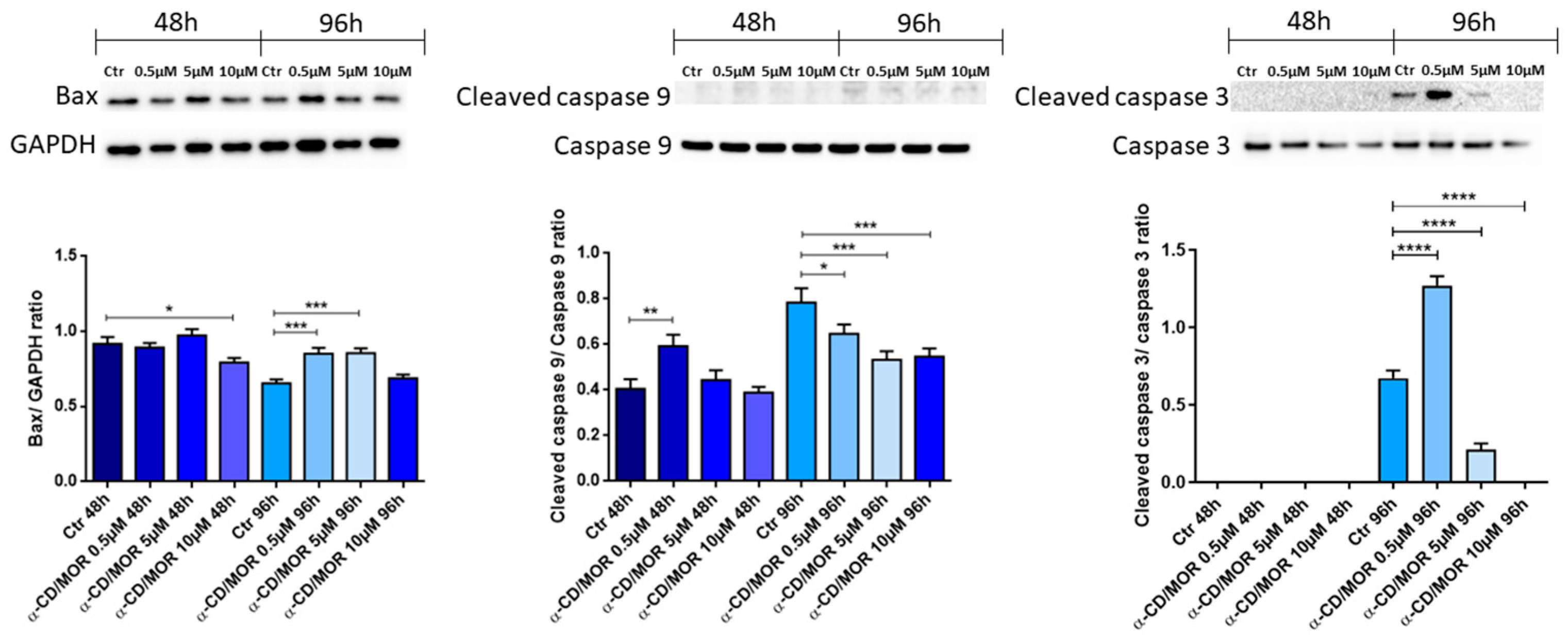
3.5. Western Blot Analysis
4. Discussion
4.1. Neuroprotective Capacity of MOR
4.2. MOR Can Activate Nrf2
4.3. BP Analysis of Nrf2 Interactors
4.4. α-CD/MOR Effects on GSH
4.5. α-CD/MOR Effects on Autophagy
4.6. α-CD/MOR Effects on Proteostasis
4.7. α-CD/MOR Effects on the Other Significant BP and Their Network
4.8. α-CD/MOR Effects on Apoptosis
4.9. Future Perspective
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Manchali, S.; Murthy, K.N.C.; Patil, B.S. Crucial facts about health benefits of popular cruciferous vegetables. J. Funct. Foods 2012, 4, 94–106. [Google Scholar] [CrossRef]
- Kou, X.; Li, B.; Olayanju, J.B.; Drake, J.M.; Chen, N. Nutraceutical or Pharmacological Potential of Moringa oleifera Lam. Nutrients 2018, 10, 343. [Google Scholar] [CrossRef] [PubMed]
- Kakarla, R.; Karuturi, P.; Siakabinga, Q.; Kasi Viswanath, M.; Dumala, N.; Guntupalli, C.; Nalluri, B.N.; Venkateswarlu, K.; Prasanna, V.S.; Gutti, G.; et al. Current understanding and future directions of cruciferous vegetables and their phytochemicals to combat neurological diseases. Phytother. Res. PTR 2024, 38, 1381–1399. [Google Scholar] [CrossRef] [PubMed]
- Na, G.; He, C.; Zhang, S.; Tian, S.; Bao, Y.; Shan, Y. Dietary Isothiocyanates: Novel Insights into the Potential for Cancer Prevention and Therapy. Int. J. Mol. Sci. 2023, 24, 1962. [Google Scholar] [CrossRef] [PubMed]
- Kamal, R.M.; Abdull Razis, A.F.; Mohd Sukri, N.S.; Perimal, E.K.; Ahmad, H.; Patrick, R.; Djedaini-Pilard, F.; Mazzon, E.; Rigaud, S. Beneficial Health Effects of Glucosinolates-Derived Isothiocyanates on Cardiovascular and Neurodegenerative Diseases. Molecules 2022, 27, 624. [Google Scholar] [CrossRef] [PubMed]
- Connolly, E.L.; Sim, M.; Travica, N.; Marx, W.; Beasy, G.; Lynch, G.S.; Bondonno, C.P.; Lewis, J.R.; Hodgson, J.M.; Blekkenhorst, L.C. Glucosinolates From Cruciferous Vegetables and Their Potential Role in Chronic Disease: Investigating the Preclinical and Clinical Evidence. Front. Pharmacol. 2021, 12, 767975. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Li, W.; Su, R.; Yang, M.; Zhang, N.; Li, X.; Li, L.; Sheng, J.; Tian, Y. Multi-Target Antibacterial Mechanism of Moringin From Moringa oleifera Seeds Against Listeria monocytogenes. Front. Microbiol. 2022, 13, 925291. [Google Scholar] [CrossRef]
- Jaafaru, M.S.; Nordin, N.; Shaari, K.; Rosli, R.; Abdull Razis, A.F. Isothiocyanate from Moringa oleifera seeds mitigates hydrogen peroxide-induced cytotoxicity and preserved morphological features of human neuronal cells. PLoS ONE 2018, 13, e0196403. [Google Scholar] [CrossRef] [PubMed]
- Manjunath, S.H.; Nataraj, P.; Swamy, V.H.; Sugur, K.; Dey, S.K.; Ranganathan, V.; Daniel, S.; Leihang, Z.; Sharon, V.; Chandrashekharappa, S.; et al. Development of Moringa oleifera as functional food targeting NRF2 signaling: Antioxidant and anti-inflammatory activity in experimental model systems. Food Funct. 2023, 14, 4734–4751. [Google Scholar] [CrossRef]
- Giacoppo, S.; Rajan, T.S.; Iori, R.; Rollin, P.; Bramanti, P.; Mazzon, E. The alpha-cyclodextrin complex of the Moringa isothiocyanate suppresses lipopolysaccharide-induced inflammation in RAW 264.7 macrophage cells through Akt and p38 inhibition. Inflamm. Res. 2017, 66, 487–503. [Google Scholar] [CrossRef]
- Rajan, T.S.; De Nicola, G.R.; Iori, R.; Rollin, P.; Bramanti, P.; Mazzon, E. Anticancer activity of glucomoringin isothiocyanate in human malignant astrocytoma cells. Fitoterapia 2016, 110, 1–7. [Google Scholar] [CrossRef]
- Silvestro, S.; Chiricosta, L.; Gugliandolo, A.; Iori, R.; Rollin, P.; Perenzoni, D.; Mattivi, F.; Bramanti, P.; Mazzon, E. The Moringin/alpha-CD Pretreatment Induces Neuroprotection in an In Vitro Model of Alzheimer’s Disease: A Transcriptomic Study. Curr. Issues Mol. Biol. 2021, 43, 197–214. [Google Scholar] [CrossRef] [PubMed]
- Giacoppo, S.; Soundara Rajan, T.; De Nicola, G.R.; Iori, R.; Bramanti, P.; Mazzon, E. Moringin activates Wnt canonical pathway by inhibiting GSK3beta in a mouse model of experimental autoimmune encephalomyelitis. Drug Des. Dev. Ther. 2016, 10, 3291–3304. [Google Scholar] [CrossRef] [PubMed]
- Giacoppo, S.; Rajan, T.S.; De Nicola, G.R.; Iori, R.; Rollin, P.; Bramanti, P.; Mazzon, E. The Isothiocyanate Isolated from Moringa oleifera Shows Potent Anti-Inflammatory Activity in the Treatment of Murine Subacute Parkinson’s Disease. Rejuvenation Res. 2017, 20, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Scarian, E.; Viola, C.; Dragoni, F.; Di Gerlando, R.; Rizzo, B.; Diamanti, L.; Gagliardi, S.; Bordoni, M.; Pansarasa, O. New Insights into Oxidative Stress and Inflammatory Response in Neurodegenerative Diseases. Int. J. Mol. Sci. 2024, 25, 2698. [Google Scholar] [CrossRef]
- Teleanu, D.M.; Niculescu, A.G.; Lungu, I.I.; Radu, C.I.; Vladacenco, O.; Roza, E.; Costachescu, B.; Grumezescu, A.M.; Teleanu, R.I. An Overview of Oxidative Stress, Neuroinflammation, and Neurodegenerative Diseases. Int. J. Mol. Sci. 2022, 23, 5938. [Google Scholar] [CrossRef] [PubMed]
- Obrador, E.; Salvador-Palmer, R.; Lopez-Blanch, R.; Jihad-Jebbar, A.; Valles, S.L.; Estrela, J.M. The Link between Oxidative Stress, Redox Status, Bioenergetics and Mitochondria in the Pathophysiology of ALS. Int. J. Mol. Sci. 2021, 22, 6352. [Google Scholar] [CrossRef] [PubMed]
- Dominko, K.; Dikic, D. Glutathionylation: A regulatory role of glutathione in physiological processes. Arh. Za Hig. Rada I Toksikol. 2018, 69, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Ngo, V.; Duennwald, M.L. Nrf2 and Oxidative Stress: A General Overview of Mechanisms and Implications in Human Disease. Antioxidants 2022, 11, 2345. [Google Scholar] [CrossRef]
- Baird, L.; Yamamoto, M. The Molecular Mechanisms Regulating the KEAP1-NRF2 Pathway. Mol. Cell. Biol. 2020, 40, e00099-20. [Google Scholar] [CrossRef]
- Houghton, C.A.; Fassett, R.G.; Coombes, J.S. Sulforaphane and Other Nutrigenomic Nrf2 Activators: Can the Clinician’s Expectation Be Matched by the Reality? Oxidative Med. Cell. Longev. 2016, 2016, 7857186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, L.; Xu, M.; Jiang, P.; Zhang, K. Moringin alleviates DSS-induced ulcerative colitis in mice by regulating Nrf2/NF-kappaB pathway and PI3K/AKT/mTOR pathway. Int. Immunopharmacol. 2024, 134, 112241. [Google Scholar] [CrossRef] [PubMed]
- Mathiron, D.; Iori, R.; Pilard, S.; Soundara Rajan, T.; Landy, D.; Mazzon, E.; Rollin, P.; Djedaini-Pilard, F. A Combined Approach of NMR and Mass Spectrometry Techniques Applied to the alpha-Cyclodextrin/Moringin Complex for a Novel Bioactive Formulation (dagger). Molecules 2018, 23, 1714. [Google Scholar] [CrossRef] [PubMed]
- Brunelli, D.; Tavecchio, M.; Falcioni, C.; Frapolli, R.; Erba, E.; Iori, R.; Rollin, P.; Barillari, J.; Manzotti, C.; Morazzoni, P.; et al. The isothiocyanate produced from glucomoringin inhibits NF-kB and reduces myeloma growth in nude mice in vivo. Biochem. Pharmacol. 2010, 79, 1141–1148. [Google Scholar] [CrossRef] [PubMed]
- Amaglo, N.K.; Bennett, R.N.; Lo Curto, R.B.; Rosa, E.A.S.; Lo Turco, V.; Giuffrida, A.; Lo Curto, A.; Crea, F.; Timpo, G.M. Profiling selected phytochemicals and nutrients in different tissues of the multipurpose tree Moringa oleifera L., grown in Ghana. Food Chem. 2010, 122, 1047–1054. [Google Scholar] [CrossRef]
- Waterman, C.; Cheng, D.M.; Rojas-Silva, P.; Poulev, A.; Dreifus, J.; Lila, M.A.; Raskin, I. Stable, water extractable isothiocyanates from Moringa oleifera leaves attenuate inflammation. Phytochemistry 2014, 103, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef] [PubMed]
- Pall, M.L.; Levine, S. Nrf2, a master regulator of detoxification and also antioxidant, anti-inflammatory and other cytoprotective mechanisms, is raised by health promoting factors. Sheng Li Xue Bao 2015, 67, 1–18. [Google Scholar] [PubMed]
- Dayalan Naidu, S.; Suzuki, T.; Yamamoto, M.; Fahey, J.W.; Dinkova-Kostova, A.T. Phenethyl Isothiocyanate, a Dual Activator of Transcription Factors NRF2 and HSF1. Mol. Nutr. Food Res. 2018, 62, e1700908. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J.; Kozumbo, W.J. The phytoprotective agent sulforaphane prevents inflammatory degenerative diseases and age-related pathologies via Nrf2-mediated hormesis. Pharmacol. Res. 2021, 163, 105283. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.; Spagnuolo, C.; Russo, G.L.; Skalicka-Wozniak, K.; Daglia, M.; Sobarzo-Sanchez, E.; Nabavi, S.F.; Nabavi, S.M. Nrf2 targeting by sulforaphane: A potential therapy for cancer treatment. Crit. Rev. Food Sci. Nutr. 2018, 58, 1391–1405. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, K. Glutathione in the Brain. Int. J. Mol. Sci. 2021, 22, 5010. [Google Scholar] [CrossRef] [PubMed]
- Iskusnykh, I.Y.; Zakharova, A.A.; Pathak, D. Glutathione in Brain Disorders and Aging. Molecules 2022, 27, 324. [Google Scholar] [CrossRef] [PubMed]
- Nishida Xavier da Silva, T.; Friedmann Angeli, J.P.; Ingold, I. GPX4: Old lessons, new features. Biochem. Soc. Trans. 2022, 50, 1205–1213. [Google Scholar] [CrossRef]
- Imai, H.; Matsuoka, M.; Kumagai, T.; Sakamoto, T.; Koumura, T. Lipid Peroxidation-Dependent Cell Death Regulated by GPx4 and Ferroptosis. Curr. Top. Microbiol. Immunol. 2017, 403, 143–170. [Google Scholar] [CrossRef]
- Liu, Y.; Wan, Y.; Jiang, Y.; Zhang, L.; Cheng, W. GPX4: The hub of lipid oxidation, ferroptosis, disease and treatment. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188890. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Ru, X.; Wen, T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Harder, B.; Rojo de la Vega, M.; Wong, P.K.; Chapman, E.; Zhang, D.D. p62 links autophagy and Nrf2 signaling. Free Radic. Biol. Med. 2015, 88, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M. p62 bodies: Phase separation, NRF2 activation, and selective autophagic degradation. IUBMB Life 2022, 74, 1200–1208. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.; Wang, X.J.; Zhao, F.; Villeneuve, N.F.; Wu, T.; Jiang, T.; Sun, Z.; White, E.; Zhang, D.D. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: Direct interaction between Keap1 and p62. Mol. Cell. Biol. 2010, 30, 3275–3285. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010, 12, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Copple, I.M.; Lister, A.; Obeng, A.D.; Kitteringham, N.R.; Jenkins, R.E.; Layfield, R.; Foster, B.J.; Goldring, C.E.; Park, B.K. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. J. Biol. Chem. 2010, 285, 16782–16788. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Lamark, T.; Sjottem, E.; Larsen, K.B.; Awuh, J.A.; Overvatn, A.; McMahon, M.; Hayes, J.D.; Johansen, T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010, 285, 22576–22591. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Liu, R.; Dong, X.; Zhong, Q. Beclin orthologs: Integrative hubs of cell signaling, membrane trafficking, and physiology. Trends Cell Biol. 2015, 25, 533–544. [Google Scholar] [CrossRef]
- Schaaf, M.B.; Keulers, T.G.; Vooijs, M.A.; Rouschop, K.M. LC3/GABARAP family proteins: Autophagy-(un)related functions. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2016, 30, 3961–3978. [Google Scholar] [CrossRef]
- Su, S.H.; Wu, Y.F.; Wang, D.P.; Hai, J. Inhibition of excessive autophagy and mitophagy mediates neuroprotective effects of URB597 against chronic cerebral hypoperfusion. Cell Death Dis. 2018, 9, 733. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Li, Q.; Liu, L.; Wu, H.; Huang, F.; Wang, C.; Lan, Y.; Zheng, F.; Xing, F.; Zhou, Q.; et al. Modafinil protects hippocampal neurons by suppressing excessive autophagy and apoptosis in mice with sleep deprivation. Br. J. Pharmacol. 2019, 176, 1282–1297. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, L.; Chen, S.; Yang, D.; Wang, Y.; Zhang, X.; Wang, Z.; Le, W. Rapamycin treatment augments motor neuron degeneration in SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Autophagy 2011, 7, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Perera, N.D.; Sheean, R.K.; Lau, C.L.; Shin, Y.S.; Beart, P.M.; Horne, M.K.; Turner, B.J. Rilmenidine promotes MTOR-independent autophagy in the mutant SOD1 mouse model of amyotrophic lateral sclerosis without slowing disease progression. Autophagy 2018, 14, 534–551. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, I.M.; Abdelmalek, D.H.; Elfiky, A.A. GRP78: A cell’s response to stress. Life Sci. 2019, 226, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Oyadomari, S.; Mori, M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004, 11, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.C. Nuclear Factor-Erythroid 2-Related Factor 2 (Nrf2) and Mitochondrial Dynamics/Mitophagy in Neurological Diseases. Antioxidants 2020, 9, 617. [Google Scholar] [CrossRef] [PubMed]
- Esteras, N.; Abramov, A.Y. Nrf2 as a regulator of mitochondrial function: Energy metabolism and beyond. Free Radic. Biol. Med. 2022, 189, 136–153. [Google Scholar] [CrossRef] [PubMed]
- Lazo-Gomez, R.; Ramirez-Jarquin, U.N.; Tovar, Y.R.L.B.; Tapia, R. Histone deacetylases and their role in motor neuron degeneration. Front. Cell. Neurosci. 2013, 7, 243. [Google Scholar] [CrossRef]
- Abbaoui, B.; Telu, K.H.; Lucas, C.R.; Thomas-Ahner, J.M.; Schwartz, S.J.; Clinton, S.K.; Freitas, M.A.; Mortazavi, A. The impact of cruciferous vegetable isothiocyanates on histone acetylation and histone phosphorylation in bladder cancer. J. Proteom. 2017, 156, 94–103. [Google Scholar] [CrossRef]
- Adebayo, O.G.; Wopara, I.; Aduema, W.; Ebo, O.T.; Umoren, E.B. Long-term consumption of Moringa oleifera-supplemented diet enhanced neurocognition, suppressed oxidative stress, acetylcholinesterase activity and neuronal degeneration in rat’s hippocampus. Drug Metab. Pers. Ther. 2021, 36, 223–231. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gugliandolo, A.; Calì, G.; Muscarà, C.; Artimagnella, O.; Rollin, P.; Perenzoni, D.; Iori, R.; Mazzon, E.; Chiricosta, L. α-Cyclodextrin/Moringin Induces an Antioxidant Transcriptional Response Activating Nrf2 in Differentiated NSC-34 Motor Neurons. Antioxidants 2024, 13, 813. https://doi.org/10.3390/antiox13070813
Gugliandolo A, Calì G, Muscarà C, Artimagnella O, Rollin P, Perenzoni D, Iori R, Mazzon E, Chiricosta L. α-Cyclodextrin/Moringin Induces an Antioxidant Transcriptional Response Activating Nrf2 in Differentiated NSC-34 Motor Neurons. Antioxidants. 2024; 13(7):813. https://doi.org/10.3390/antiox13070813
Chicago/Turabian StyleGugliandolo, Agnese, Gabriella Calì, Claudia Muscarà, Osvaldo Artimagnella, Patrick Rollin, Daniele Perenzoni, Renato Iori, Emanuela Mazzon, and Luigi Chiricosta. 2024. "α-Cyclodextrin/Moringin Induces an Antioxidant Transcriptional Response Activating Nrf2 in Differentiated NSC-34 Motor Neurons" Antioxidants 13, no. 7: 813. https://doi.org/10.3390/antiox13070813
APA StyleGugliandolo, A., Calì, G., Muscarà, C., Artimagnella, O., Rollin, P., Perenzoni, D., Iori, R., Mazzon, E., & Chiricosta, L. (2024). α-Cyclodextrin/Moringin Induces an Antioxidant Transcriptional Response Activating Nrf2 in Differentiated NSC-34 Motor Neurons. Antioxidants, 13(7), 813. https://doi.org/10.3390/antiox13070813





