Delta-9-Tetrahydrocannabinol Blocks Bone Marrow-Derived Macrophage Differentiation through Elimination of Reactive Oxygen Species
Abstract
:1. Introduction
2. Methods
2.1. Mice
2.2. Reagents
2.3. Cell Preparation
2.4. Flow Cytometry
2.5. RNA-Seq
2.6. Fluorescent Assays
2.7. Statistical Analyses
3. Results
3.1. THC Blocks Macrophage Differentiation in a Dose-Dependent Manner
3.2. Identifying the Receptors through Which THC Suppresses Macrophage Differentiation
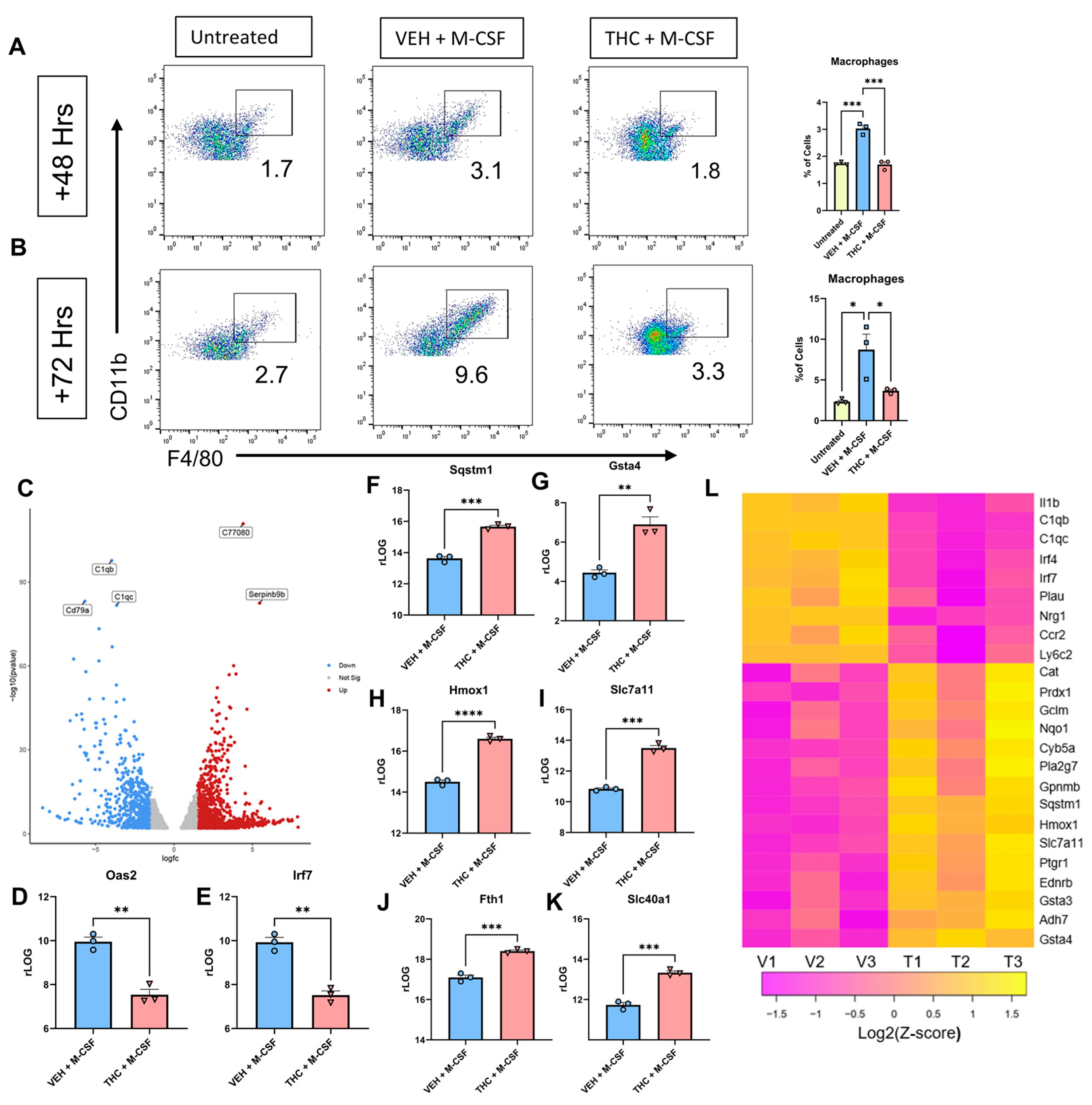
3.3. RNA-Seq Analysis of BMDCs
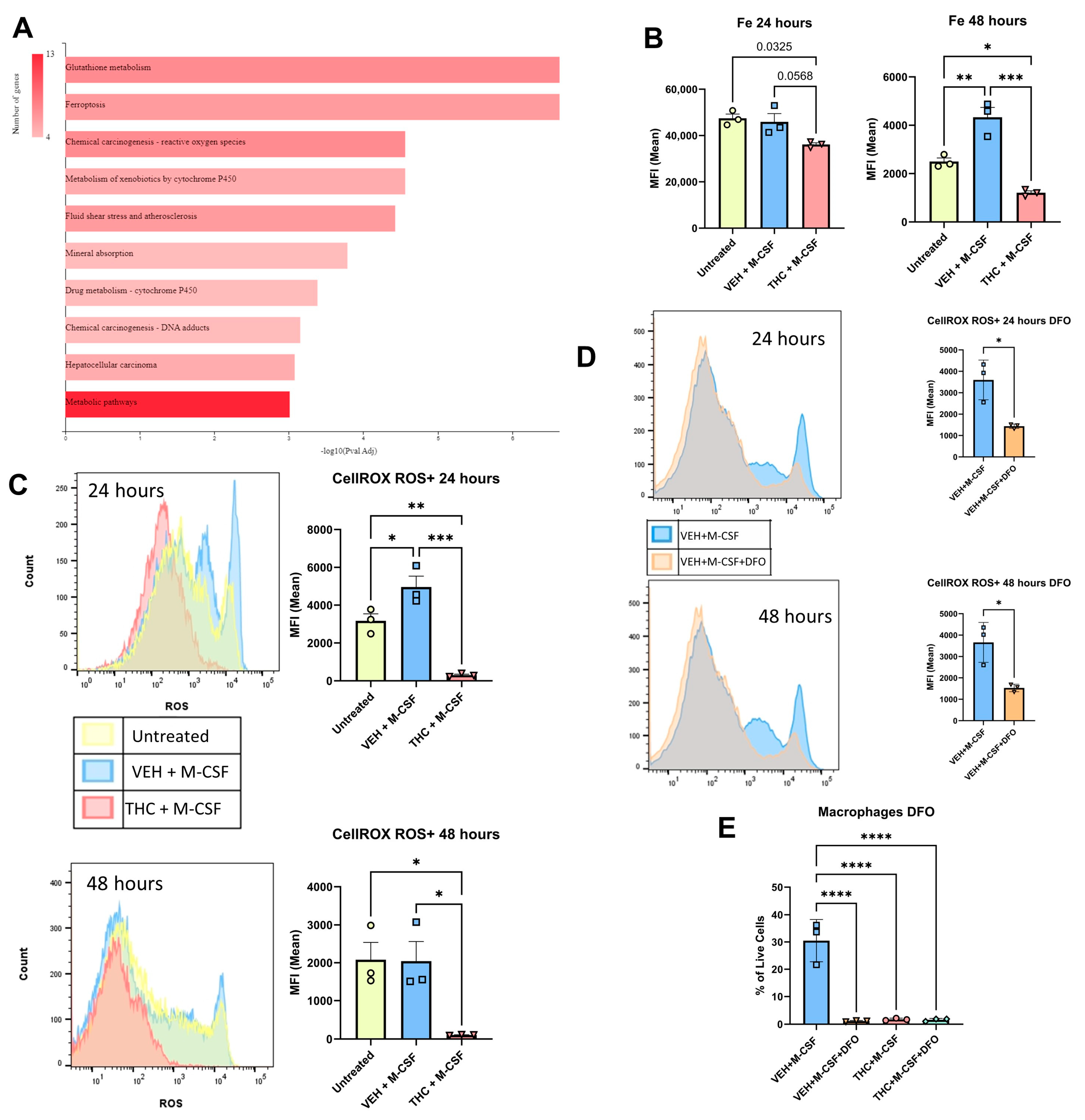
3.4. Reactive Oxygen Species and Fenton Reaction
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Naito, M. Macrophage differentiation and function in health and disease. Pathol. Int. 2008, 58, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Ushach, I.; Zlotnik, A. Biological role of granulocyte macrophage colony-stimulating factor (GM-CSF) and macrophage colony-stimulating factor (M-CSF) on cells of the myeloid lineage. J. Leukoc. Biol. 2016, 100, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Toh, M.L.; Bonnefoy, J.Y.; Accart, N.; Cochin, S.; Pohle, S.; Haegel, H.; De Meyer, M.; Zemmour, C.; Preville, X.; Guillen, C.; et al. Bone-and cartilage-protective effects of a monoclonal antibody against colony-stimulating factor 1 receptor in experimental arthritis. Arthritis Rheumatol. 2014, 66, 2989–3000. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.K.; Miikeda, A.; Fouladian, Z.; Mehrabian, M.; Edillor, C.; Shih, D.; Zhou, Z.; Paul, M.K.; Charugundla, S.; Davis, R.C.; et al. Local M-CSF (macrophage colony-stimulating factor) expression regulates macrophage proliferation and apoptosis in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Le Meur, Y.; Tesch, G.H.; Hill, P.A.; Mu, W.; Foti, R.; Nikolic-Paterson, D.J.; Atkins, R.C. Macrophage accumulation at a site of renal inflammation is dependent on the M-CSF/c-fms pathway. J. Leukoc. Biol. 2002, 72, 530–537. [Google Scholar] [CrossRef]
- Menke, J.; Iwata, Y.; Rabacal, W.A.; Basu, R.; Stanley, E.R.; Kelley, V.R. Distinct roles of CSF-1 isoforms in lupus nephritis. J. Am. Soc. Nephrol. 2011, 22, 1821. [Google Scholar] [CrossRef]
- Pixley, F.J.; Stanley, E.R. CSF-1 regulation of the wandering macrophage: Complexity in action. Trends Cell Biol. 2004, 14, 628–638. [Google Scholar] [CrossRef]
- Lawrence, T.; Natoli, G. Transcriptional regulation of macrophage polarization: Enabling diversity with identity. Nat. Rev. Immunol. 2011, 11, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.; Li, H.; Tang, W. Targeting CSF-1R represents an effective strategy in modulating inflammatory diseases. Pharmacol. Res. 2023, 187, 106566. [Google Scholar] [CrossRef]
- Zhang, Y.; Choksi, S.; Chen, K.; Pobezinskaya, Y.; Linnoila, I.; Liu, Z.G. ROS play a critical role in the differentiation of alternatively activated macrophages and the occurrence of tumor-associated macrophages. Cell Res. 2013, 23, 898–914. [Google Scholar] [CrossRef]
- Owusu-Ansah, E.; Banerjee, U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature 2009, 461, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Brüne, B.; Dehne, N.; Grossmann, N.; Jung, M.; Namgaladze, D.; Schmid, T.; von Knethen, A.; Weigert, A. Redox control of inflammation in macrophages. Antioxid. Redox Signal. 2013, 19, 595–637. [Google Scholar] [CrossRef] [PubMed]
- Sukhbaatar, N.; Weichhart, T. Iron regulation: Macrophages in control. Pharmaceuticals 2018, 11, 137. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, Y.; Guo, L.; Gao, W.; Tang, T.L.; Yan, M. Interaction between macrophages and ferroptosis. Cell Death Dis. 2022, 13, 355. [Google Scholar] [CrossRef] [PubMed]
- Preteroti, M.W.; Traboulsi, H.; Greiss, P.; Lapohos, O.; Fonseca, G.J.; Eidelman, D.H.; Baglole, C.J. Receptor-mediated effects of Δ9-tetrahydrocannabinol and cannabidiol on the inflammatory response of alveolar macrophages. Immunol. Cell Biol. 2023, 101, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, J.M.; Minogue, E.; Curham, L.; Tyrrell, H.; Gavigan, P.; Hind, W.; Downer, E.J. MyD88-dependent and-independent signalling via TLR3 and TLR4 are differentially modulated by Δ9-tetrahydrocannabinol and cannabidiol in human macrophages. J. Neuroimmunol. 2020, 343, 577217. [Google Scholar] [CrossRef] [PubMed]
- Becker, W.; Alrafas, H.R.; Wilson, K.; Miranda, K.; Culpepper, C.; Chatzistamou, I.; Cai, G.; Nagarkatti, M.; Nagarkatti, P.S. Activation of cannabinoid receptor 2 prevents colitis-associated colon cancer through myeloid cell de-activation upstream of IL-22 production. iScience 2020, 23, 101504. [Google Scholar] [CrossRef] [PubMed]
- Cohen, G.; Gover, O.; Schwartz, B. Phytocannabinoids Reduce Inflammation of Primed Macrophages and Enteric Glial Cells: An In Vitro Study. Int. J. Mol. Sci. 2023, 24, 14628. [Google Scholar] [CrossRef]
- Springs, A.E.; Karmaus, P.W.; Crawford, R.B.; Kaplan, B.L.; Kaminski, N.E. Effects of targeted deletion of cannabinoid receptors CB1 and CB2 on immune competence and sensitivity to immune modulation by Δ9-tetrahydrocannabinol. J. Leucoc. Biol. 2008, 84, 1574–1584. [Google Scholar] [CrossRef]
- Miranda, K.; Becker, W.; Busbee, P.B.; Dopkins, N.; Abdulla, O.A.; Zhong, Y.; Zhang, J.; Nagarkatti, M.; Nagarkatti, P.S. Yin and yang of cannabinoid CB1 receptor: CB1 deletion in immune cells causes exacerbation while deletion in non-immune cells attenuates obesity. iScience 2022, 25, 104994. [Google Scholar] [CrossRef]
- Lauckner, J.E.; Jensen, J.B.; Chen, H.Y.; Lu, H.C.; Hille, B.; Mackie, K. GPR55 is a cannabinoid receptor that increases intracellular calcium and inhibits M current. Proc. Natl. Acad. Sci. USA 2008, 105, 2699–2704. [Google Scholar] [CrossRef] [PubMed]
- McHugh, D.; Page, J.; Dunn, E.; Bradshaw, H.B. Δ9-Tetrahydrocannabinol and N-arachidonyl glycine are full agonists at GPR18 receptors and induce migration in human endometrial HEC-1B cells. Br. J. Pharmacol. 2012, 165, 2414–2424. [Google Scholar] [CrossRef] [PubMed]
- Qin, N.; Neeper, M.P.; Liu, Y.; Hutchinson, T.L.; Lubin, M.L.; Flores, C.M. TRPV2 is activated by cannabidiol and mediates CGRP release in cultured rat dorsal root ganglion neurons. J. Neurosci. 2008, 28, 6231–6238. [Google Scholar] [CrossRef] [PubMed]
- Aso, E.; Fernández-Dueñas, V.; López-Cano, M.; Taura, J.; Watanabe, M.; Ferrer, I.; Luján, R.; Ciruela, F. Adenosine A 2A-Cannabinoid CB 1 Receptor Heteromers in the Hippocampus: Cannabidiol Blunts Δ 9-Tetrahydrocannabinol-Induced Cognitive Impairment. Mol. Neurobiol. 2019, 56, 5382–5391. [Google Scholar] [CrossRef] [PubMed]
- Morales, P.; Reggio, P.H. An update on non-CB1, non-CB2 cannabinoid related G-protein-coupled receptors. Cannabis Cannabinoid Res. 2017, 2, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Hilairet, S.; Bouaboula, M.; Carrière, D.; Le Fur, G.; Casellas, P. Hypersensitization of the Orexin 1 receptor by the CB1 receptor: Evidence for cross-talk blocked by the specific CB1 antagonist, SR141716. J. Biol. Chem. 2003, 278, 23731–23737. [Google Scholar] [CrossRef]
- Reyes-Resina, I.; Navarro, G.; Aguinaga, D.; Canela, E.I.; Schoeder, C.T.; Załuski, M.; Kieć-Kononowicz, K.; Saura, C.A.; Müller, C.E.; Franco, R. Molecular and functional interaction between GPR18 and cannabinoid CB2 G-protein-coupled receptors. Relevance in neurodegenerative diseases. Biochem. Pharmacol. 2018, 157, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Borges, R.S.; Batista, J., Jr.; Viana, R.B.; Baetas, A.C.; Orestes, E.; Andrade, M.A.; Honório, K.M.; Da Silva, A.B. Understanding the molecular aspects of tetrahydrocannabinol and cannabidiol as antioxidants. Molecules 2013, 18, 12663–12674. [Google Scholar] [CrossRef] [PubMed]
- Hampson, A.J.; Grimaldi, M.; Axelrod, J.; Wink, D. Cannabidiol and (−) Δ9-tetrahydrocannabinol are neuroprotective antioxidants. Proc. Natl. Acad. Sci. USA 1998, 95, 8268–8273. [Google Scholar] [CrossRef]
- Lacey, D.C.; Achuthan, A.; Fleetwood, A.J.; Dinh, H.; Roiniotis, J.; Scholz, G.M.; Chang, M.W.; Beckman, S.K.; Cook, A.D.; Hamilton, J.A. Defining GM-CSF–and macrophage-CSF–dependent macrophage responses by in vitro models. J. Immunol. 2012, 188, 5752–5765. [Google Scholar] [CrossRef]
- Fleetwood, A.J.; Dinh, H.; Cook, A.D.; Hertzog, P.J.; Hamilton, J.A. GM-CSF-and M-CSF-dependent macrophage phenotypes display differential dependence on type I interferon signaling. J. Leukoc. Biol. 2009, 86, 411–421. [Google Scholar] [CrossRef] [PubMed]
- The Galaxy Community. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022, 50, W345–W351. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.C.; Appelberg, S.; Goldberger, B.A.; Klein, T.W.; Sleasman, J.W.; Goodenow, M.M. Δ 9-Tetrahydrocannabinol Treatment during Human Monocyte Differentiation Reduces Macrophage Susceptibility to HIV-1 Infection. J. Neuroimmune Pharmacol. 2014, 9, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Pertwee, R. The diverse CB1 and CB2 receptor pharmacology of three plant cannabinoids: Δ9-tetrahydrocannabinol, cannabidiol and Δ9-tetrahydrocannabivarin. Br. J. Pharmacol. 2008, 153, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Hegde, V.L.; Nagarkatti, M.; Nagarkatti, P.S. Cannabinoid receptor activation leads to massive mobilization of myeloid-derived suppressor cells with potent immunosuppressive properties. Eur. J. Immunol. 2010, 40, 3358–3371. [Google Scholar] [CrossRef] [PubMed]
- Al-Ghezi, Z.Z.; Miranda, K.; Nagarkatti, M.; Nagarkatti, P.S. Combination of cannabinoids, Δ9-tetrahydrocannabinol and cannabidiol, ameliorates experimental multiple sclerosis by suppressing neuroinflammation through regulation of miRNA-mediated signaling pathways. Front. Immunol. 2019, 10, 466430. [Google Scholar] [CrossRef] [PubMed]
- Chiang, N.; Dalli, J.; Colas, R.A.; Serhan, C.N. Identification of resolvin D2 receptor mediating resolution of infections and organ protection. J. Exp. Med. 2015, 212, 1203–1217. [Google Scholar] [CrossRef] [PubMed]
- Schoeder, C.T.; Kaleta, M.; Mahardhika, A.B.; Olejarz-Maciej, A.; Łażewska, D.; Kieć-Kononowicz, K.; Müller, C.E. Structure-activity relationships of imidazothiazinones and analogs as antagonists of the cannabinoid-activated orphan G protein-coupled receptor GPR18. Eur. J. Med. Chem. 2018, 155, 381–397. [Google Scholar] [CrossRef]
- Raup-Konsavage, W.M.; Johnson, M.; Legare, C.A.; Yochum, G.S.; Morgan, D.J.; Vrana, K.E. Synthetic cannabinoid activity against colorectal cancer cells. Cannabis Cannabinoid Res. 2018, 3, 272–281. [Google Scholar] [CrossRef]
- Ferré, S.; Lluís, C.; Justinova, Z.; Quiroz, C.; Orru, M.; Navarro, G.; Canela, E.I.; Franco, R.; Goldberg, S.R. Adenosine–cannabinoid receptor interactions. Implications for striatal function. Br. J. Pharmacol. 2010, 160, 443–453. [Google Scholar] [CrossRef]
- Stollenwerk, T.M.; Pollock, S.; Hillard, C.J. Contribution of the adenosine 2A receptor to behavioral effects of tetrahydrocannabinol, cannabidiol and PECS-101. Molecules 2021, 26, 5354. [Google Scholar] [CrossRef] [PubMed]
- Justinová, Z.; Redhi, G.H.; Goldberg, S.R.; Ferré, S. Differential effects of presynaptic versus postsynaptic adenosine A2A receptor blockade on Δ9-tetrahydrocannabinol (THC) self-administration in squirrel monkeys. J. Neurosci. 2014, 34, 6480–6484. [Google Scholar] [CrossRef]
- Mangmool, S.; Kurose, H. Gi/o protein-dependent and-independent actions of pertussis toxin (PTX). Toxins 2011, 3, 884–899. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.; Takeuchi, O.; Vandenbon, A.; Yasuda, K.; Tanaka, Y.; Kumagai, Y.; Miyake, T.; Matsushita, K.; Okazaki, T.; Saitoh, T.; et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 2010, 11, 936–944. [Google Scholar] [CrossRef] [PubMed]
- Knights, A.J.; Farrell, E.C.; Ellis, O.M.; Song, M.J.; Appleton, C.T.; Maerz, T. Synovial macrophage diversity and activation of M-CSF signaling in post-traumatic osteoarthritis. eLife 2023, 12, RP93283. [Google Scholar]
- Forman, H.J.; Zhang, H. Targeting oxidative stress in disease: Promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021, 20, 689–709. [Google Scholar] [CrossRef] [PubMed]
- Rybarczyk, A.; Majchrzak-Celińska, A.; Krajka-Kuźniak, V. Targeting Nrf2 signaling pathway in cancer prevention and treatment: The role of cannabis compounds. Antioxidants 2023, 12, 2052. [Google Scholar] [CrossRef] [PubMed]
- Baird, L.; Yamamoto, M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol. Cell Biol. 2020, 40, e00099-20. [Google Scholar] [CrossRef]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010, 12, 213–223. [Google Scholar] [CrossRef]
- Keller, C.; Rading, S.; Bindila, L.; Karsak, M. Behavioral Studies of p62 KO Animals with Implications of a Modulated Function of the Endocannabinoid System. Cells 2022, 11, 1517. [Google Scholar] [CrossRef]
- Shi, F.; Zhang, Z.; Cui, H.; Wang, J.; Wang, Y.; Tang, Y.; Yang, W.; Zou, P.; Ling, X.; Han, F.; et al. Analysis by transcriptomics and metabolomics for the proliferation inhibition and dysfunction through redox imbalance-mediated DNA damage response and ferroptosis in male reproduction of mice and TM4 Sertoli cells exposed to PM2.5. Ecotoxicol. Environ. Saf. 2022, 238, 113569. [Google Scholar] [CrossRef] [PubMed]
- Ade, N.; Leon, F.; Pallardy, M.; Peiffer, J.L.; Kerdine-Romer, S.; Tissier, M.H.; Bonnet, P.A.; Fabre, I.; Ourlin, J.C. HMOX1 and NQO1 genes are upregulated in response to contact sensitizers in dendritic cells and THP-1 cell line: Role of the Keap1/Nrf2 pathway. Toxicol. Sci. 2009, 107, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Gunderstofte, C.; Iversen, M.B.; Peri, S.; Thielke, A.; Balachandran, S.; Holm, C.K.; Olagnier, D. Nrf2 Negatively regulates Type I interferon responses and increases susceptibility to herpes genital infection in mice. Front. Immunol. 2019, 10, 2101. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.C.; Almeida, J.I.; Lima, I.S.; Kapitao, A.S.; Gozzelino, R. Iron metabolism and the inflammatory response. IUBMB Life 2017, 69, 442–450. [Google Scholar] [CrossRef]
- Goncharov, I.; Weiner, L.; Vogel, Z. Δ9-tetrahydrocannabinol increases C6 glioma cell death produced by oxidative stress. Neuroscience 2005, 134, 567–574. [Google Scholar] [CrossRef]
- Szabó, E.; Türk, D.; Telbisz, Á.; Kucsma, N.; Horváth, T.; Szakács, G.; Homolya, L.; Sarkadi, B.; Várady, G. A new fluorescent dye accumulation assay for parallel measurements of the ABCG2, ABCB1 and ABCC1 multidrug transporter functions. PLoS ONE 2018, 13, e0190629. [Google Scholar] [CrossRef] [PubMed]
- Nagai, Y.; Matsuoka, T.A.; Shimo, N.; Miyatsuka, T.; Miyazaki, S.; Tashiro, F.; Miyazaki, J.I.; Katakami, N.; Shimomura, I. Glucotoxicity-induced suppression of Cox6a2 expression provokes β-cell dysfunction via augmented ROS production. Biochem. Biophys. Res. Commun. 2021, 556, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Ransy, C.; Vaz, C.; Lombès, A.; Bouillaud, F. Use of H2O2 to cause oxidative stress, the catalase issue. Int. J. Mol. Sci. 2020, 21, 9149. [Google Scholar] [CrossRef]
- Xie, W.; Lorenz, S.; Dolder, S.; Hofstetter, W. Extracellular iron is a modulator of the differentiation of osteoclast lineage cells. Calcif. Tissue Int. 2016, 98, 275–283. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carter, T.H.; Weyer-Nichols, C.E.; Garcia-Sanchez, J.I.; Wilson, K.; Nagarkatti, P.; Nagarkatti, M. Delta-9-Tetrahydrocannabinol Blocks Bone Marrow-Derived Macrophage Differentiation through Elimination of Reactive Oxygen Species. Antioxidants 2024, 13, 887. https://doi.org/10.3390/antiox13080887
Carter TH, Weyer-Nichols CE, Garcia-Sanchez JI, Wilson K, Nagarkatti P, Nagarkatti M. Delta-9-Tetrahydrocannabinol Blocks Bone Marrow-Derived Macrophage Differentiation through Elimination of Reactive Oxygen Species. Antioxidants. 2024; 13(8):887. https://doi.org/10.3390/antiox13080887
Chicago/Turabian StyleCarter, Taylor H., Chloe E. Weyer-Nichols, Jeffrey I. Garcia-Sanchez, Kiesha Wilson, Prakash Nagarkatti, and Mitzi Nagarkatti. 2024. "Delta-9-Tetrahydrocannabinol Blocks Bone Marrow-Derived Macrophage Differentiation through Elimination of Reactive Oxygen Species" Antioxidants 13, no. 8: 887. https://doi.org/10.3390/antiox13080887





