Multi-Omics Reveals the Effects of Spirulina platensis Powder Replacement of Fish Meal on Intestinal Metabolism and Stress in Zig-Zag Eel (Mastacembelus armatus)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Statement of Ethics in Animal Experimentation
2.2. Diet and Nutrition
2.3. Sample Collection
2.4. Biochemical Indices Assay
2.5. Transcriptomics Testing
2.6. Metabolomics Assays
2.7. RT-qPCR to Detect the Expression Pattern of Related Genes
2.8. Statistical Analysis
3. Results
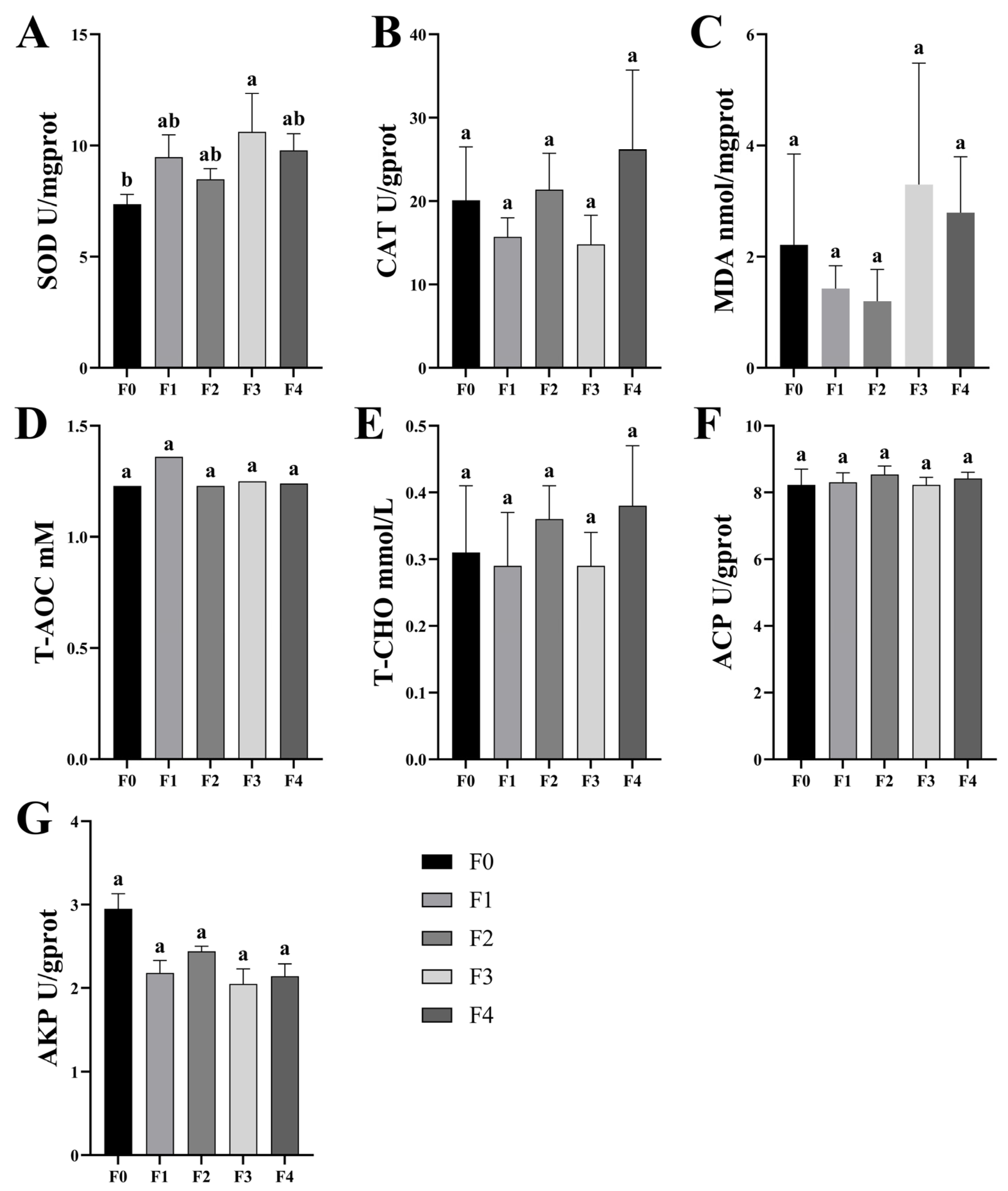
3.1. Effects of Different Ratios of SPP on the Antioxidant Capacity of MA’s Intestinal and Serum
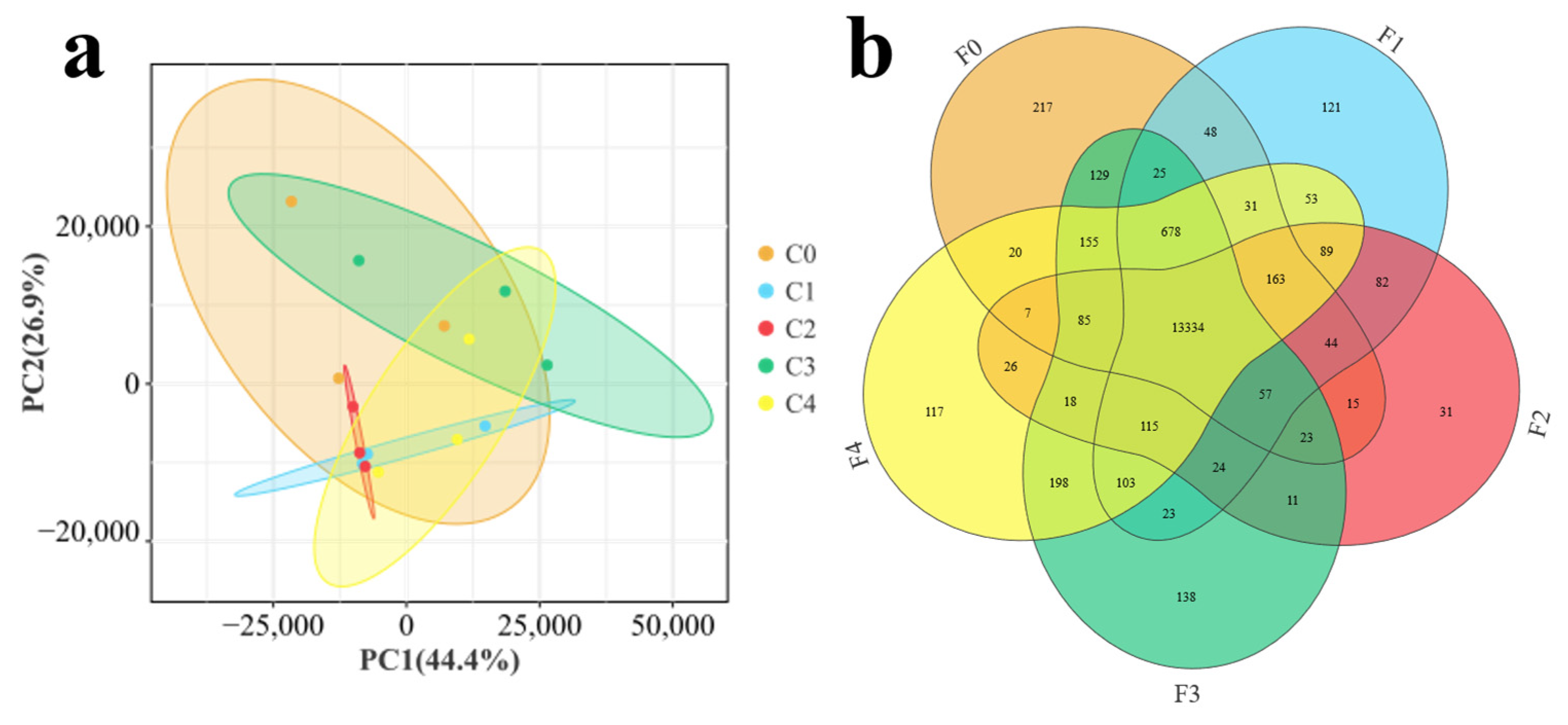
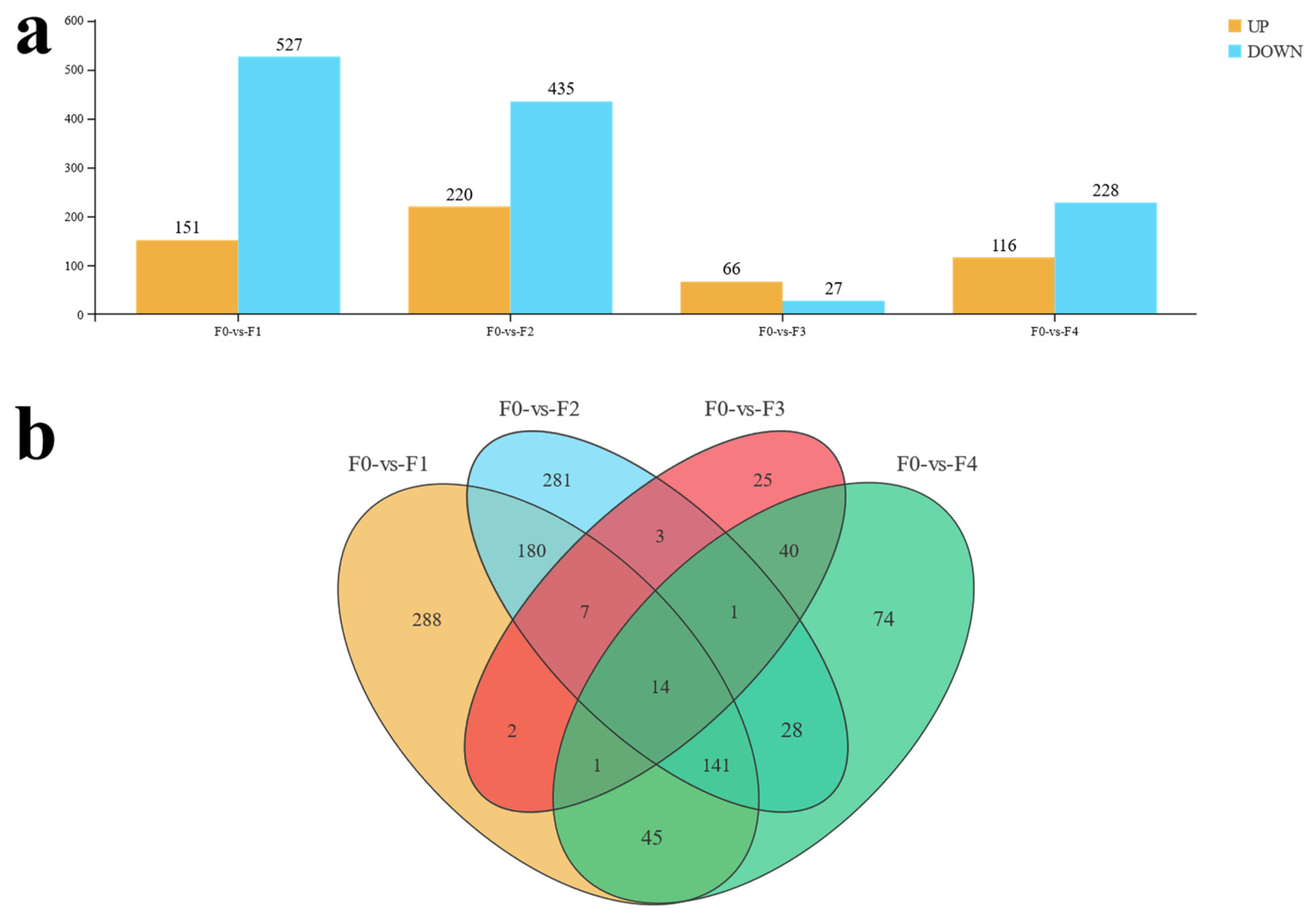
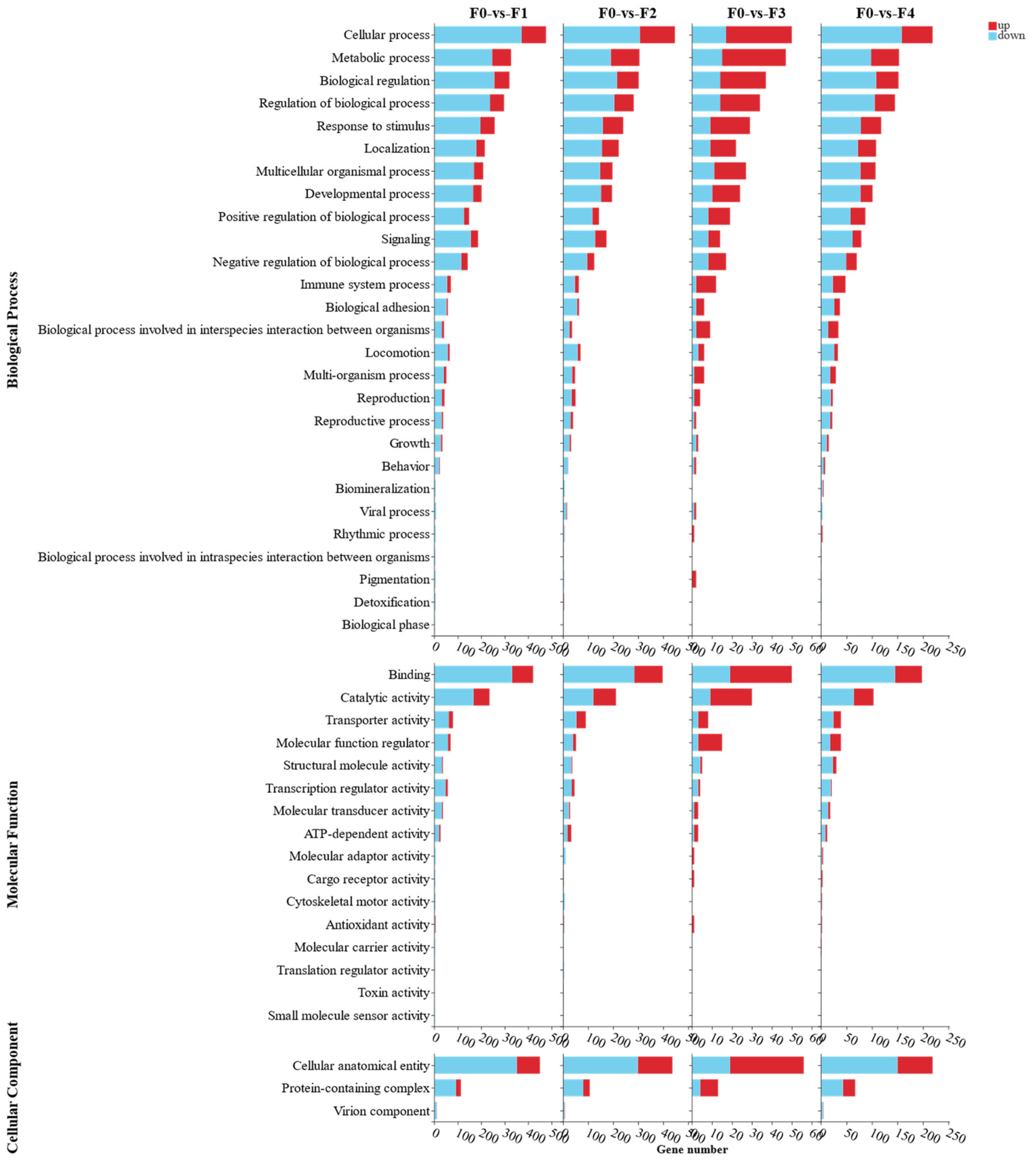
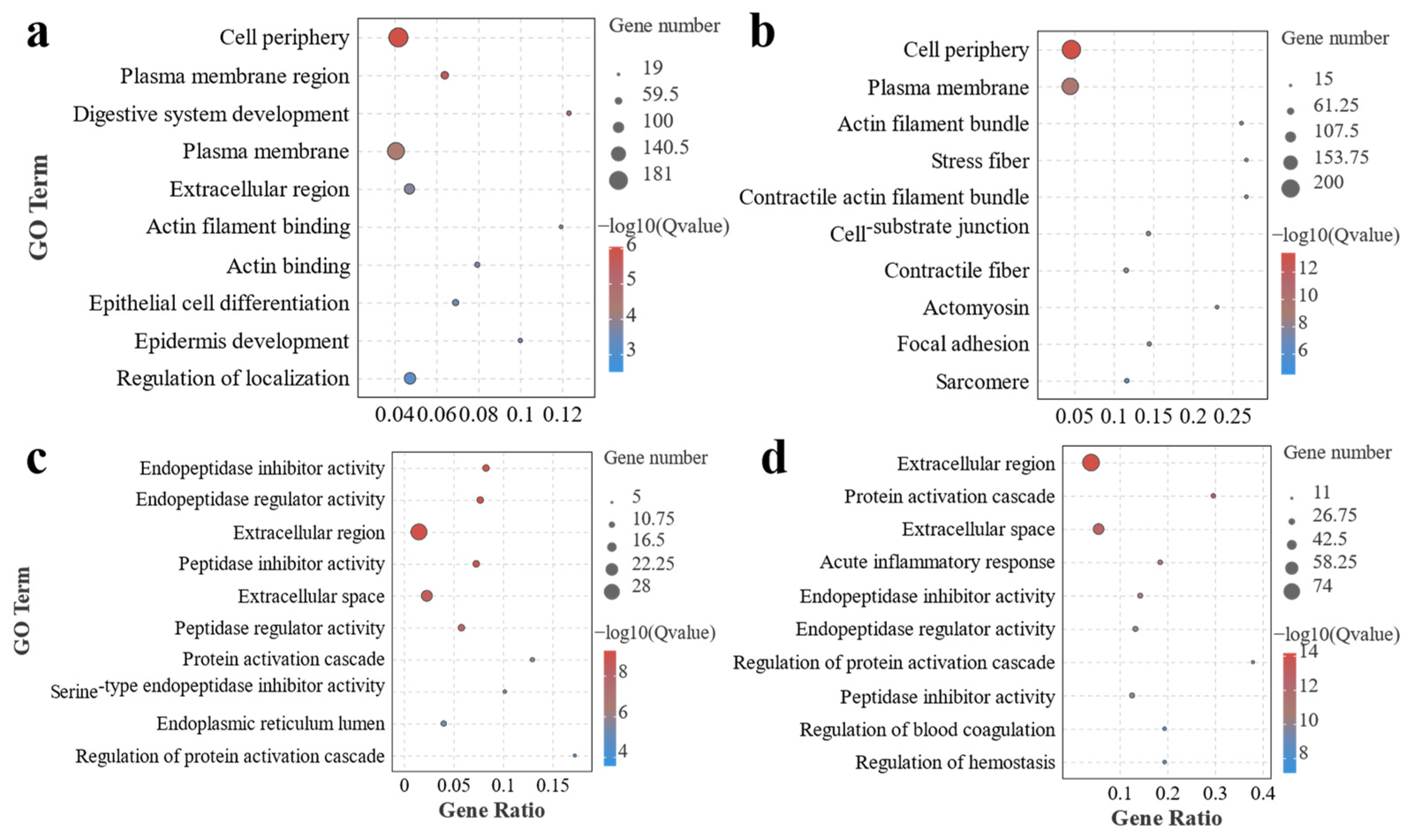
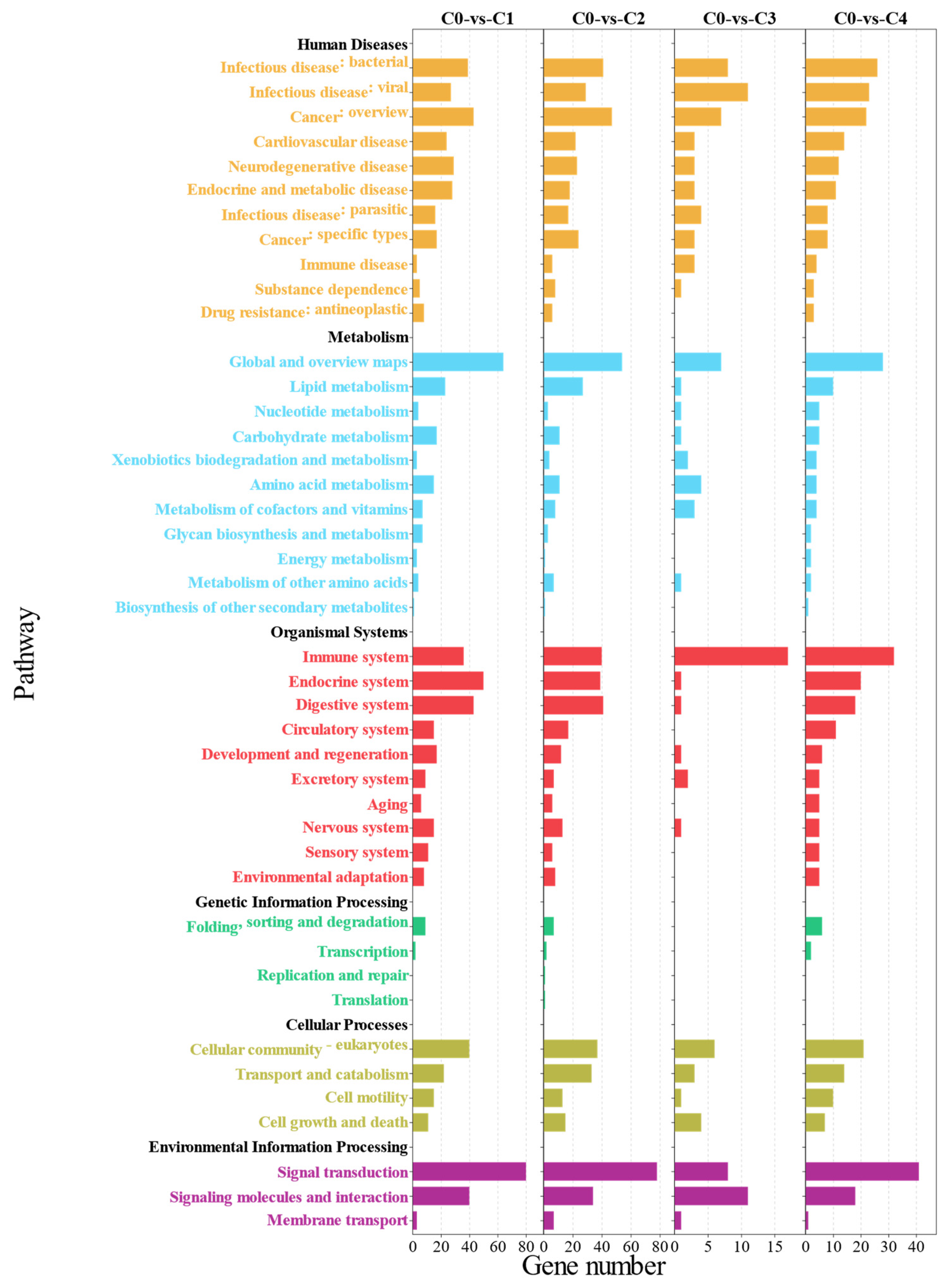
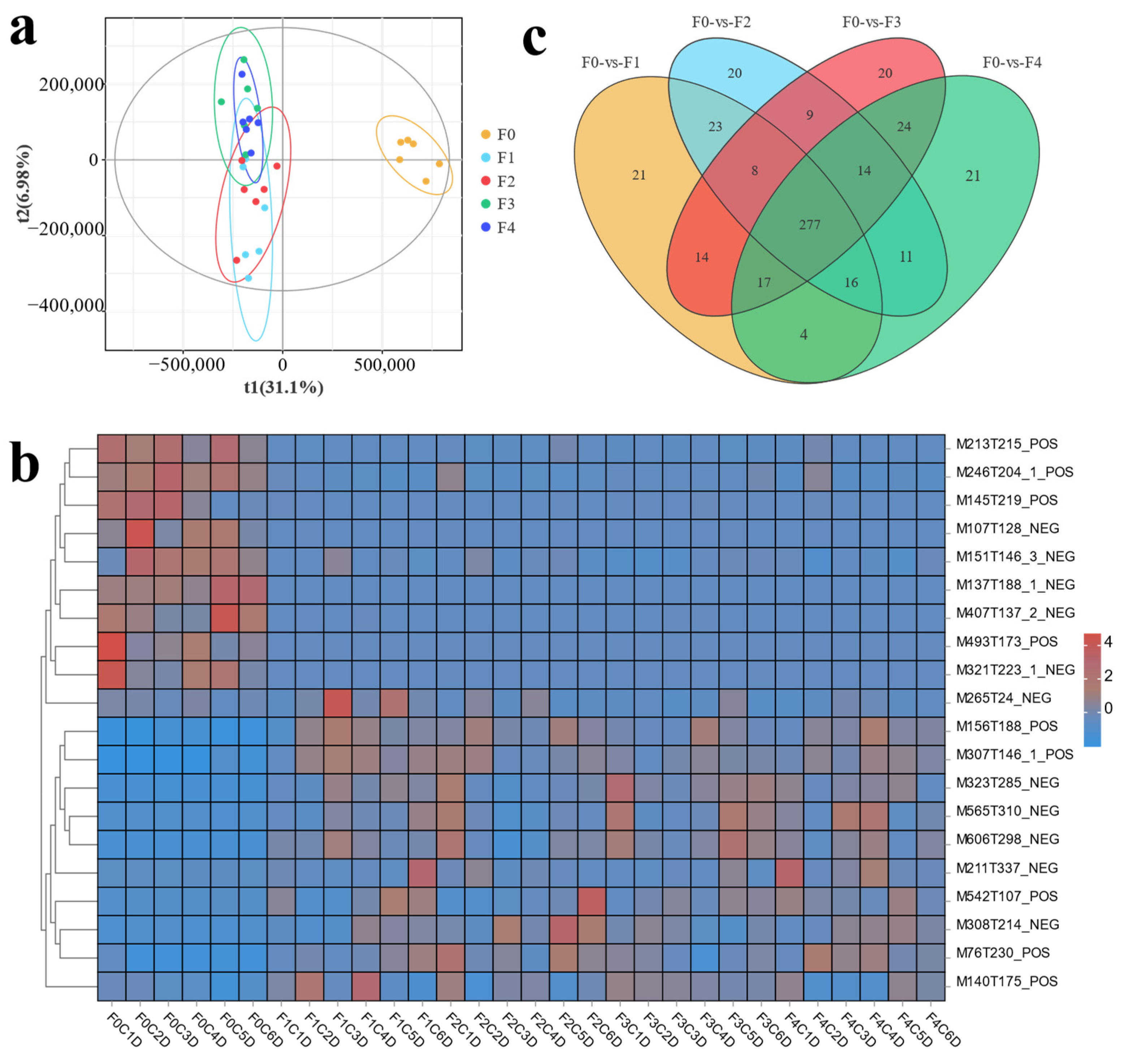
3.2. Transcriptome Analysis of Effects of Different Ratios of SPP on MA’s Intestine
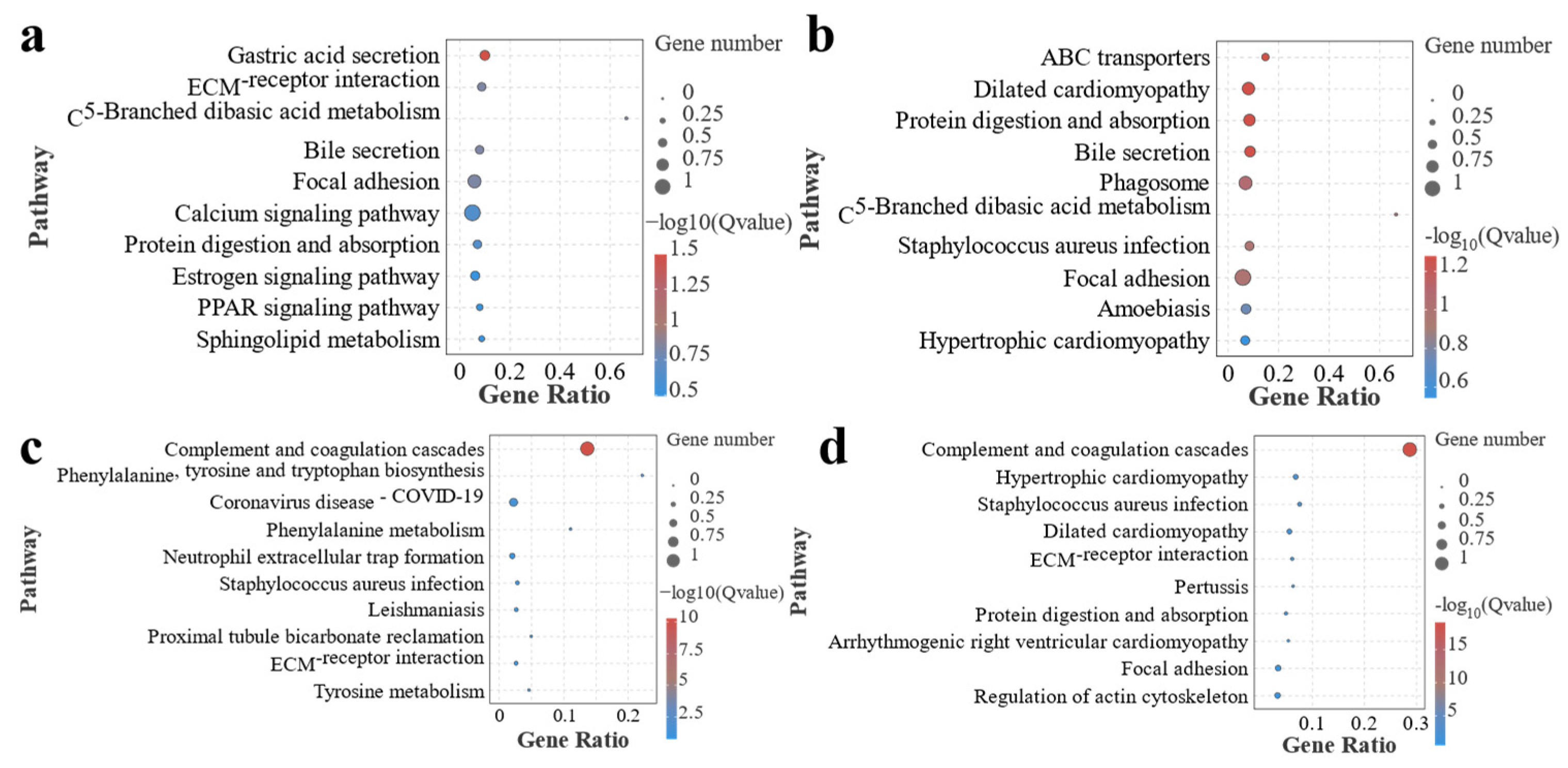
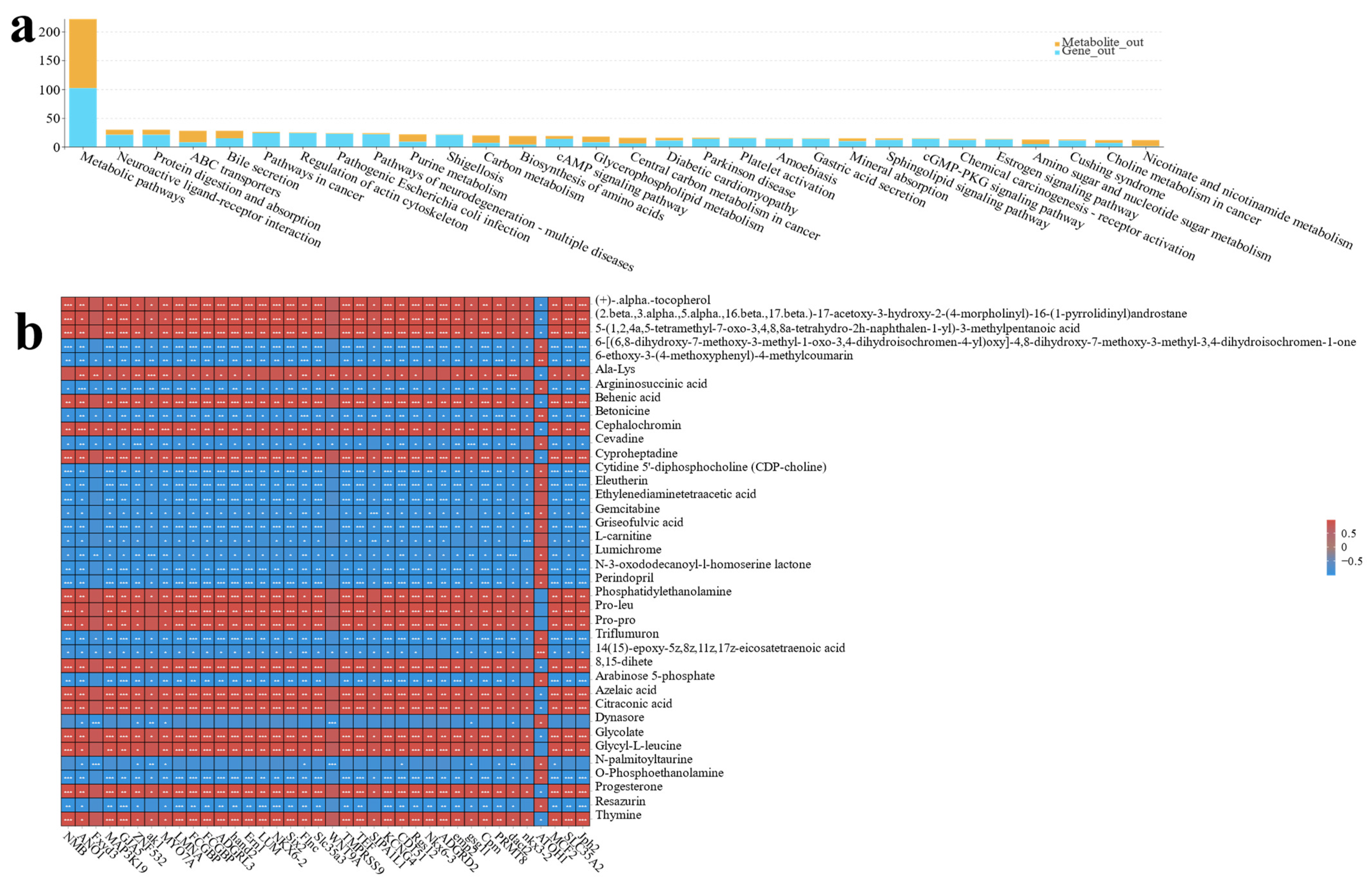
3.3. Metabolome Analysis of Effects of Different Ratios of SPP on MA’s Intestine
3.4. Co-Analysis of Transcriptome and Metabolome Reveals the Effects of Different Proportions of Spirulina on MA’s Intestine
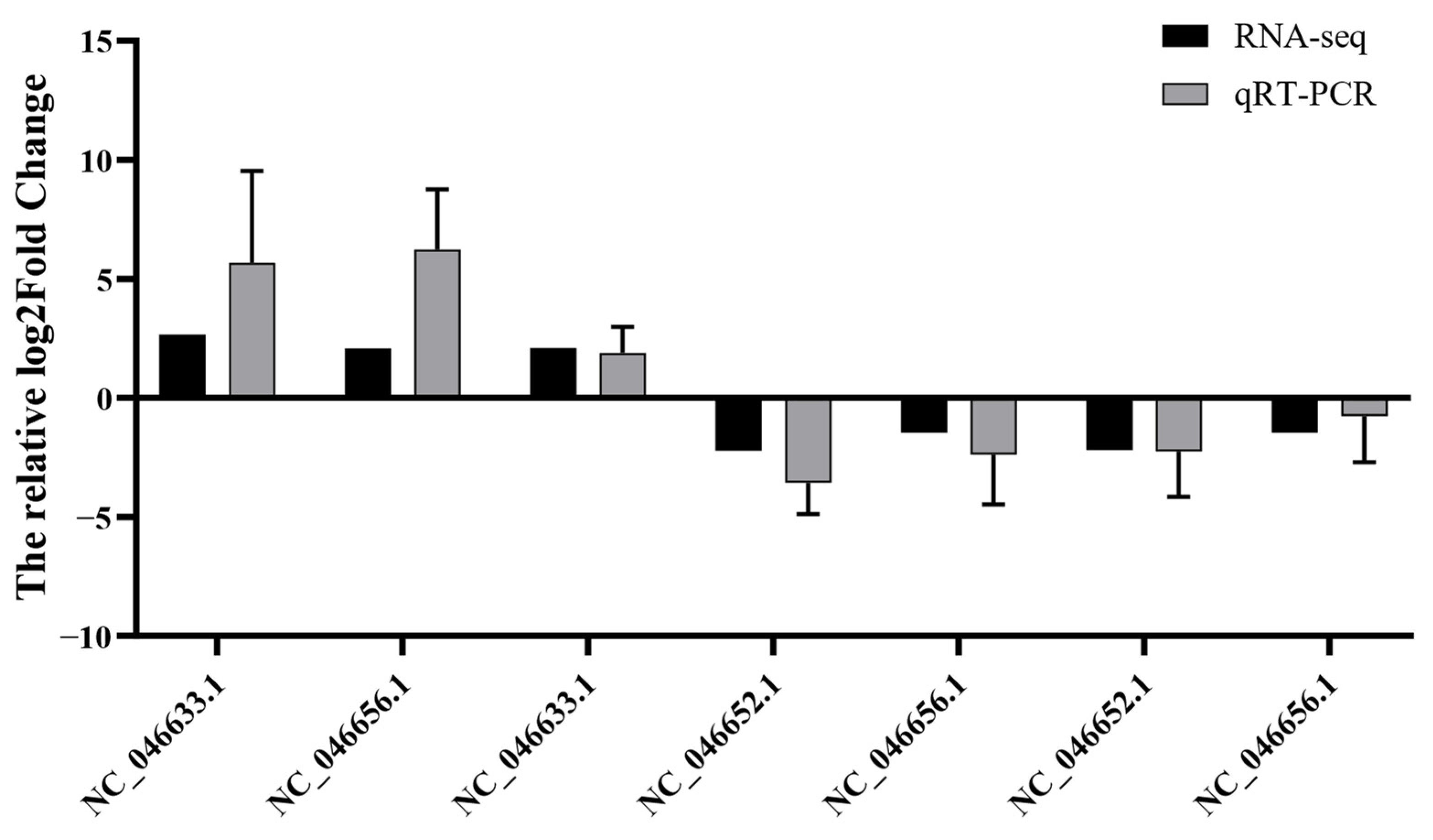
3.5. Validation of the Screened mRNAs by RT-qPCR
4. Discussion
4.1. Effect of Different Ratios of SPP on Intestinal and Serum Antioxidant Capacity of MA
4.2. Transcriptomics Reveals the Effects of Different Ratios of SPP on the Intestine of MA
4.3. Metabolomics Reveals the Effects of Different Ratios of SPP on the Intestine in MA
4.4. Combined Transcriptomics and Metabolomics Analysis Reveals the Effects of Different Ratios of SPP on the Intestine of MA
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pereira, A.G.; Fraga-Corral, M.; Garcia-Oliveira, P.; Otero, P.; Soria-Lopez, A.; Cassani, L.; Cao, H.; Xiao, J.; Prieto, M.A.; Simal-Gandara, J. Single-Cell Proteins Obtained by Circular Economy Intended as a Feed Ingredient in Aquaculture. Foods 2022, 11, 2831. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Yao, W.; Leng, X. Dietary effects of cottonseed protein concentrate substituting fishmeal on the growth and flesh quality of Pacific white shrimp (Litopenaeus vannamei). Aquac. Rep. 2023, 29, 101482. [Google Scholar] [CrossRef]
- Hua, K.; Cobcroft, J.M.; Cole, A.; Condon, K.; Jerry, D.R.; Mangott, A.; Praeger, C.; Vucko, M.J.; Zeng, C.; Zenger, K.; et al. The Future of Aquatic Protein: Implications for Protein Sources in Aquaculture Diets. One Earth 2019, 1, 316–329. [Google Scholar] [CrossRef]
- Savonitto, G.; Barkan, R.; Harpaz, S.; Neori, A.; Chernova, H.; Terlizzi, A.; Guttman, L. Fishmeal replacement by periphyton reduces the fish in fish out ratio and alimentation cost in gilthead sea bream Sparus aurata. Sci. Rep. 2021, 11, 20990. [Google Scholar] [CrossRef]
- Siddik, M.A.B.; Sørensen, M.; Islam, S.M.M.; Saha, N.; Rahman, M.A.; Francis, D.S. Expanded utilisation of microalgae in global aquafeeds. Rev. Aquac. 2024, 16, 6–33. [Google Scholar] [CrossRef]
- Trevi, S.; Uren Webster, T.; Consuegra, S.; Garcia de Leaniz, C. Benefits of the microalgae Spirulina and Schizochytrium in fish nutrition: A meta-analysis. Sci. Rep. 2023, 13, 2208. [Google Scholar] [CrossRef] [PubMed]
- Sim, Y.J.; Cho, S.H.; Kim, K.; Jeong, S. Effect of substituting fish meal with various by-product meals of swine-origin in diet on olive flounder (Paralichthys olivaceus). Aquac. Rep. 2023, 33, 101844. [Google Scholar] [CrossRef]
- Aragão, C.; Gonçalves, A.T.; Costas, B.; Azeredo, R.; Xavier, M.J.; Engrola, S. Alternative Proteins for Fish Diets: Implications beyond Growth. Animals 2022, 12, 1211. [Google Scholar] [CrossRef]
- Mugwanya, M.; Dawood, M.A.O.; Kimera, F.; Sewilam, H. Replacement of fish meal with fermented plant proteins in the aquafeed industry: A systematic review and meta-analysis. Rev. Aquac. 2023, 15, 62–88. [Google Scholar] [CrossRef]
- Langi, S.; Panana, E.; Alloo, C.; Van Stappen, G.; Meeus, W. Replacement of fishmeal using poultry-based protein sources in feeds for pikeperch (Sander lucioperca, Linnaeus, 1758) during grow out phase. Aquac. Int. 2023, 31, 65–80. [Google Scholar] [CrossRef]
- Flores-Moreno, S.; Vizcaíno, A.J.; Sáez, M.I.; Macías-Vidal, J.; Martínez, T.F.; Martos-Sitcha, J.A.; Alarcón-López, F.J. Effects of Phytase and Microalgae Supplementation on the Utilization of Aquafeeds for European Seabass (Dicentrarchus labrax) Formulated with a High Inclusion Level of Plant Protein. Aquac. Res. 2024, 2024, 4775004. [Google Scholar] [CrossRef]
- Marchi, A.; Benini, E.; Dondi, F.; Ferrari, M.G.; Scicchitano, D.; Palladino, G.; Candela, M.; Cerri, R.; Di Biase, A.; Vizcaíno, A.J.; et al. The use of fishery and aquaculture by-products with Nannochloropsis sp. allows total dietary replacement of wild-caught fishmeal, fish oil and soy protein in European sea bass juveniles. Aquaculture 2024, 590, 741015. [Google Scholar] [CrossRef]
- Kim, S.W.; Less, J.F.; Wang, L.; Yan, T.; Kiron, V.; Kaushik, S.J.; Lei, X.G. Meeting Global Feed Protein Demand: Challenge, Opportunity, and Strategy. Annu. Rev. Anim. Biosci. 2019, 7, 221–243. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Pang, A.; Tan, B.; Xin, Y.; Liu, Y.; Xie, R.; Zhang, H.; Yang, Q.; Deng, J.; Chi, S. Tryptophan metabolism and gut flora profile in different soybean protein induced enteritis of pearl gentian groupers. Front. Nutr. 2022, 9, 1014502. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Zhang, S.; Dong, X.; Chi, S.; Yang, Q.; Liu, H.; Tan, B.; Xie, S. Effects of fishmeal replacement by black soldier fly on growth performance, digestive enzyme activity, intestine morphology, intestinal flora and immune response of pearl gentian grouper (Epinephelus fuscoguttatus ♀ × Epinephelus lanceolatus ♂). Fish Shellfish Immunol. 2022, 120, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Dixit, R.B.; Sagaram, U.S.; Gocher, C.; Krishna Kumar, G.R.; Dasgupta, S. Biomolecular characterisation of marine microalga in comparison to fishmeal and soymeal as an alternative feed ingredient. Phytochem. Anal. PCA 2022, 33, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, H.; Zhang, P. Effect of Spirulina Meal Supplementation on Growth Performance and Feed Utilization in Fish and Shrimp: A Meta-Analysis. Aquac. Nutr. 2022, 2022, 8517733. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Sun, D.; Wang, D.; Ye, B.; Wang, S.; Zhou, A.; Dong, Z.; Zou, J. Effect of dietary koumine on the immune and antioxidant status of carp (Cyprinus carpio) after Aeromonas hydrophila infection. Microb. Pathog. 2024, 186, 106464. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Cai, M.; Xie, S.; Hu, Y.; Zhang, J. Evaluation of soybean meal replacement by Clostridium autoethanogenum protein in diets for juvenile grass carp (Ctenopharyngodon idellus). Aquaculture 2024, 592, 741218. [Google Scholar] [CrossRef]
- Al Mamun, M.; Hossain, M.; Saha, J.; Khan, S.; Akter, T.; Banu, M. Effects of spirulina Spirulina platensis meal as a feed additive on growth performance and immunological response of Gangetic mystus Mystus cavasius. Aquac. Rep. 2023, 30, 101553. [Google Scholar] [CrossRef]
- Awad, L.Z.; El-Mahallawy, H.S.; Abdelnaeim, N.S.; Mahmoud, M.M.A.; Dessouki, A.A.; ElBanna, N.I. Role of dietary Spirulina platensis and betaine supplementation on growth, hematological, serum biochemical parameters, antioxidant status, immune responses, and disease resistance in Nile tilapia. Fish Shellfish Immunol. 2022, 126, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Dam, C.T.M.; Ventura, T.; Booth, M.; Pirozzi, I.; Salini, M.; Smullen, R.; Elizur, A. Intestinal Transcriptome Analysis Highlights Key Differentially Expressed Genes Involved in Nutrient Metabolism and Digestion in Yellowtail Kingfish (Seriola lalandi) Fed Terrestrial Animal and Plant Proteins. Genes 2020, 11, 621. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhao, Z.; Lu, S.; Liu, Y.; Han, S.; Jiang, H.; Yang, Y.; Liu, H. Physiological, Nutritional and Transcriptomic Responses of Sturgeon (Acipenser schrenckii) to Complete Substitution of Fishmeal with Cottonseed Protein Concentrate in Aquafeed. Biology 2023, 12, 490. [Google Scholar] [CrossRef] [PubMed]
- Galafat, A.; Sáez, M.I.; Rodríguez, C.; Hernández de Rojas, A.; Vizcaíno, A.J.; Martínez, T.F.; Alarcón-López, F.J. In vitro evaluation of algae and their effect as dietary ingredient on growth, chemical composition and intestinal functionality in juvenile turbot (Scophthalmus maximus). Aquaculture 2024, 592, 741208. [Google Scholar] [CrossRef]
- Zhao, H.; Huang, Y.; Shu, S.; Wang, G.; Fu, C.; Huang, R.; Zhang, J.; Su, H.; He, Y.; Lei, C.; et al. Transcriptomics and metabolomics of blood, urine and ovarian follicular fluid of yak at induced estrus stage. BMC Genom. 2024, 25, 201. [Google Scholar] [CrossRef] [PubMed]
- de Jong, E.; Bosco, A. Unlocking immune-mediated disease mechanisms with transcriptomics. Biochem. Soc. Trans. 2021, 49, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Dixon, E.E.; Wu, H.; Sulvarán-Guel, E.; Guo, J.; Humphreys, B.D. Spatially resolved transcriptomics and the kidney: Many opportunities. Kidney Int. 2022, 102, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Teng, Y.; Han, C.; Wei, S.; Li, L.; Liu, H.; Hu, S.; Kang, B.; Xu, H. Multi-omics reveals goose fatty liver formation from metabolic reprogramming. Front. Vet. Sci. 2024, 11, 1122904. [Google Scholar] [CrossRef]
- Liang, X.; Wang, H.; Xu, W.; Liu, X.; Zhao, C.; Chen, J.; Wang, D.; Xu, S.; Cao, J.; Sun, C.; et al. Metabolome and Transcriptome Analysis Revealed the Basis of the Difference in Antioxidant Capacity in Different Tissues of Citrus reticulata ‘Ponkan’. Antioxidants 2024, 13, 243. [Google Scholar] [CrossRef]
- van Gelderen, T.A.; Ladisa, C.; Salazar-Moscoso, M.; Folgado, C.; Habibi, H.R.; Ribas, L. Metabolomic and transcriptomic profiles after immune stimulation in the zebrafish testes. Genomics 2023, 115, 110581. [Google Scholar] [CrossRef]
- Ye, B.; Wang, Q.; Ye, Q.; Wang, D.; Wang, Z.; Dong, Z.; Zou, J. Effects of different combinations of koumine and gelsemine on growth performance, intestinal health, and transcriptome of Cyprinus carpio. J. Hazard. Mater. 2024, 465, 133130. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Schriml, L.M.; Arze, C.; Nadendla, S.; Chang, Y.W.; Mazaitis, M.; Felix, V.; Feng, G.; Kibbe, W.A. Disease Ontology: A backbone for disease semantic integration. Nucleic Acids Res. 2012, 40, D940–D946. [Google Scholar] [CrossRef] [PubMed]
- Croft, D.; O’Kelly, G.; Wu, G.; Haw, R.; Gillespie, M.; Matthews, L.; Caudy, M.; Garapati, P.; Gopinath, G.; Jassal, B.; et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2011, 39, D691–D697. [Google Scholar] [CrossRef] [PubMed]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liu, H.; Zhu, X.; Han, D.; Jin, J.; Yang, Y.; Xie, S. The Effects of Dietary Arthrospira platensis on Oxidative Stress Response and Pigmentation in Yellow Catfish Pelteobagrus fulvidraco. Antioxidants 2022, 11, 1100. [Google Scholar] [CrossRef] [PubMed]
- Hoseinifar, S.H.; Yousefi, S.; Van Doan, H.; Ashouri, G.; Gioacchini, G.; Maradonna, F.; Carnevali, O. Oxidative Stress and Antioxidant Defense in Fish: The Implications of Probiotic, Prebiotic, and Synbiotics. Rev. Fish Sci. Aquac. 2020, 29, 198–217. [Google Scholar] [CrossRef]
- Rohani, M.F.; Islam, S.M.; Hossain, M.K.; Ferdous, Z.; Siddik, M.A.; Nuruzzaman, M.; Padeniya, U.; Brown, C.; Shahjahan, M. Probiotics, prebiotics and synbiotics improved the functionality of aquafeed: Upgrading growth, reproduction, immunity and disease resistance in fish. Fish Shellfish Immunol. 2022, 120, 569–589. [Google Scholar] [CrossRef] [PubMed]
- He, S.; You, J.; Liang, X.; Zhang, Z.; Zhang, Y. Transcriptome sequencing and metabolome analysis of food habits domestication from live prey fish to artificial diets in mandarin fish (Siniperca chuatsi). BMC Genom. 2021, 22, 129. [Google Scholar] [CrossRef] [PubMed]
- Naiel, M.A.E.; Negm, S.S.; Ghazanfar, S.; Shukry, M.; Abdelnour, S.A. The risk assessment of high-fat diet in farmed fish and its mitigation approaches: A review. J. Anim. Physiol. Anim. Nutr. 2023, 107, 948–969. [Google Scholar] [CrossRef]
- Altmann, B.A.; Rosenau, S. Spirulina as Animal Feed: Opportunities and Challenges. Foods 2022, 11, 965. [Google Scholar] [CrossRef]
- Akter, T.; Hossain, A.; Rabiul Islam, M.; Hossain, M.A.; Das, M.; Rahman, M.M.; Aye, A.T.; Abdel-Tawwab, M. Effects of spirulina (Arthrospira platensis) as a fishmeal replacer in practical diets on growth performance, proximate composition, and amino acids profile of pabda catfish (Ompok pabda). J. Appl. Aquac. 2023, 35, 69–82. [Google Scholar] [CrossRef]
- Rosas, V.T.; Monserrat, J.M.; Bessonart, M.; Magnone, L.; Romano, L.A.; Tesser, M.B. Fish oil and meal replacement in mullet (Mugil liza) diet with Spirulina (Arthrospira platensis) and linseed oil. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2018, 218, 46–54. [Google Scholar] [CrossRef]
- Liu, C.; Liu, H.; Xu, W.; Han, D.; Xie, S.; Jin, J.; Yang, Y.; Zhu, X. Effects of dietary Arthrospira platensis supplementation on the growth, pigmentation, and antioxidation in yellow catfish (Pelteobagrus fulvidraco). Aquaculture 2019, 510, 267–275. [Google Scholar] [CrossRef]
- Gou, N.; Ji, H.; Wu, W.; Zhong, M.; Zhang, B. Transcriptional response to cold and fasting acclimation in Onychostoma macrolepis during the overwintering stage. Comp. Biochem. Physiology. Part D Genom. Proteom. 2021, 40, 100901. [Google Scholar] [CrossRef]
- Dou, X.; Liu, Y.; Cao, Y.; Zhang, Y.; Fu, X.; Deng, J.; Tan, B. Effects of Dietary Lysine Level on Growth Performance and Protein Metabolism in Juvenile Leopard Coral Grouper (Plectropomus leopardus). Aquac. Nutr. 2023, 2023, 1017222. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Kos, J. Peptidases: Role and Function in Health and Disease. Int. J. Mol. Sci. 2023, 24, 7823. [Google Scholar] [CrossRef]
- Abdel-Latif, H.M.R.; Soliman, A.A.; Khaled, A.A.; Kord, M.; Abdel-Tawwab, M.; Darwish, S.; Grana, Y.S.; Zaki, M.; Nour, A.; Ali, E.; et al. Growth performance, antioxidant activities, and immunological responses of hapa-reared thinlip mullet (Liza ramada) juveniles fed on diets supplemented with spirulina (Arthrospira platensis). Fish Shellfish Immunol. 2022, 130, 359–367. [Google Scholar] [CrossRef]
- Adel, M.; Yeganeh, S.; Dadar, M.; Sakai, M.; Dawood, M.A.O. Effects of dietary Spirulina platensis on growth performance, humoral and mucosal immune responses and disease resistance in juvenile great sturgeon (Huso huso Linnaeus, 1754). Fish Shellfish Immunol. 2016, 56, 436–444. [Google Scholar] [CrossRef]
- Qin, C.; Xie, J.; Shi, X.; Tan, J. Cold stress after swimming fatigue decreases immunity-related gene expression in the spleen of the Chinese sucker. Fish Shellfish Immunol. 2022, 122, 455–464. [Google Scholar] [CrossRef]
- Kim, S.; Rahimnejad, S.; Kim, K.; Lee, B.; Lee, K. Effects of Dietary Supplementation of Spirulina and Quercetin on Growth, Innate Immune Responses, Disease Resistance Against Edwardsiella tarda, and Dietary Antioxidant Capacity in the Juvenile Olive Flounder Paralichthys olivaceus. Fish. Aquat. Sci. 2013, 16, 7–14. [Google Scholar] [CrossRef]
- Mohammadiazarm, H.; Maniat, M.; Ghorbanijezeh, K.; Ghotbeddin, N. Effects of spirulina powder (Spirulina platensis) as a dietary additive on Oscar fish, Astronotus ocellatus: Assessing growth performance, body composition, digestive enzyme activity, immune-biochemical parameters, blood indices and total pigmentation. Aquac. Nutr. 2021, 27, 252–260. [Google Scholar] [CrossRef]
- Harikrishnan, R.; Balasundaram, C.; Heo, M. Impact of plant products on innate and adaptive immune system of cultured finfish and shellfish. Aquaculture 2011, 317, 1–15. [Google Scholar] [CrossRef]
- Yu, W.; Wen, G.; Lin, H.; Yang, Y.; Huang, X.; Zhou, C.; Zhang, Z.; Duan, Y.; Huang, Z.; Li, T. Effects of dietary Spirulina platensis on growth performance, hematological and serum biochemical parameters, hepatic antioxidant status, immune responses and disease resistance of Coral trout Plectropomus leopardus (Lacepede, 1802). Fish Shellfish Immunol. 2018, 74, 649–655. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, C.; Sun, Y.; Chen, Y.; Chen, S.; Han, T.; Wang, J. Excessive Substitution of Fish Meal with Fermented Soybean Meal Induces Oxidative Stress by Impairing Glutathione Metabolism in Largemouth Bass (Micropterus salmoides). Antioxidants 2023, 12, 2096. [Google Scholar] [CrossRef]
- Li, L.; Wang, Y.; Huang, Y.; Wang, C. Multi-omics approach to study the dual effects of novel proteins on the intestinal health of juvenile largemouth bass (Micropterus salmoides) under an alternate feeding strategy. Front. Immunol. 2023, 14, 1110696. [Google Scholar] [CrossRef]
- Xu, X.; Li, X.; Xu, Z.; Yang, H.; Lin, X.; Leng, X. Replacing fishmeal with cottonseed protein concentrate in practical diet of largemouth bass (Micropterus salmoides): Growth, flesh quality and metabolomics. Aquaculture 2024, 579, 740164. [Google Scholar] [CrossRef]
- Orlandi, I.; Alberghina, L.; Vai, M. Nicotinamide, Nicotinamide Riboside and Nicotinic Acid-Emerging Roles in Replicative and Chronological Aging in Yeast. Biomolecules 2020, 10, 604. [Google Scholar] [CrossRef]
- Lo, C.; Liu, Z.; Chen, S.; Lin, F.; Berneshawi, A.R.; Yu, C.Q.; Koo, E.B.; Kowal, T.J.; Ning, K.; Hu, Y.; et al. Primary cilia formation requires the Leigh syndrome-associated mitochondrial protein NDUFAF2. J. Clin. Investig. 2024, 134, e175560. [Google Scholar] [CrossRef]
- Dhuguru, J.; Dellinger, R.W.; Migaud, M.E. Defining NAD(P)(H) Catabolism. Nutrients 2023, 15, 3064. [Google Scholar] [CrossRef]
- Dong, Y.; Wang, X.; Wei, L.; Liu, Z.; Chu, X.; Xiong, W.; Liu, W.; Li, X. The Effectiveness of Four Nicotinamide Adenine Dinucleotide (NAD(+)) Precursors in Alleviating the High-Glucose-Induced Damage to Hepatocytes in Megalobrama amblycephala: Evidence in NAD(+) Homeostasis, Sirt1/3 Activation, Redox Defense, Inflammatory Response, Apoptosis, and Glucose Metabolism. Antioxidants 2024, 13, 385. [Google Scholar] [CrossRef]
- AlFadhly, N.K.Z.; Alhelfi, N.; Altemimi, A.B.; Verma, D.K.; Cacciola, F.; Narayanankutty, A. Trends and Technological Advancements in the Possible Food Applications of Spirulina and Their Health Benefits: A Review. Molecules 2022, 27, 5584. [Google Scholar] [CrossRef]
- Guy, A.T.; Nagatsuka, Y.; Ooashi, N.; Inoue, M.; Nakata, A.; Greimel, P.; Inoue, A.; Nabetani, T.; Murayama, A.; Ohta, K.; et al. NEURONAL DEVELOPMENT. Glycerophospholipid regulation of modality-specific sensory axon guidance in the spinal cord. Science 2015, 349, 974–977. [Google Scholar] [CrossRef]
- Zhu, Q.; Wu, Y.; Mai, J.; Guo, G.; Meng, J.; Fang, X.; Chen, X.; Liu, C.; Zhong, S. Comprehensive Metabolic Profiling of Inflammation Indicated Key Roles of Glycerophospholipid and Arginine Metabolism in Coronary Artery Disease. Front. Immunol. 2022, 13, 829425. [Google Scholar] [CrossRef]
- Zhao, Y.; Qiao, R.; Zhang, S.; Wang, G. Metabolomic profiling reveals the intestinal toxicity of different length of microplastic fibers on zebrafish (Danio rerio). J. Hazard. Mater. 2021, 403, 123663. [Google Scholar] [CrossRef]
- Luo, Q.; Zhou, Z.; Zhao, J.; Xu, H.; Limbu, S.M.; Xu, Q. Dietary β-conglycinin induces intestinal enteritis and affects glycerophospholipid and arginine metabolism in mirror carp (Cyprinus carpio). Aquaculture 2023, 567, 739257. [Google Scholar] [CrossRef]
- Yoshinaga, H.; Yasuike, M.; Mekuchi, M.; Soma, S.; Yamamoto, T.; Murashita, K.; Matsunari, H.; Oku, H.; Furuita, H. Multi-omics analysis of hepatopancreas of red seabream (Pagrus major) fed a soybean meal-based diet. Aquaculture 2023, 574, 739631. [Google Scholar] [CrossRef]
- An, W.; Xu, J.; Chen, F.; Ma, Y.; Su, Z.; Guang, J.; Su, N.; Zhang, G.; Xu, C.; Li, Y.; et al. Effects of dietary n-3 LC-PUFA levels on the growth, immunity, and lipid metabolism of freshwater carnivorous teleost largemouth bass (Micropterus salmoides) juveniles. Aquac. Rep. 2023, 32, 101704. [Google Scholar] [CrossRef]
- Sui, Z.; Wang, X.; Sun, Y.; Zhou, H.; Liu, C.; Mai, K.; He, G. Methionine deficiency affects myogenesis and muscle macronutrient metabolism in juvenile turbot Scophthalmus maximus. Aquaculture 2024, 578, 740013. [Google Scholar] [CrossRef]
- Yao, C.; Feng, L.; Wu, P.; Liu, Y.; Jiang, J.; Zhang, L.; Mi, H.; Zhou, X.; Jiang, W. Promotion of fatty acid metabolism and glucose metabolism in the muscle of sub-adult grass carp (Ctenopharyngodon idella): The role of alpha-linoleic acid/linoleic acid (ALA/LNA) ratios. Food Chem. X 2023, 19, 100752. [Google Scholar] [CrossRef]



| Primer | Forward | Sequences (5′ to 3′) |
|---|---|---|
| β-actin | F | TGAGCGTGAGATTGTGCGT |
| R | AAGAGGGCTGGAAAAGGGA | |
| NC_046633.1 | F | TCACTCGTGCTGTTGACTTAC |
| R | TAGAAGAAGGAGAGGAGGAAGG | |
| NC_046656.1 | F | CCTGGAGTGTGTAAATGGAGTAG |
| R | GGACTGTGTGGGAACTTTGA | |
| NC_046636.1 | F | CGTTGTGCTGTGGGAGATAA |
| R | GTGGATGGAAGGAGCAGAAA | |
| NC_046633.1 | F | TGCTGCTGTGTCTGTCTATG |
| R | GTTACCTACTCGGGCATCTATTT | |
| NC_046652.1 | F | ACTAGGCTGTCAAGTGCTAATG |
| R | GCATCGCTGGAAGGATAACT | |
| NC_046656.1 | F | CCTAACCTCAACCCACCAATTA |
| R | GCAGAGAGGAAAGGAGAGAATG | |
| NC_046652.1 | F | TGGTGATCGGTCGTCTTTATTC |
| R | TGGGCTCTTTGTGGTCTTTAC | |
| NC_046656.1 | F | CTCCACTCTCCATTCGCTTATC |
| R | CAAGGCTATCTCCACAACTACC |
| Sample | Raw Data (G) | Clean Data (G) | Q20 (%) | Q30 (%) | Total Reads | Total Mapped (%) |
|---|---|---|---|---|---|---|
| Z0C1 | 6.27 | 6.15 | 97.71 | 95.74 | 44,534,846 | 80.28 |
| Z0C2 | 5.66 | 5.54 | 97.39 | 95.23 | 40,183,868 | 82.54 |
| Z0C3 | 6.32 | 6.17 | 97.72 | 95.75 | 44,853,084 | 79.93 |
| Z1C1 | 6.4 | 6.29 | 97.76 | 95.84 | 45,431,520 | 77.74 |
| Z1C2 | 6.25 | 6.12 | 97.73 | 95.78 | 44,361,932 | 77.74 |
| Z1C3 | 6.02 | 5.91 | 97.43 | 95.3 | 42,725,926 | 77.07 |
| Z2C1 | 5.48 | 5.37 | 97.8 | 95.91 | 38,839,870 | 78.89 |
| Z2C2 | 5.79 | 5.62 | 97.53 | 95.49 | 41,011,036 | 80.93 |
| Z2C3 | 5.22 | 5.12 | 97.71 | 95.72 | 36,995,762 | 78.32 |
| Z3C1 | 5.12 | 4.98 | 96.73 | 94.11 | 36,307,444 | 79.85 |
| Z3C2 | 5.38 | 5.26 | 97.91 | 95.98 | 38,208,560 | 78.76 |
| Z3C3 | 5.39 | 5.3 | 97.99 | 96.14 | 38,301,174 | 82.47 |
| Z4C1 | 5.47 | 5.36 | 96.88 | 94.37 | 38,794,138 | 79.61 |
| Z4C2 | 5.55 | 5.45 | 98.06 | 96.27 | 39,386,622 | 79.37 |
| Z4C3 | 5.81 | 5.69 | 97.06 | 94.75 | 41,189,022 | 78.64 |
| Z5C1 | 5.57 | 5.46 | 97.66 | 95.64 | 39,503,432 | 79.36 |
| Z5C2 | 6.67 | 6.54 | 97.68 | 95.67 | 47,359,346 | 79.38 |
| Z5C3 | 5.64 | 5.53 | 97.44 | 95.33 | 40,076,522 | 78.84 |
| Type | All | Known | Unknown |
|---|---|---|---|
| POS | 6981 | 1500 | 5481 |
| NEG | 5770 | 1317 | 4453 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, D.; Hou, D.; Zheng, Y.; Xiang, W.; Huang, Y.; Wu, H.; Zou, J. Multi-Omics Reveals the Effects of Spirulina platensis Powder Replacement of Fish Meal on Intestinal Metabolism and Stress in Zig-Zag Eel (Mastacembelus armatus). Antioxidants 2024, 13, 851. https://doi.org/10.3390/antiox13070851
Sun D, Hou D, Zheng Y, Xiang W, Huang Y, Wu H, Zou J. Multi-Omics Reveals the Effects of Spirulina platensis Powder Replacement of Fish Meal on Intestinal Metabolism and Stress in Zig-Zag Eel (Mastacembelus armatus). Antioxidants. 2024; 13(7):851. https://doi.org/10.3390/antiox13070851
Chicago/Turabian StyleSun, Di, Dongqiang Hou, Yushun Zheng, Wenzhou Xiang, Yingshi Huang, Hualian Wu, and Jixing Zou. 2024. "Multi-Omics Reveals the Effects of Spirulina platensis Powder Replacement of Fish Meal on Intestinal Metabolism and Stress in Zig-Zag Eel (Mastacembelus armatus)" Antioxidants 13, no. 7: 851. https://doi.org/10.3390/antiox13070851







