Isolation and Characterization of Antioxidant Peptides from Dairy Cow (Bos taurus) Placenta and Their Antioxidant Activities
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Virtual Screening Method to Isolate AOP
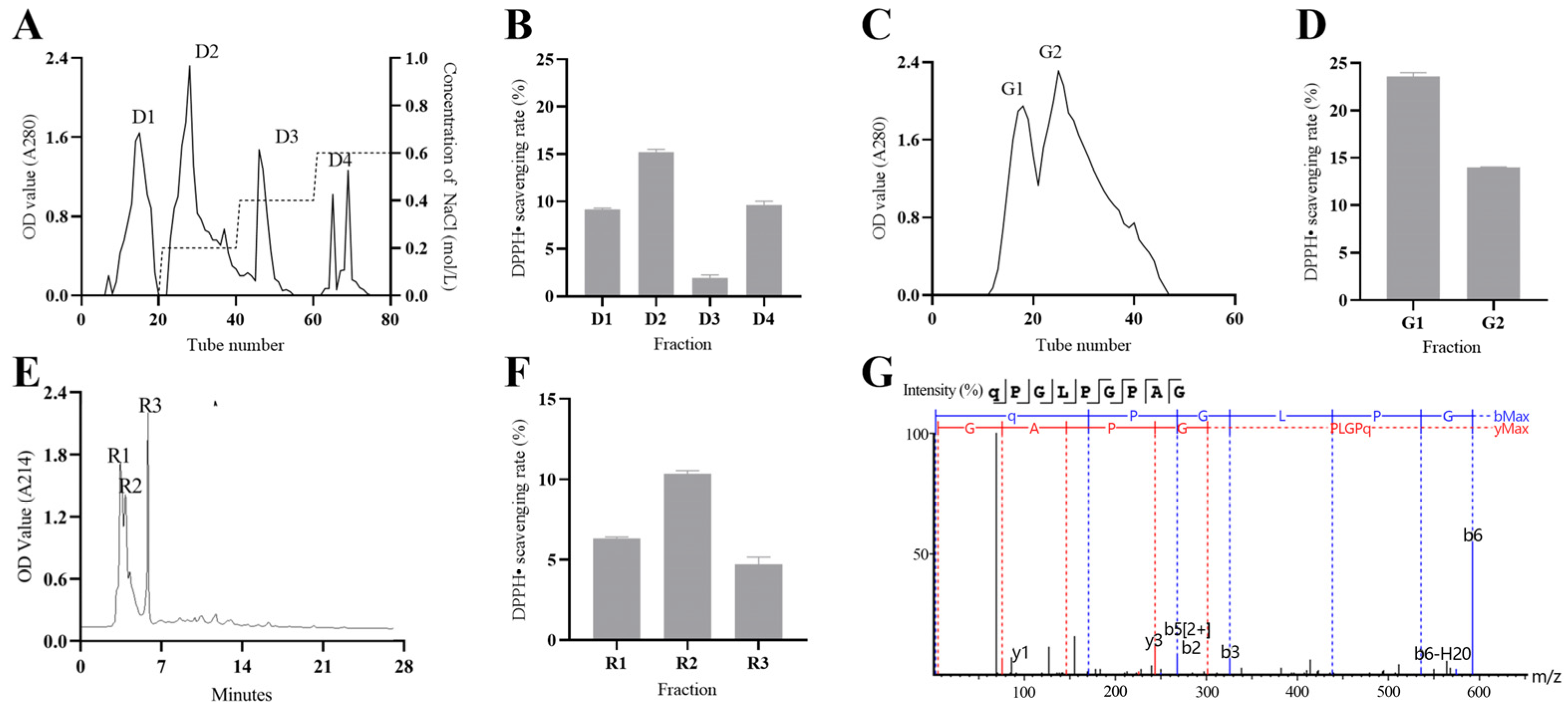
2.3. Chromatography to Isolate AOPs
2.3.1. Ion Exchange Chromatography
2.3.2. Molecular Sieve Chromatographic Separation
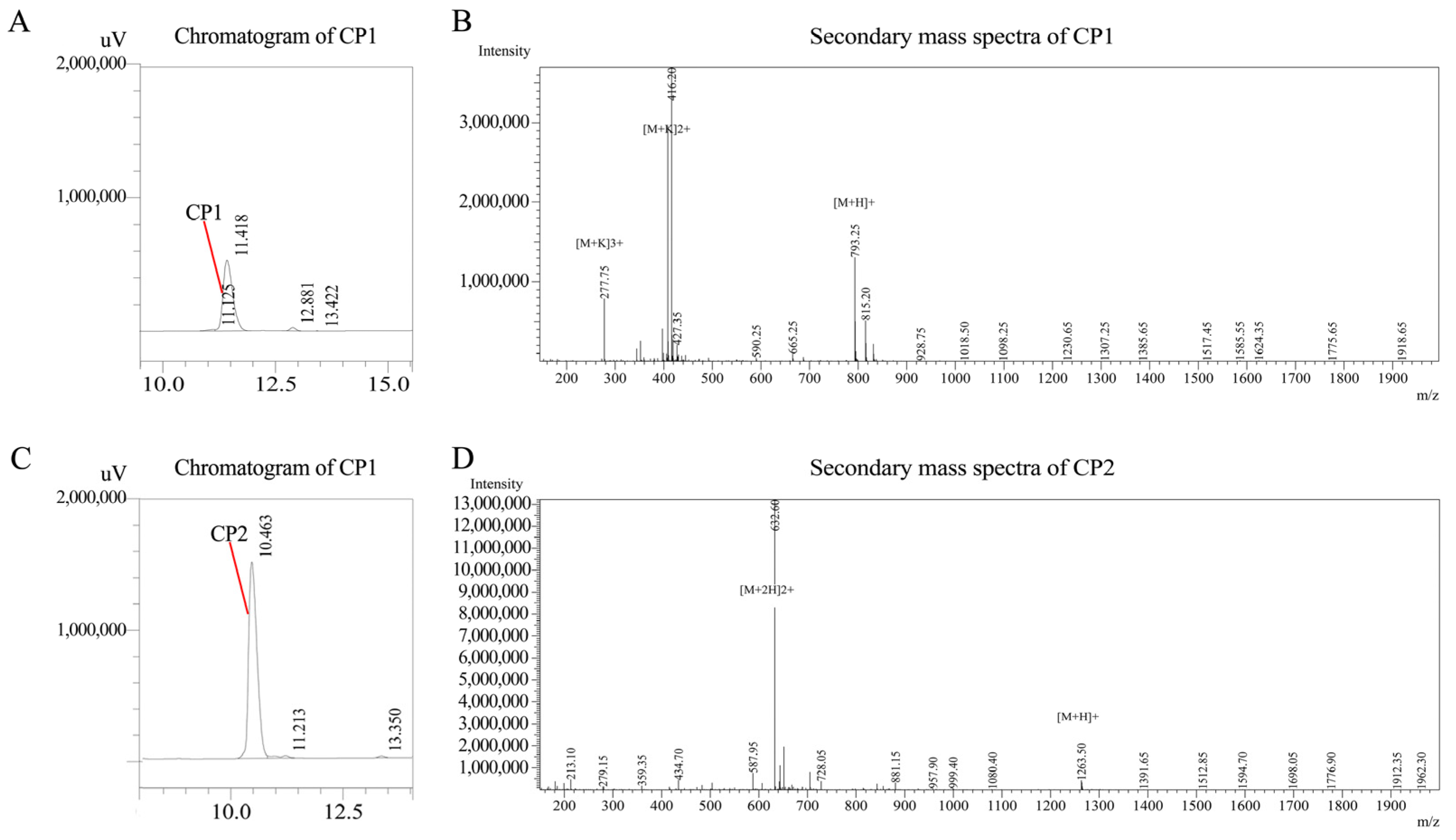
2.3.3. RP-HPLC Separation
2.3.4. LC–MS/MS Identification
2.4. Synthesize CP1 and CP2
2.5. In Vitro Assay of CP1 and CP2 Antioxidant Activities
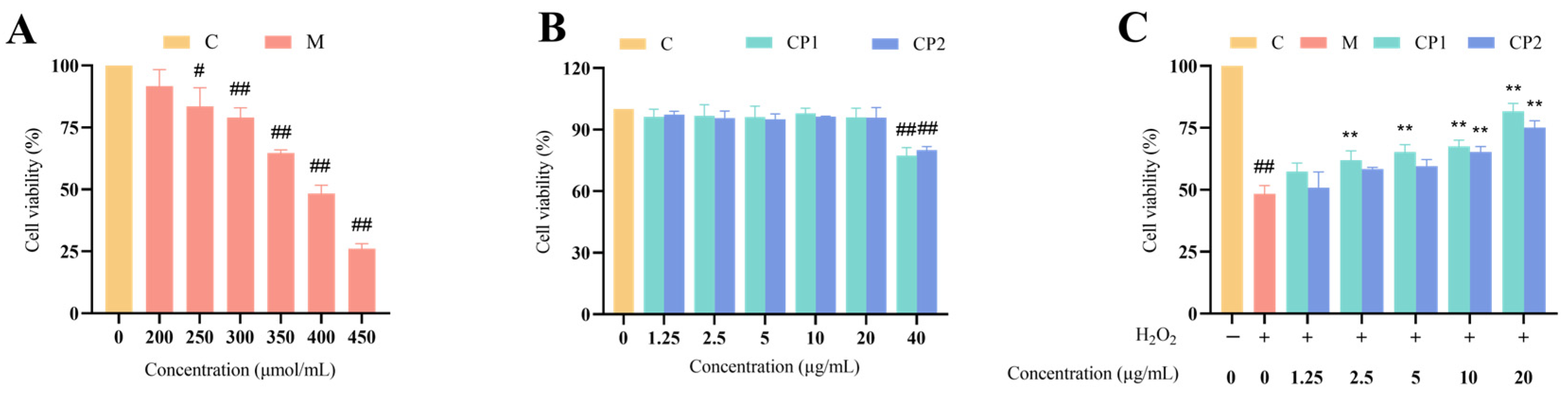
2.6. Establishment of the H2O2-Induced Oxidative Stress RAW264.7 Cell Model
2.7. Effects of CP1 and CP2 on the Survival of Normal and Oxidatively Stressed Cells
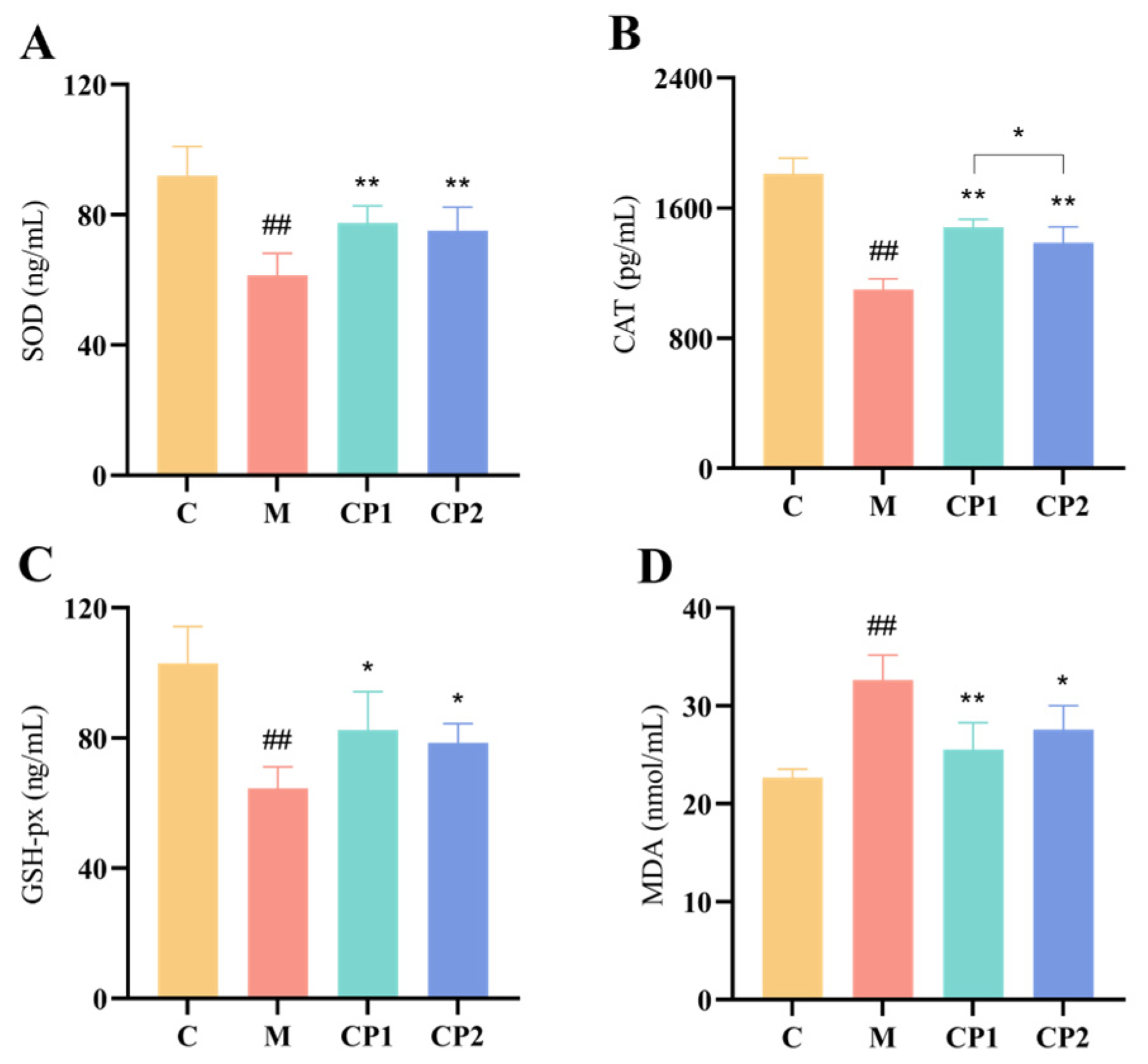
2.8. RAW264.7 Detection of Cellular Antioxidant Enzymes and MDA
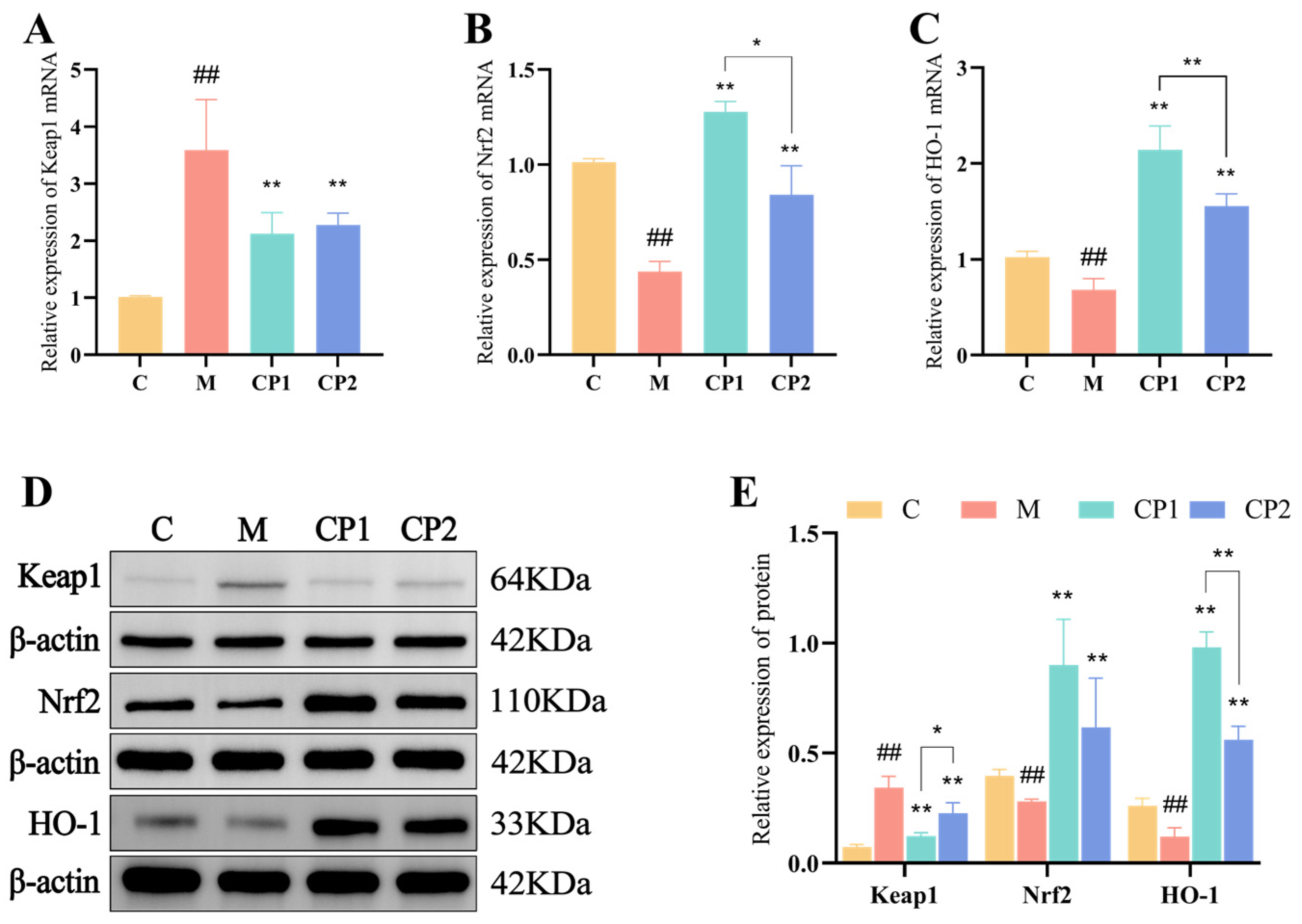
2.9. RAW264.7 Detection of Relative Expression of Keap1, Nrf2, and HO-1 Genes in Cells
2.10. RAW264.7 Detection of Relative Expression of Keap1, Nrf2, and HO-1 Proteins in Cells
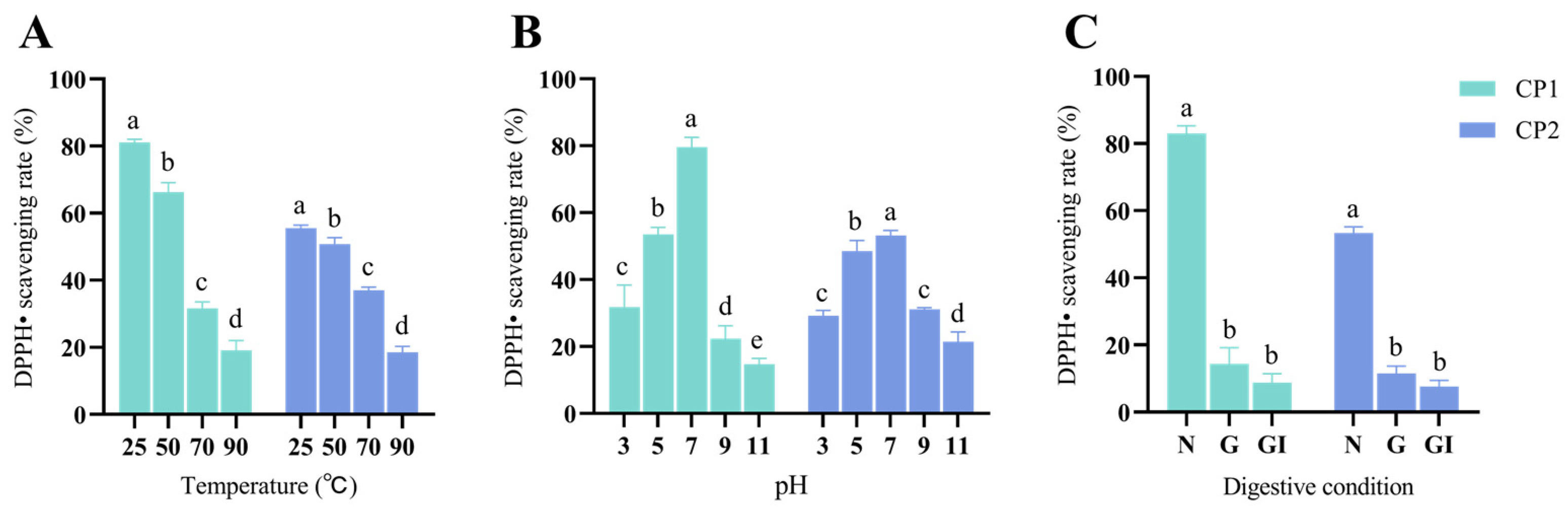
2.11. Detecting CP1 and CP2 Stability
2.12. Statistical Analysis
3. Results
3.1. Virtual Screening Method to Isolate CP1
3.2. Chromatography to Isolate CP2
3.3. Purity Analysis of Synthetic AOPs
3.4. In Vitro Antioxidant Activity of CP1 and CP2
3.5. Effect of H2O2, CP1, and CP2 on the Survival of RAW264.7 Cells
3.6. Effects of CP1 and CP2 on Antioxidant Enzymes and MDA in Oxidatively Stressed RAW264.7 Cells
3.7. Effects of CP1 and CP2 on Antioxidant Pathways in Oxidatively Stressed RAW264.7 Cells
3.8. Stability of CP1 and CP2
4. Discussion
4.1. Effects of Separation Method on the Separation of AOPs
4.2. Evaluation of the Antioxidant Activity of CP1 and CP2
4.3. Comparative Analysis of the Antioxidant Activity of CP1 and CP2
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, W.; Ren, J.; Wu, H.; Zhang, X.; Han, L.; Gu, R. Inhibitory effects and action mechanism of five antioxidant peptides derived from wheat gluten on cells oxidative stress injury. Food Biosci. 2023, 56, 103236. [Google Scholar] [CrossRef]
- Lv, R.; Dong, Y.; Bao, Z.; Zhang, S.; Lin, S.; Sun, N. Advances in the activity evaluation and cellular regulation pathways of food-derived antioxidant peptides. Trends Food Sci. Technol. 2022, 122, 171–186. [Google Scholar] [CrossRef]
- Zhu, L.; Xiong, H.; Huang, X.; Guyonnet, V.; Ma, M.; Chen, X.; Zheng, Y.; Wang, L.; Hu, G. Identification and molecular mechanisms of novel antioxidant peptides from two sources of eggshell membrane hydrolysates showing cytoprotection against oxidative stress: A combined in silico and in vitro study. Food Res. Int. 2022, 157, 111266. [Google Scholar] [CrossRef]
- Zou, L.; Yu, X.; Cai, K.; Xu, B.; Chen, C.; Xiao, G. Identification of antioxidant peptides targeting Keap1–Nrf2–ARE pathway from in vitro digestion of pork sausage with partial substitution of NaCl by KCl. Food Res. Int. 2023, 174, 113585. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, Y.; Liu, M.; Li, X.; Xu, H.; Liu, L. Application of virtual screening technology in the research of traditional Chinese medicine. Chin. J. New Drugs 2022, 31, 1676–1683. [Google Scholar] [CrossRef]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Bharadvaja, N. Treasuring the computational approach in medicinal plant research. Prog. Biophys. Mol. Biol. 2021, 164, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, D.; Caballero, J. Is It Reliable to Use Common Molecular Docking Methods for Comparing the Binding Affinities of Enantiomer Pairs for Their Protein Target? Int. J. Mol. Sci. 2016, 17, 525. [Google Scholar] [CrossRef]
- Wen, C.; Zhang, J.; Zhang, H.; Duan, Y.; Ma, H. Plant protein-derived antioxidant peptides: Isolation, identification, mechanism of action and application in food systems: A review. Trends Food Sci. Technol. 2020, 105, 308–322. [Google Scholar] [CrossRef]
- Jaradat, D. Thirteen decades of peptide synthesis: Key developments in solid phase peptide synthesis and amide bond formation utilized in peptide ligation. Amino Acids 2018, 50, 39–68. [Google Scholar] [CrossRef]
- Liu, R.; Xing, L.; Fu, Q.; Zhou, G.H.; Zhang, W.G. A Review of Antioxidant Peptides Derived from Meat Muscle and By-Products. Antioxidants 2016, 5, 32. [Google Scholar] [CrossRef] [PubMed]
- Shahidi, F.; Zhong, Y. Measurement of antioxidant activity. J. Funct. Foods 2015, 18, 757–781. [Google Scholar] [CrossRef]
- Zhu, Y.; Lao, F.; Pan, X.; Wu, J. Food Protein-Derived Antioxidant Peptides: Molecular Mechanism, Stability and Bioavailability. Biomolecules 2022, 12, 1622. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z. Chemical Methods to Evaluate Antioxidant Ability. Chem. Rev. 2010, 110, 5675–5691. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.N.; Bristi, N.J.; Rafiquzzaman, M. Review on in vivo and in vitro methods evaluation of antioxidant activity. Saudi Pharm. J. 2013, 21, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.H.; Fan, L.; Zhang, Y.; Zhu, Y.K.; Zong, X.L.; Peng, G.N.; Cao, S.Z. Protective effect and mechanism of placenta extract on liver. Nutrients 2022, 14, 5071. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.H.; Zhang, Y.; You, L.C.; Zhu, Y.K.; Shi, T.; Dong, K.; Cao, S.Z. Establishment and optimization by different enzymolysis methods of reducing polypeptide preparation from dairy cows placenta. J. Northeast Agric. Univ. 2021, 52, 54–63. [Google Scholar] [CrossRef]
- Shen, L.H.; Fan, L.; Luo, H.; Li, W.; Cao, S.Z.; Yu, S.M. Cow placenta extract ameliorates d-galactose-induced liver damage by regulating BAX/CASP3 and p53/p21/p16 pathways. J. Ethnopharmacol. 2024, 323, 117685–117701. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.H.; Fan, L.; Zhang, Y.; Shen, Y.; Su, Z.T.; Peng, G.N.; Deng, J.L.; Zhong, Z.J.; Wu, X.; Yu, S.M.; et al. Antioxidant capacity and protective effect of cow placenta extract on d-galactose-induced skin aging in mice. Nutrients 2022, 14, 4659. [Google Scholar] [CrossRef]
- Zhang, Y.; You, L.C.; Zhu, Y.K.; Shen, Y.; Lv, S.K.; Pei, M.L.; Fu, H.Q.; Shen, L.H. LC-MS/MS based comparative analysis and antioxidant activity prediction of cow placenta hydrolyzed by different proteases. J. China Agric. Univ. 2021, 26, 133–141. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v.2—A server for in silico prediction of allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Consortium, D.; Raghava, G.P. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Jiao, L.; Wang, R.; Zhao, Y.; Hao, Y.; Liang, G. Prediction of antioxidant peptides using a quantitative structure–activity relationship predictor (AnOxPP) based on bidirectional long short-term memory neural network and interpretable amino acid descriptors. Comput. Biol. Med. 2023, 154, 106591. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Li, X.; Wang, D.; Zhao, B.; Shi, Y.; Huang, A. Discovery of specific antioxidant peptide from Chinese Dahe black pig and hybrid pig dry-cured hams based on peptidomics strategy. Food Res. Int. 2023, 166, 112610. [Google Scholar] [CrossRef]
- Fu, J.; Hu, X.; Ma, S.; Wang, F.; Guo, G.; Huang, K.; Yang, Y.; He, X. Preparation Technology and Antioxidant Activities of Different Molecular Weight Macadamia Nut Polypeptides. Sci. Technol. Food Ind. 2023, 44, 414–421. [Google Scholar] [CrossRef]
- Hogan, S.; Zhang, L.; Li, J.; Wang, H.; Zhou, K. Development of antioxidant rich peptides from milk protein by microbial proteases and analysis of their effects on lipid peroxidation in cooked beef. Food Chem. 2009, 117, 438–443. [Google Scholar] [CrossRef]
- Gu, F.; Kim, J.M.; Hayat, K.; Xia, S.; Feng, B.; Zhang, X. Characteristics and antioxidant activity of ultrafiltrated Maillard reaction products from a casein–glucose model system. Food Chem. 2009, 117, 48–54. [Google Scholar] [CrossRef]
- Chai, T.; Xiao, J.; Mohana Dass, S.; Teoh, J.; Ee, K.; Ng, W.; Wong, F. Identification of antioxidant peptides derived from tropical jackfruit seed and investigation of the stability profiles. Food Chem. 2021, 340, 127876. [Google Scholar] [CrossRef] [PubMed]
- Gallego, M.; Mora, L.; Toldrá, F. Characterisation of the antioxidant peptide AEEEYPDL and its quantification in Spanish dry-cured ham. Food Chem. 2018, 258, 8–15. [Google Scholar] [CrossRef]
- Li, L.; Liu, J.; Nie, S.; Ding, L.; Wang, L.; Liu, J.; Liu, W.; Zhang, T. Direct inhibition of Keap1–Nrf2 interaction by egg-derived peptides DKK and DDW revealed by molecular docking and fluorescence polarization. RSC Adv. 2017, 7, 34963–34971. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Ren, T.; Xiao, P.; Duan, J.A. Novel and efficient techniques in the discovery of antioxidant peptides. Crit. Rev. Food Sci. Nutr. 2023, 10, 1–15. [Google Scholar] [CrossRef]
- Zhang, Q.; Yu, Z.; Zhao, W. Identification and action mechanism of novel antioxidative peptides from copra meal protein. Food Sci. Technol. 2023, 188, 115425. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, Z.; Zhuang, J.; Zhang, J.; Shen, F.; Yu, P.; Zhong, H.; Feng, F. Antiaging function of Chinese pond turtle (Chinemys reevesii) peptide through activation of the Nrf2/Keap1 signaling pathway and its structure-activity relationship. Front. Nutr. 2022, 9, 961922. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Guan, K.; Liu, M.; Liu, D.; Wang, W.; Zhu, A. Novel antioxidant peptides from MFGM protein Hydrolysates: Separation, identification and effects on Dexamethasone-induced mitochondrial dysfunction. Food Chem. 2023, 403, 134473. [Google Scholar] [CrossRef]
- Ren, L.; Yang, Y.; Ma, C.; Fan, J.; Bian, X.; Liu, B.; Wang, D.; Zhu, P.; Fu, Y.; Zhang, N. Identification and in silico analysis of novel antioxidant peptides in broken rice protein hydrolysate and its cytoprotective effect against H2O2-induced 2BS cell model. Food Res. Int. 2022, 162, 112108. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Huang, Z.; Tang, S.; Lu, C.; Wan, H.; Zhou, J.; Li, Y.; Ming, T.; Jim, W.Z.; Su, X. The novel peptides ICRD and LCGEC screened from tuna roe show antioxidative activity via Keap1/Nrf2-ARE pathway regulation and gut microbiota modulation. Food Chem. 2020, 327, 127094. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Li, L.; Chi, C.; Wang, B. Twelve Antioxidant Peptides from Protein Hydrolysate of Skipjack Tuna (Katsuwonus pelamis) Roe Prepared by Flavourzyme: Purification, Sequence Identification, and Activity Evaluation. Front. Nutr. 2022, 8, 813780. [Google Scholar] [CrossRef] [PubMed]
- Nwachukwu, I.D.; Sarteshnizi, R.A.; Udenigwe, C.C.; Aluko, R.E. A Concise Review of Current In Vitro Chemical and Cell-Based Antioxidant Assay Methods. Molecules 2021, 26, 4865. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, X.; Zhuang, Y.; Li, Y.; Tian, H.; Shi, P.; Li, G. Isolation of Novel ACE-Inhibitory and Antioxidant Peptides from Quinoa Bran Albumin Assisted with an In Silico Approach: Characterization, In Vivo Antihypertension, and Molecular Docking. Molecules 2019, 24, 4562. [Google Scholar] [CrossRef]
- Yang, J.; Huang, J.; Dong, X.; Zhang, Y.; Zhou, X.; Huang, M.; Zhou, G. Purification and identification of antioxidant peptides from duck plasma proteins. Food Chem. 2020, 319, 126534. [Google Scholar] [CrossRef]
- Guo, S.; Ding, W.; Pang, X.; Cheng, Z. Preparation of Collagen Antioxidant Peptides from Sheepskin Scraps. China Leather 2023, 52, 1–7. [Google Scholar] [CrossRef]
- He, Y.; Pan, X.; Chi, C.; Sun, K.; Wang, B. Ten new pentapeptides from protein hydrolysate of miiuy croaker (Miichthys miiuy) muscle: Preparation, identification, and antioxidant activity evaluation. LWT 2019, 105, 1–8. [Google Scholar] [CrossRef]
- Chai, T.T.; Law, Y.C.; Wong, F.C.; Kim, S.K. Enzyme-Assisted Discovery of Antioxidant Peptides from Edible Marine Invertebrates: A Review. Mar. Drugs 2017, 15, 42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, G.; Suo, S.; Wang, Y.; Chi, C.; Wang, B. Purification, Identification, Activity Evaluation, and Stability of Antioxidant Peptides from Alcalase Hydrolysate of Antarctic Krill (Euphausia superba) Proteins. Mar. Drugs 2021, 19, 347. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zheng, X.; Kopparapu, N.; Cong, W.; Deng, Y.; Sun, X.; Liu, X. Purification and evaluation of a novel antioxidant peptide from corn protein hydrolysate. Process Biochem. 2014, 49, 1562–1569. [Google Scholar] [CrossRef]
- Rahman, M.S.; Hee Choi, Y.; Seok Choi, Y.; Alam, M.B.; Han Lee, S.; Cheol Yoo, J. A novel antioxidant peptide, purified from Bacillus amyloliquefaciens, showed strong antioxidant potential via Nrf-2 mediated heme oxygenase-1 expression. Food Chem. 2018, 239, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Rajapakse, N.; Mendis, E.; Jung, W.; Je, J.; Kim, S. Purification of a radical scavenging peptide from fermented mussel sauce and its antioxidant properties. Food Res. Int. 2005, 38, 175–182. [Google Scholar] [CrossRef]
- Qian, Z.; Jung, W.; Kim, S. Free radical scavenging activity of a novel antioxidative peptide purified from hydrolysate of bullfrog skin, Rana catesbeiana Shaw. Bioresour. Technol. 2008, 99, 1690–1698. [Google Scholar] [CrossRef] [PubMed]
- Saiga, A.; Tanabe, S.; Nishimura, T. Antioxidant Activity of Peptides Obtained from Porcine Myofibrillar Proteins by Protease Treatment. J. Agric. Food Chem. 2003, 51, 3661–3667. [Google Scholar] [CrossRef]
- Li, Y.; Yu, J. Research Progress in Structure-Activity Relationship of Bioactive Peptides. J. Med. Food 2015, 18, 147–156. [Google Scholar] [CrossRef]
- Saadi, S.; Saari, N.; Anwar, F.; Abdul Hamid, A.; Ghazali, H.M. Recent advances in food biopeptides: Production, biological functionalities and therapeutic applications. Biotechnol. Adv. 2015, 33, 80–116. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, S.; Rong, Y.; Zeng, W.; Hu, Z.; Ma, X.; Feng, S. Identification and evaluation of antioxidant peptides from highland barley distiller’s grains protein hydrolysate assisted by molecular docking. Food Chem. 2024, 434, 137441. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Feng, M.; Sun, J. Novel antioxidant peptides in fermented pork sausage: Purification, characterization, and cytoprotective functions on Caco-2 cells. Food Chem. 2023, 426, 136566. [Google Scholar] [CrossRef] [PubMed]

| Physicochemical | Websites |
|---|---|
| Antioxidant activity | AnOxPP (http://www.cqudfbp.net/AnOxPP/index.jsp), accessed on 8 August 2023 |
| Stability | ProtParam (http://web.expasy.org/protparam/), accessed on 13 August 2023 |
| Water solubility | Innovagen (http://www.innovagen.com/proteomics-tools/), accessed on 26 July 2023 |
| Sensitization | AllerTOP (v.2.0) (https://www.ddg-pharmfac.net/AllerTOP/), accessed on 30 July 2023 |
| Toxicity | ToxinPred (http://crdd.osdd.net/raghava/toxinpred/), accessed on 8 August 2023 |
| Peptide | Antioxidant Activity | Stability | Water Solubility | Sensitization | Toxicity |
|---|---|---|---|---|---|
| DLFENTNHTQVQ | + | −29.28 | −1.23 | – | – |
| ANNGKQWAEVF | + | −4.43 | −0.78 | – | – |
| WELTDDKNQRFF | + | −4.19 | −1.51 | – | – |
| RDNLLDDLQRLK | + | 29.26 | −1.27 | – | – |
| DGRHDPRDDDLNLR | + | 31.41 | −2.29 | – | – |
| Peptide | Molecular Weight | −CIE |
|---|---|---|
| ANNGKQWAEVF | 1263.38 Da | 109.87 KJ/mol |
| RDNLLDDLQRLK | 1498.68 Da | 109.32 KJ/mol |
| DGRHDPRDDDLNLR | 1693.72 Da | 105.85 KJ/mol |
| DLFENTNHTQVQ | 1445.48 Da | 105.81 KJ/mol |
| WELTDDKNQRFF | 1598.70 Da | 51.94 KJ/mol |
| TX6 | 331.40 Da | 25.58 KJ/mol |
| Antioxidant Ability | CP1 (mg/mL) | CP2 (mg/mL) |
|---|---|---|
| Hydroxyl radical scavenging rate (IC50) | 1.00 | 1.34 |
| DPPH free-radical scavenging rate (IC50) | 0.85 | 1.71 |
| Transition metal chelation rate (IC50) | 4.67 | 5.20 |
| Lipid peroxidation inhibition rate (IC50) | 3.49 | 3.90 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, X.; Zhang, Z.; Zhao, Y.; Tang, A.; Zeng, Z.; Zheng, W.; Zhang, H.; Luo, Y.; Lu, W.; Fan, L.; et al. Isolation and Characterization of Antioxidant Peptides from Dairy Cow (Bos taurus) Placenta and Their Antioxidant Activities. Antioxidants 2024, 13, 913. https://doi.org/10.3390/antiox13080913
Tian X, Zhang Z, Zhao Y, Tang A, Zeng Z, Zheng W, Zhang H, Luo Y, Lu W, Fan L, et al. Isolation and Characterization of Antioxidant Peptides from Dairy Cow (Bos taurus) Placenta and Their Antioxidant Activities. Antioxidants. 2024; 13(8):913. https://doi.org/10.3390/antiox13080913
Chicago/Turabian StyleTian, Xinyu, Zeru Zhang, Yuquan Zhao, Anguo Tang, Zhi Zeng, Weijian Zheng, Hanwen Zhang, Yuxin Luo, Wei Lu, Lei Fan, and et al. 2024. "Isolation and Characterization of Antioxidant Peptides from Dairy Cow (Bos taurus) Placenta and Their Antioxidant Activities" Antioxidants 13, no. 8: 913. https://doi.org/10.3390/antiox13080913
APA StyleTian, X., Zhang, Z., Zhao, Y., Tang, A., Zeng, Z., Zheng, W., Zhang, H., Luo, Y., Lu, W., Fan, L., & Shen, L. (2024). Isolation and Characterization of Antioxidant Peptides from Dairy Cow (Bos taurus) Placenta and Their Antioxidant Activities. Antioxidants, 13(8), 913. https://doi.org/10.3390/antiox13080913






