Gut Microbiota Dysbiosis, Oxidative Stress, Inflammation, and Epigenetic Alterations in Metabolic Diseases
Abstract
1. Introduction
2. Gut Microbiota Structure and Gut Microbiome Development in Mammals
3. The Gut Microbiome and Diet Interaction
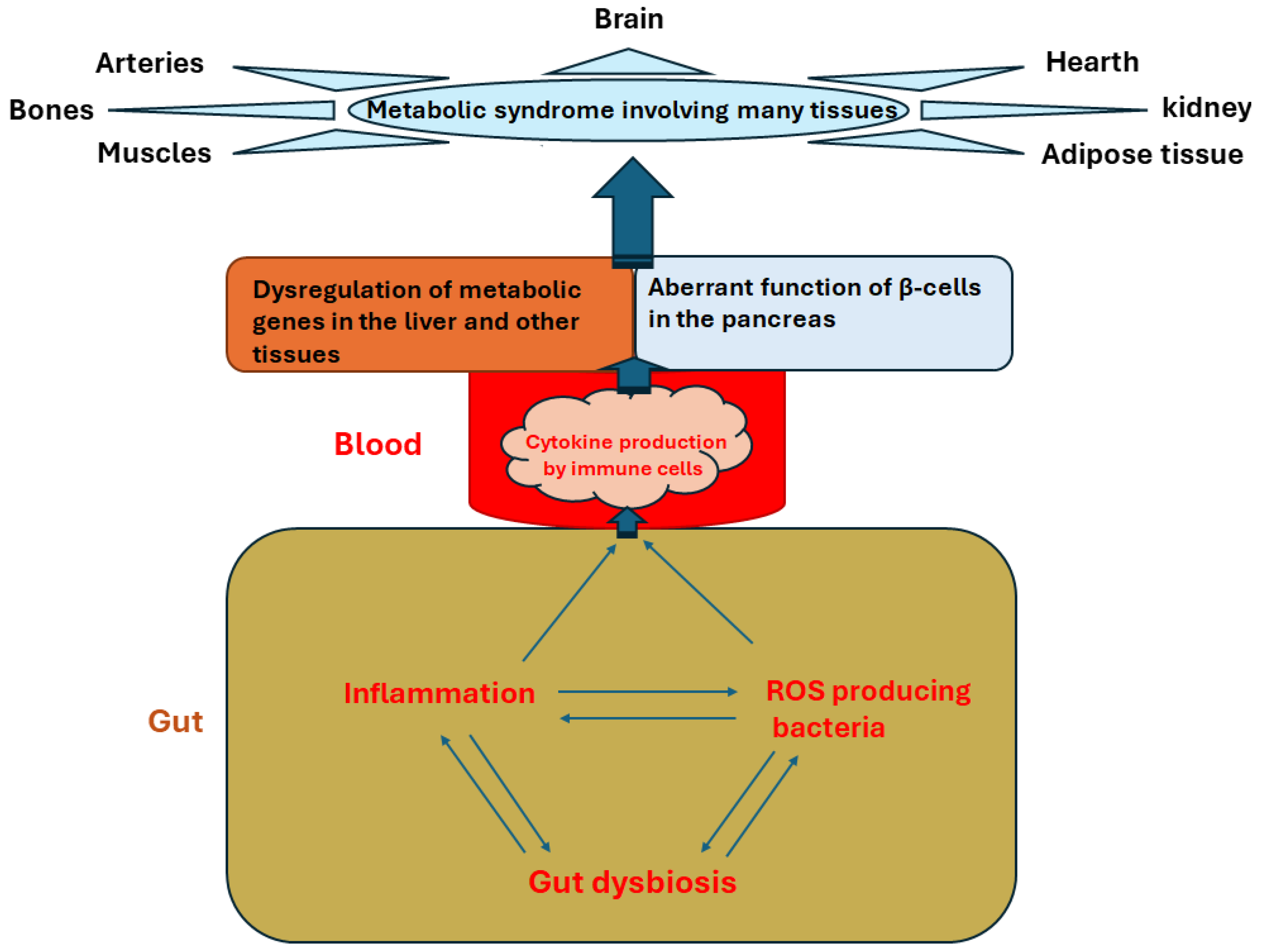
4. Gut Dysbiosis and ROS Production
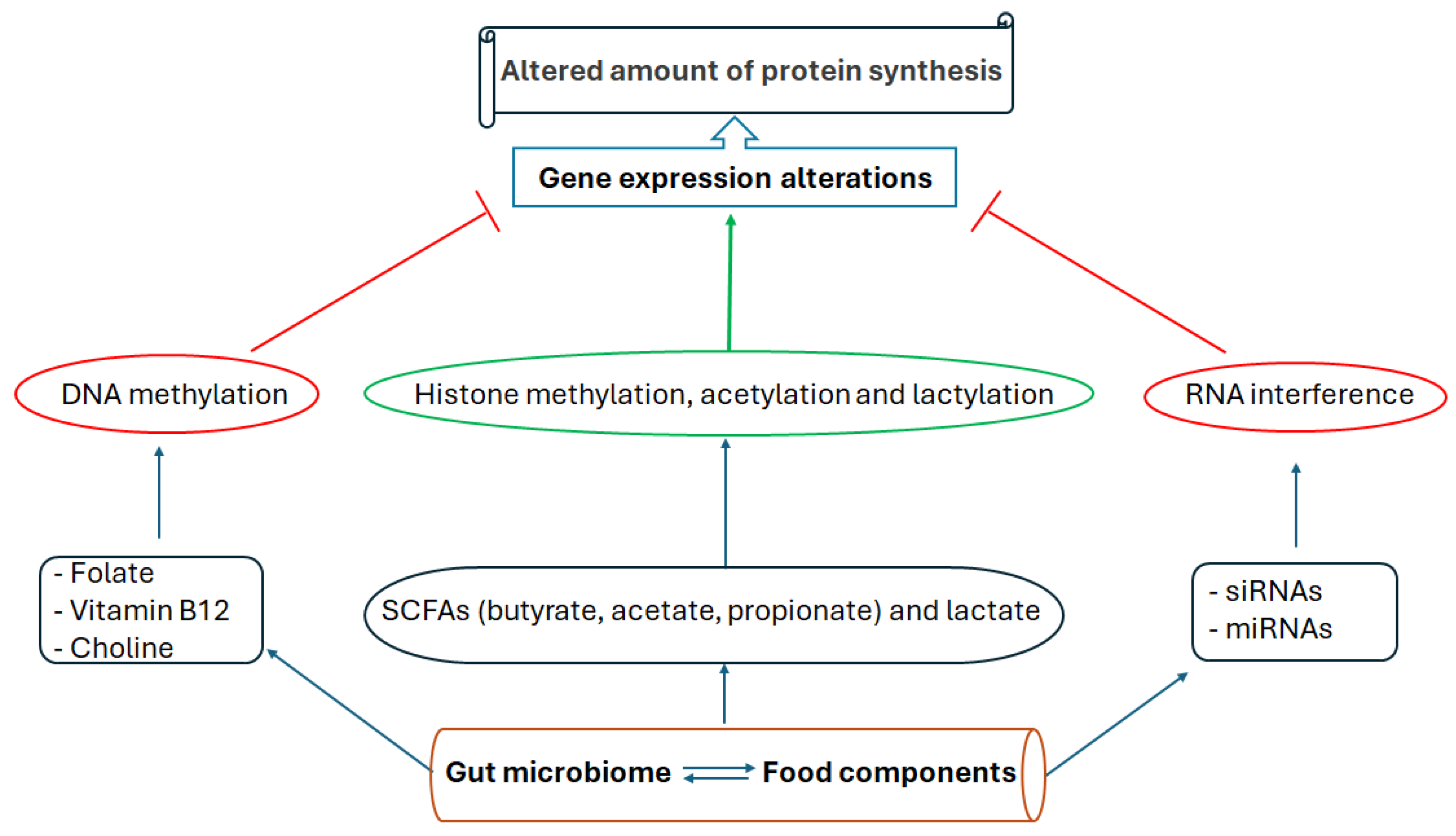
5. Metabolic Impacts of Gut Dysbiosis Involving Epigenetic Mechanisms
6. Gut Microbiome, Inflammation, ROS, and DNA Methylome Interactions
7. Transfer of Gut Microbiota-Related Metabolic Diseases to the Next Generation through Epigenetic Mechanism
8. Dietary and Probiotic Interventions to Modulate Gut Microbiome, ROS, and Metabolic Diseases
9. Dietary and Microbiome-Induced Health Benefits Mediated by Epigenetic Modifications
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alemany, M. The metabolic syndrome, a human disease. Int. J. Mol. Sci. 2024, 25, 2251. [Google Scholar] [CrossRef] [PubMed]
- Kivimäki, M.; Bartolomucci, A.; Kawachi, I. The multiple roles of life stress in metabolic disorders. Nat. Rev. Endocrinol. 2023, 19, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Defesche, J.C.; Gidding, S.S.; Harada-Shiba, M.; Hegele, R.A.; Santos, R.D.; Wierzbicki, A.S. Familial hypercholesterolaemia. Nat. Rev. Dis. Primers 2017, 3, 17093. [Google Scholar] [CrossRef] [PubMed]
- Portincasa, P.; Khalil, M.; Graziani, A.; Frühbeck, G.; Baffy, G.; Garruti, G.; Di Ciaula, A.; Bonfrate, L. Gut microbes in metabolic disturbances. Promising role for therapeutic manipulations? Eur. J. Intern. Med. 2023, 119, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, J.; Seok, J.W.; Park, C.G.; Jun, J. Comprehensive lifestyle modification interventions for metabolic syndrome: A systematic review and meta-analysis. J. Nurs. Scholarsh. 2024, 56, 249–259. [Google Scholar] [CrossRef]
- Stilling, R.M.; Dinan, T.G.; Cryan, J.F. Microbial genes, brain & behaviour–epigenetic regulation of the gut–brain axis. Genes Brain Behav. 2014, 13, 69–86. [Google Scholar]
- Hugon, P.; Dufour, J.-C.; Colson, P.; Fournier, P.-E.; Sallah, K.; Raoult, D. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect. Dis. 2015, 15, 1211–1219. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef]
- Parker, A.; Fonseca, S.; Carding, S.R. Gut microbes and metabolites as modulators of blood-brain barrier integrity and brain health. Gut Microbes 2020, 11, 135–157. [Google Scholar] [CrossRef]
- Martel, J.; Chang, S.-H.; Ko, Y.-F.; Hwang, T.-L.; Young, J.D.; Ojcius, D.M. Gut barrier disruption and chronic disease. Trends Endocrinol. Metab. 2022, 33, 247–265. [Google Scholar] [CrossRef]
- Rescigno, M. Dendritic cell–epithelial cell crosstalk in the gut. Immunol. Rev. 2014, 260, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Rescigno, M.; Di Sabatino, A. Dendritic cells in intestinal homeostasis and disease. J. Clin. Investig. 2009, 119, 2441–2450. [Google Scholar] [CrossRef] [PubMed]
- Stagg, A.J. Intestinal dendritic cells in health and gut inflammation. Front. Immunol. 2018, 9, 2883. [Google Scholar] [CrossRef] [PubMed]
- Castro Dopico, X.; Guryleva, M.; Mandolesi, M.; Corcoran, M.; Coquet, J.M.; Murrell, B.; Karlsson Hedestam, G.B. Maintenance of caecal homeostasis by diverse adaptive immune cells in the rhesus macaque. Clin. Transl. Immunol. 2024, 13, e1508. [Google Scholar] [CrossRef] [PubMed]
- Ruder, B.; Becker, C. At the forefront of the mucosal barrier: The role of macrophages in the intestine. Cells 2020, 9, 2162. [Google Scholar] [CrossRef] [PubMed]
- Koleva, P.T.; Kim, J.S.; Scott, J.A.; Kozyrskyj, A.L. Microbial programming of health and disease starts during fetal life. Birth Defects Res. Part C Embryo Today Rev. 2015, 105, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, K.M.; Gerlach, M.J.; Adam, T.; Heimesaat, M.M.; Rossi, L.; Surette, M.G.; Sloboda, D.M.; Braun, T. Fetal meconium does not have a detectable microbiota before birth. Nat. Microbiol. 2021, 6, 865–873. [Google Scholar] [CrossRef]
- Blaser, M.J.; Devkota, S.; McCoy, K.D.; Relman, D.A.; Yassour, M.; Young, V.B. Lessons learned from the prenatal microbiome controversy. Microbiome 2021, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Qin, Y.; Chen, M.; Li, X.; Wang, R.; Huang, Z.; Xu, Q.; Yu, M.; Zhang, Y.; Han, X. Prenatal low-dose DEHP exposure induces metabolic adaptation and obesity: Role of hepatic thiamine metabolism. J. Hazard. Mater. 2020, 385, 121534. [Google Scholar] [CrossRef]
- Shukla, S.; Srivastava, A.; Verma, D.; Gangopadhyay, S.; Chauhan, A.; Srivastava, V.; Budhwar, S.; Tyagi, D.; Sharma, D.C. Analysis of gut bacteriome of in utero arsenic-exposed mice using 16S rRNA-based metagenomic approach. Front. Microbiol. 2023, 14, 1147505. [Google Scholar] [CrossRef]
- Bittinger, K.; Zhao, C.; Li, Y.; Ford, E.; Friedman, E.; Ni, J.; Kulkarni, C.; Cai, J.; Tian, Y.; Liu, Q. Bacterial colonization reprograms the neonatal gut metabolome. Nat. Microbiol. 2020, 5, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Shi, Y.; Meng, L.; Fan, H.; Tang, X.; Luo, H.; Wang, D.; Zhou, J.; Xiao, X. Factors affecting the early establishment of neonatal intestinal flora and its intervention measures. Front. Cell. Infect. Microbiol. 2023, 13, 1295111. [Google Scholar] [CrossRef] [PubMed]
- Willers, M.; Ulas, T.; Völlger, L.; Vogl, T.; Heinemann, A.S.; Pirr, S.; Pagel, J.; Fehlhaber, B.; Halle, O.; Schöning, J. S100A8 and S100A9 are important for postnatal development of gut microbiota and immune system in mice and infants. Gastroenterology 2020, 159, 2130–2145.e2135. [Google Scholar] [CrossRef] [PubMed]
- Thaiss, C.A.; Levy, M.; Korem, T.; Dohnalová, L.; Shapiro, H.; Jaitin, D.A.; David, E.; Winter, D.R.; Gury-BenAri, M.; Tatirovsky, E. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 2016, 167, 1495–1510.e1412. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.G.; Sato, T.; Shimaji, K.; Koronowski, K.B.; Petrus, P.; Cervantes, M.; Kinouchi, K.; Lutter, D.; Dyar, K.A.; Sassone-Corsi, P. Antibiotic-induced microbiome depletion remodels daily metabolic cycles in the brain. Life Sci. 2022, 303, 120601. [Google Scholar] [CrossRef] [PubMed]
- Fawad, J.A.; Luzader, D.H.; Hanson, G.F.; Moutinho Jr, T.J.; McKinney, C.A.; Mitchell, P.G.; Brown-Steinke, K.; Kumar, A.; Park, M.; Lee, S. Histone deacetylase inhibition by gut microbe-generated short-chain fatty acids entrains intestinal epithelial circadian rhythms. Gastroenterology 2022, 163, 1377–1390.e1311. [Google Scholar] [CrossRef] [PubMed]
- Shon, J.; Han, Y.; Song, S.; Kwon, S.Y.; Na, K.; Lindroth, A.M.; Park, Y.J. Anti-obesity effect of butyrate links to modulation of gut microbiome and epigenetic regulation of muscular circadian clock. J. Nutr. Biochem. 2024, 127, 109590. [Google Scholar] [CrossRef] [PubMed]
- Smits, S.A.; Leach, J.; Sonnenburg, E.D.; Gonzalez, C.G.; Lichtman, J.S.; Reid, G.; Knight, R.; Manjurano, A.; Changalucha, J.; Elias, J.E. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 2017, 357, 802–806. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Gao, X.; Huang, X.; Fan, Y.; Wang, Y.-E.; Zhang, Y.; Chen, X.; Wen, J.; He, H.; Hong, Y. Moderate altitude exposure impacts host fasting blood glucose and serum metabolome by regulation of the intestinal flora. Sci. Total Environ. 2023, 905, 167016. [Google Scholar] [CrossRef]
- Spinelli, R.; Parrillo, L.; Longo, M.; Florese, P.; Desiderio, A.; Zatterale, F.; Miele, C.; Raciti, G.A.; Beguinot, F. Molecular basis of ageing in chronic metabolic diseases. J. Endocrinol. Investig. 2020, 43, 1373–1389. [Google Scholar] [CrossRef]
- Bosco, N.; Noti, M. The aging gut microbiome and its impact on host immunity. Genes Immun. 2021, 22, 289–303. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Huang, X.; Dou, L.; Yan, M.; Shen, T.; Tang, W.; Li, J. Aging and aging-related diseases: From molecular mechanisms to interventions and treatments. Signal Transduct. Target. Ther. 2022, 7, 391. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, T.S.; Das, M.; Jeffery, I.B.; O’Toole, P.W. Adjusting for age improves identification of gut microbiome alterations in multiple diseases. elife 2020, 9, e50240. [Google Scholar] [CrossRef] [PubMed]
- Wilmanski, T.; Diener, C.; Rappaport, N.; Patwardhan, S.; Wiedrick, J.; Lapidus, J.; Earls, J.C.; Zimmer, A.; Glusman, G.; Robinson, M. Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat. Metab. 2021, 3, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Brunt, V.E.; Gioscia-Ryan, R.A.; Richey, J.J.; Zigler, M.C.; Cuevas, L.M.; Gonzalez, A.; Vázquez-Baeza, Y.; Battson, M.L.; Smithson, A.T.; Gilley, A.D. Suppression of the gut microbiome ameliorates age-related arterial dysfunction and oxidative stress in mice. J. Physiol. 2019, 597, 2361–2378. [Google Scholar] [CrossRef] [PubMed]
- Torma, F.; Kerepesi, C.; Jókai, M.; Babszki, G.; Koltai, E.; Ligeti, B.; Kalcsevszki, R.; McGreevy, K.M.; Horvath, S.; Radák, Z. Alterations of the gut microbiome are associated with epigenetic age acceleration and physical fitness. Aging Cell 2024, 23, e14101. [Google Scholar] [CrossRef] [PubMed]
- Boytar, A.N.; Skinner, T.L.; Wallen, R.E.; Jenkins, D.G.; Dekker Nitert, M. The effect of exercise prescription on the human gut microbiota and comparison between clinical and apparently healthy populations: A systematic review. Nutrients 2023, 15, 1534. [Google Scholar] [CrossRef] [PubMed]
- Quiroga, R.; Nistal, E.; Estébanez, B.; Porras, D.; Juárez-Fernández, M.; Martínez-Flórez, S.; García-Mediavilla, M.V.; de Paz, J.A.; González-Gallego, J.; Sánchez-Campos, S. Exercise training modulates the gut microbiota profile and impairs inflammatory signaling pathways in obese children. Exp. Mol. Med. 2020, 52, 1048–1061. [Google Scholar] [CrossRef] [PubMed]
- Tschopp, J.; Schroder, K. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat. Rev. Immunol. 2010, 10, 210–215. [Google Scholar] [CrossRef]
- Carmody, R.N.; Gerber, G.K.; Luevano, J.M.; Gatti, D.M.; Somes, L.; Svenson, K.L.; Turnbaugh, P.J. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 2015, 17, 72–84. [Google Scholar] [CrossRef]
- Júnior, R.E.M.; de Carvalho, L.M.; Dos Reis, D.C.; Cassali, G.D.; Faria, A.M.C.; Maioli, T.U.; Brunialti-Godard, A.L. Diet-induced obesity leads to alterations in behavior and gut microbiota composition in mice. J. Nutr. Biochem. 2021, 92, 108622. [Google Scholar]
- Morrison, K.E.; Jašarević, E.; Howard, C.D.; Bale, T.L. It’s the fiber, not the fat: Significant effects of dietary challenge on the gut microbiome. Microbiome 2020, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, J.G.; Milani, C.; De Giori, G.S.; Sesma, F.; Van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Kunst, C.; Schmid, S.; Michalski, M.; Tümen, D.; Buttenschön, J.; Müller, M.; Gülow, K. The influence of gut microbiota on oxidative stress and the immune system. Biomedicines 2023, 11, 1388. [Google Scholar] [CrossRef] [PubMed]
- Ballard, J.W.O.; Towarnicki, S.G. Mitochondria, the gut microbiome and ROS. Cell. Signal. 2020, 75, 109737. [Google Scholar] [CrossRef]
- Li, L.; Peng, P.; Ding, N.; Jia, W.; Huang, C.; Tang, Y. Oxidative stress, inflammation, gut dysbiosis: What can polyphenols do in inflammatory bowel disease? Antioxidants 2023, 12, 967. [Google Scholar] [CrossRef]
- Yardeni, T.; Tanes, C.E.; Bittinger, K.; Mattei, L.M.; Schaefer, P.M.; Singh, L.N.; Wu, G.D.; Murdock, D.G.; Wallace, D.C. Host mitochondria influence gut microbiome diversity: A role for ROS. Sci. Signal. 2019, 12, eaaw3159. [Google Scholar] [CrossRef]
- Singh, V.; Ahlawat, S.; Mohan, H.; Gill, S.S.; Sharma, K.K. Balancing reactive oxygen species generation by rebooting gut microbiota. J. Appl. Microbiol. 2022, 132, 4112–4129. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, X.; Li, L.; Zhong, C.; Zhang, Y.; Yang, X.; Li, M.; Yang, C. The role of gut microbiota in intestinal disease: From an oxidative stress perspective. Front. Microbiol. 2024, 15, 1328324. [Google Scholar] [CrossRef] [PubMed]
- Van Buiten, C.B.; Seitz, V.A.; Metcalf, J.L.; Raskin, I. Dietary Polyphenols Support Akkermansia muciniphila Growth via Mediation of the Gastrointestinal Redox Environment. Antioxidants 2024, 13, 304. [Google Scholar] [CrossRef] [PubMed]
- van der Post, S.; Birchenough, G.M.; Held, J.M. NOX1-dependent redox signaling potentiates colonic stem cell proliferation to adapt to the intestinal microbiota by linking EGFR and TLR activation. Cell Rep. 2021, 35, 108949. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Rosen, C.E.; González-Hernández, J.A.; Song, D.; Potempa, J.; Ring, A.M.; Palm, N.W. Highly multiplexed bioactivity screening reveals human and microbiota metabolome-GPCRome interactions. Cell 2023, 186, 3095–3110.e3019. [Google Scholar] [CrossRef]
- Pardella, E.; Ippolito, L.; Giannoni, E.; Chiarugi, P. Nutritional and metabolic signalling through GPCRs. FEBS Lett. 2022, 596, 2364–2381. [Google Scholar] [CrossRef]
- Jin, C.; Chen, H.; Xie, L.; Zhou, Y.; Liu, L.-l.; Wu, J. GPCRs involved in metabolic diseases: Pharmacotherapeutic development updates. Acta Pharmacol. Sin. 2024, 45, 1321–1336. [Google Scholar] [CrossRef]
- Singh, R.; Zogg, H.; Wei, L.; Bartlett, A.; Ghoshal, U.C.; Rajender, S.; Ro, S. Gut microbial dysbiosis in the pathogenesis of gastrointestinal dysmotility and metabolic disorders. J. Neurogastroenterol. Motil. 2021, 27, 19. [Google Scholar] [CrossRef]
- Vallianou, N.; Christodoulatos, G.S.; Karampela, I.; Tsilingiris, D.; Magkos, F.; Stratigou, T.; Kounatidis, D.; Dalamaga, M. Understanding the role of the gut microbiome and microbial metabolites in non-alcoholic fatty liver disease: Current evidence and perspectives. Biomolecules 2021, 12, 56. [Google Scholar] [CrossRef]
- Bandopadhyay, P.; Ganguly, D. Gut dysbiosis and metabolic diseases. Prog. Mol. Biol. Transl. Sci. 2022, 191, 153–174. [Google Scholar]
- Dabke, K.; Hendrick, G.; Devkota, S. The gut microbiome and metabolic syndrome. J. Clin. Investig. 2019, 129, 4050–4057. [Google Scholar] [CrossRef]
- Lakshmanan, A.P.; Al Zaidan, S.; Bangarusamy, D.K.; Al-Shamari, S.; Elhag, W.; Terranegra, A. Increased relative abundance of ruminoccocus is associated with reduced cardiovascular risk in an obese population. Front. Nutr. 2022, 9, 849005. [Google Scholar] [CrossRef] [PubMed]
- Noureldein, M.H.; Bitar, S.; Youssef, N.; Azar, S.; Eid, A.A. Butyrate modulates diabetes-linked gut dysbiosis: Epigenetic and mechanistic modifications. J. Mol. Endocrinol. 2020, 64, 29–42. [Google Scholar] [CrossRef]
- Thomas, S.P.; Denu, J.M. Short-chain fatty acids activate acetyltransferase p300. Elife 2021, 10, e72171. [Google Scholar] [CrossRef] [PubMed]
- Olaniyi, K.S.; Amusa, O.A.; Ajadi, I.O.; Alabi, B.Y.; Agunbiade, T.B.; Ajadi, M.B. Repression of HDAC5 by acetate restores hypothalamic-pituitary-ovarian function in type 2 diabetes mellitus. Reprod. Toxicol. 2021, 106, 69–81. [Google Scholar] [CrossRef]
- Saiman, Y.; Shen, T.C.D.; Lund, P.J.; Gershuni, V.M.; Jang, C.; Patel, S.; Jung, S.; Furth, E.E.; Friedman, E.S.; Chau, L. Global microbiota-dependent histone acetylation patterns are irreversible and independent of short chain fatty acids. Hepatology 2021, 74, 3427–3440. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, R.; Han, B.; Sun, C.; Chen, R.; Wei, H.; Chen, L.; Du, H.; Li, G.; Yang, Y. Functional and metabolic alterations of gut microbiota in children with new-onset type 1 diabetes. Nat. Commun. 2022, 13, 6356. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, A.; Agrawala, P.K.; Dutta, A. Mitigative and anti-inflammatory effects of Trichostatin A against radiation-induced gastrointestinal toxicity and gut microbiota alteration in mice. Int. J. Radiat. Biol. 2023, 99, 1865–1878. [Google Scholar] [CrossRef]
- Song, M.; Zhang, S.; Tao, Z.; Li, J.; Shi, Y.; Xiong, Y.; Zhang, W.; Liu, C.; Chen, S. MMP-12 siRNA improves the homeostasis of the small intestine and metabolic dysfunction in high-fat diet feeding-induced obese mice. Biomaterials 2021, 278, 121183. [Google Scholar] [CrossRef]
- Rossi, M.; Amaretti, A.; Raimondi, S. Folate production by probiotic bacteria. Nutrients 2011, 3, 118–134. [Google Scholar] [CrossRef]
- Miro-Blanch, J.; Yanes, O. Epigenetic regulation at the interplay between gut microbiota and host metabolism. Front. Genet. 2019, 10, 638. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, Y.; Yang, S.; Lu, J.; Jin, X.; Wu, M. Diet-gut microbiota-epigenetics in metabolic diseases: From mechanisms to therapeutics. Biomed. Pharmacother. 2022, 153, 113290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Li, X.; Zhang, S.; Qi, C.; Zhang, Z.; Ma, R.; Xiang, L.; Chen, L.; Zhu, Y.; Tang, C. Oxidative stress gene expression, DNA methylation, and gut microbiota interaction trigger Crohn’s disease: A multi-omics Mendelian randomization study. BMC Med. 2023, 21, 179. [Google Scholar] [CrossRef]
- Papachristoforou, E.; Lambadiari, V.; Maratou, E.; Makrilakis, K. Association of glycemic indices (hyperglycemia, glucose variability, and hypoglycemia) with oxidative stress and diabetic complications. J. Diabetes Res. 2020, 2020, 7489795. [Google Scholar] [CrossRef]
- Wurster, J.I.; Peterson, R.L.; Brown, C.E.; Penumutchu, S.; Guzior, D.V.; Neugebauer, K.; Sano, W.H.; Sebastian, M.M.; Quinn, R.A.; Belenky, P. Streptozotocin-induced hyperglycemia alters the cecal metabolome and exacerbates antibiotic-induced dysbiosis. Cell Rep. 2021, 37, 110113. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.-H.; Gadkari, M.; Zhou, Q.; Yu, S.; Gao, N.; Guan, Y.; Schady, D.; Roshan, T.N.; Chen, M.-H.; Laritsky, E. Postnatal epigenetic regulation of intestinal stem cells requires DNA methylation and is guided by the microbiome. Genome Biol. 2015, 16, 211. [Google Scholar] [CrossRef] [PubMed]
- Ansari, I.; Raddatz, G.; Gutekunst, J.; Ridnik, M.; Cohen, D.; Abu-Remaileh, M.; Tuganbaev, T.; Shapiro, H.; Pikarsky, E.; Elinav, E. The microbiota programs DNA methylation to control intestinal homeostasis and inflammation. Nat. Microbiol. 2020, 5, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Kline, K.T.; Lian, H.; Zhong, X.S.; Luo, X.; Winston, J.H.; Cong, Y.; Savidge, T.C.; Dashwood, R.H.; Powell, D.W.; Li, Q. Neonatal injury increases gut permeability by epigenetically suppressing E-cadherin in adulthood. J. Immunol. 2020, 204, 980–989. [Google Scholar] [CrossRef]
- Lonati, E.; Sala, G.; Corbetta, P.; Pagliari, S.; Cazzaniga, E.; Botto, L.; Rovellini, P.; Bruni, I.; Palestini, P.; Bulbarelli, A. Digested Cinnamon (Cinnamomum verum J. Presl) bark extract modulates claudin-2 gene expression and protein levels under TNFα/IL-1β inflammatory stimulus. Int. J. Mol. Sci. 2023, 24, 9201. [Google Scholar] [CrossRef]
- Shi, C.; Yue, F.; Shi, F.; Qin, Q.; Wang, L.; Wang, G.; Mu, L.; Liu, D.; Li, Y.; Yu, T. Selenium-containing amino acids protect dextran sulfate sodium-induced colitis via ameliorating oxidative stress and intestinal inflammation. J. Inflamm. Res. 2020, 14, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Hill, E.B.; Konigsberg, I.R.; Ir, D.; Frank, D.N.; Jambal, P.; Litkowski, E.M.; Lange, E.M.; Lange, L.A.; Ostendorf, D.M.; Scorsone, J.J. The microbiome, epigenome, and diet in adults with obesity during behavioral weight loss. Nutrients 2023, 15, 3588. [Google Scholar] [CrossRef] [PubMed]
- van der Vossen, E.W.; Bastos, D.; Stols-Gonçalves, D.; de Goffau, M.C.; Davids, M.; Pereira, J.P.; Li Yim, A.Y.; Henneman, P.; Netea, M.G.; de Vos, W.M. Effects of fecal microbiota transplant on DNA methylation in subjects with metabolic syndrome. Gut Microbes 2021, 13, 1993513. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Ge, J.; Li, X.; Jiao, R.; Li, Y.; Quan, H.; Li, J.; Guo, Q.; Wang, W. Integrated metabolome analysis reveals novel connections between maternal fecal metabolome and the neonatal blood metabolome in women with gestational diabetes mellitus. Sci. Rep. 2020, 10, 3660. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Zhang, C.; Chen, Y.; Duan, R.; Zhang, Y.; Zheng, H.; Zhang, J.; Zhang, T.; Xu, J.; Li, K. Preconception Maternal Gut Dysbiosis Affects Enteric Nervous System Development and Disease Susceptibility in Offspring. 2024. Preprint. Available online: https://www.researchsquare.com/article/rs-4408084/v1 (accessed on 10 August 2024).
- Drokhlyansky, E.; Smillie, C.S.; Van Wittenberghe, N.; Ericsson, M.; Griffin, G.K.; Eraslan, G.; Dionne, D.; Cuoco, M.S.; Goder-Reiser, M.N.; Sharova, T. The human and mouse enteric nervous system at single-cell resolution. Cell 2020, 182, 1606–1622.e1623. [Google Scholar] [CrossRef] [PubMed]
- Bon-Frauches, A.C.; Boesmans, W. The enteric nervous system: The hub in a star network. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 717–718. [Google Scholar] [CrossRef]
- Takahashi, Y.; Valencia, M.M.; Yu, Y.; Ouchi, Y.; Takahashi, K.; Shokhirev, M.N.; Lande, K.; Williams, A.E.; Fresia, C.; Kurita, M. Transgenerational inheritance of acquired epigenetic signatures at CpG islands in mice. Cell 2023, 186, 715–731.e719. [Google Scholar] [CrossRef]
- Romano, K.A.; Martinez-del Campo, A.; Kasahara, K.; Chittim, C.L.; Vivas, E.I.; Amador-Noguez, D.; Balskus, E.P.; Rey, F.E. Metabolic, epigenetic, and transgenerational effects of gut bacterial choline consumption. Cell Host Microbe 2017, 22, 279–290.e277. [Google Scholar] [CrossRef] [PubMed]
- Sugino, K.Y.; Mandala, A.; Janssen, R.C.; Gurung, S.; Trammell, M.; Day, M.W.; Brush, R.S.; Papin, J.F.; Dyer, D.W.; Agbaga, M.-P. Western diet-induced shifts in the maternal microbiome are associated with altered microRNA expression in baboon placenta and fetal liver. Front. Clin. Diabetes Healthc. 2022, 3, 945768. [Google Scholar] [CrossRef]
- Kimura, I.; Miyamoto, J.; Ohue-Kitano, R.; Watanabe, K.; Yamada, T.; Onuki, M.; Aoki, R.; Isobe, Y.; Kashihara, D.; Inoue, D. Maternal gut microbiota in pregnancy influences offspring metabolic phenotype in mice. Science 2020, 367, eaaw8429. [Google Scholar] [CrossRef]
- Kaye, D.M.; Shihata, W.A.; Jama, H.A.; Tsyganov, K.; Ziemann, M.; Kiriazis, H.; Horlock, D.; Vijay, A.; Giam, B.; Vinh, A. Deficiency of prebiotic fiber and insufficient signaling through gut metabolite-sensing receptors leads to cardiovascular disease. Circulation 2020, 141, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.-J.; Li, L.; Li, Q.-N.; Xu, K.; Yue, W.; Qiao, J.-Y.; Meng, T.-G.; Dong, M.-Z.; Lei, W.-L.; Guo, J.-N. The transgenerational effects of maternal low-protein diet during lactation on offspring. J. Genet. Genom. 2024, 51, 824–835. [Google Scholar] [CrossRef] [PubMed]
- Chleilat, F.; Schick, A.; Deleemans, J.M.; Ma, K.; Alukic, E.; Wong, J.; Noye Tuplin, E.W.; Nettleton, J.E.; Reimer, R.A. Paternal high protein diet modulates body composition, insulin sensitivity, epigenetics, and gut microbiota intergenerationally in rats. FASEB J. 2021, 35, e21847. [Google Scholar] [CrossRef]
- Chleilat, F.; Schick, A.; Deleemans, J.M.; Reimer, R.A. Paternal methyl donor supplementation in rats improves fertility, physiological outcomes, gut microbial signatures and epigenetic markers altered by high fat/high sucrose diet. Int. J. Mol. Sci. 2021, 22, 689. [Google Scholar] [CrossRef]
- Chleilat, F.; Schick, A.; Reimer, R.A. Microbiota changes in fathers consuming a high prebiotic fiber diet have minimal effects on male and female offspring in rats. Nutrients 2021, 13, 820. [Google Scholar] [CrossRef] [PubMed]
- Argaw-Denboba, A.; Schmidt, T.S.; Di Giacomo, M.; Ranjan, B.; Devendran, S.; Mastrorilli, E.; Lloyd, C.T.; Pugliese, D.; Paribeni, V.; Dabin, J. Paternal microbiome perturbations impact offspring fitness. Nature 2024, 629, 652–659. [Google Scholar] [CrossRef]
- Harris, J.C.; Trigg, N.A.; Goshu, B.; Yokoyama, Y.; Dohnalová, L.; White, E.K.; Harman, A.; Murga-Garrido, S.M.; Pan, J.T.-C.; Bhanap, P. The microbiota and T cells non-genetically modulate inherited phenotypes transgenerationally. Cell Rep. 2024, 43, 114029. [Google Scholar] [CrossRef]
- Bodden, C.; Pang, T.Y.; Feng, Y.; Mridha, F.; Kong, G.; Li, S.; Watt, M.J.; Reichelt, A.C.; Hannan, A.J. Intergenerational effects of a paternal Western diet during adolescence on offspring gut microbiota, stress reactivity and social behavior. bioRxiv 2022, 1, e21981. [Google Scholar] [CrossRef]
- Chakraborty, A.; Ghosh, S.; Biswas, B.; Pramanik, S.; Nriagu, J.; Bhowmick, S. Epigenetic modifications from arsenic exposure: A comprehensive review. Sci. Total Environ. 2022, 810, 151218. [Google Scholar] [CrossRef]
- Gong, Y.; Xue, Y.; Li, X.; Zhang, Z.; Zhou, W.; Marcolongo, P.; Benedetti, A.; Mao, S.; Han, L.; Ding, G. Inter-and Transgenerational Effects of Paternal Exposure to Inorganic Arsenic. Adv. Sci. 2021, 8, 2002715. [Google Scholar] [CrossRef]
- Aleksandrova, K.; Romero-Mosquera, B.; Hernandez, V. Diet, gut microbiome and epigenetics: Emerging links with inflammatory bowel diseases and prospects for management and prevention. Nutrients 2017, 9, 962. [Google Scholar] [CrossRef]
- Al Theyab, A.; Almutairi, T.; Al-Suwaidi, A.M.; Bendriss, G.; McVeigh, C.; Chaari, A. Epigenetic effects of gut metabolites: Exploring the path of dietary prevention of type 1 diabetes. Front. Nutr. 2020, 7, 563605. [Google Scholar] [CrossRef] [PubMed]
- Lapatto, H.A.; Kuusela, M.; Heikkinen, A.; Muniandy, M.; van der Kolk, B.W.; Gopalakrishnan, S.; Pöllänen, N.; Sandvik, M.; Schmidt, M.S.; Heinonen, S. Nicotinamide riboside improves muscle mitochondrial biogenesis, satellite cell differentiation, and gut microbiota in a twin study. Sci. Adv. 2023, 9, eadd5163. [Google Scholar] [CrossRef] [PubMed]
- Dou, X.; Shen, C.; Wang, Z.; Li, S.; Zhang, X.; Song, Z. Protection of nicotinic acid against oxidative stress-induced cell death in hepatocytes contributes to its beneficial effect on alcohol-induced liver injury in mice. J. Nutr. Biochem. 2013, 24, 1520–1528. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.; Gevezova, M.; Sarafian, V.; Maes, M. Redox regulation of the immune response. Cell. Mol. Immunol. 2022, 19, 1079–1101. [Google Scholar] [CrossRef] [PubMed]
- Amabebe, E.; Robert, F.O.; Agbalalah, T.; Orubu, E.S. Microbial dysbiosis-induced obesity: Role of gut microbiota in homoeostasis of energy metabolism. Br. J. Nutr. 2020, 123, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Marques, F.Z.; Nelson, E.; Chu, P.-Y.; Horlock, D.; Fiedler, A.; Ziemann, M.; Tan, J.K.; Kuruppu, S.; Rajapakse, N.W.; El-Osta, A. High-fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation 2017, 135, 964–977. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Pellegrini, N.; Vannini, L.; Jeffery, I.B.; La Storia, A.; Laghi, L.; Serrazanetti, D.I.; Di Cagno, R.; Ferrocino, I.; Lazzi, C. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 2016, 65, 1812–1821. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, T.S.; Rampelli, S.; Jeffery, I.B.; Santoro, A.; Neto, M.; Capri, M.; Giampieri, E.; Jennings, A.; Candela, M.; Turroni, S. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: The NU-AGE 1-year dietary intervention across five European countries. Gut 2020, 69, 1218–1228. [Google Scholar] [CrossRef]
- Whelan, K.; Bancil, A.S.; Lindsay, J.O.; Chassaing, B. Ultra-processed foods and food additives in gut health and disease. Nat. Rev. Gastroenterol. Hepatol. 2024, 21, 406–427. [Google Scholar] [CrossRef]
- Murga-Garrido, S.M.; Hong, Q.; Cross, T.-W.L.; Hutchison, E.R.; Han, J.; Thomas, S.P.; Vivas, E.I.; Denu, J.; Ceschin, D.G.; Tang, Z.-Z. Gut microbiome variation modulates the effects of dietary fiber on host metabolism. Microbiome 2021, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- Mars, R.A.; Yang, Y.; Ward, T.; Houtti, M.; Priya, S.; Lekatz, H.R.; Tang, X.; Sun, Z.; Kalari, K.R.; Korem, T. Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome. Cell 2020, 182, 1460–1473.e1417. [Google Scholar] [CrossRef]
- Fan, Y.; Qian, H.; Zhang, M.; Tao, C.; Li, Z.; Yan, W.; Huang, Y.; Zhang, Y.; Xu, Q.; Wang, X. Caloric restriction remodels the hepatic chromatin landscape and bile acid metabolism by modulating the gut microbiota. Genome Biol. 2023, 24, 98. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, N.; Yamashita, T.; Osone, T.; Hosooka, T.; Shinohara, M.; Kitahama, S.; Sasaki, K.; Sasaki, D.; Yoneshiro, T.; Suzuki, T. Bacteroides spp. promotes branched-chain amino acid catabolism in brown fat and inhibits obesity. iScience 2021, 24, 103342. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Liu, Y.; Zhang, L.; Zhao, S.; Wang, Y.; Bian, R.; Xu, B.; Chen, X.; Xu, X. Potential Value of Probiotics on Lipid Profiles in Hyperlipidemia and Healthy Participants: Systematic Review and Meta-Analysis. Altern. Ther. Health Med. 2023, 30, 84–89. [Google Scholar]
- Van Syoc, E.P.; Damani, J.; DiMattia, Z.; Ganda, E.; Rogers, C.J. The effects of Bifidobacterium probiotic supplementation on blood glucose: A systematic review and meta-analysis of preclinical animal models and clinical evidence. Adv. Nutr. 2023, 15, 100137. [Google Scholar] [CrossRef]
- Chen, T.; Jing, W.; Fei, G.; Liu, Z. Effect of supplementation with probiotics or synbiotics on cardiovascular risk factors in patients with me tabolic syndrome: A systematic review and meta-analysis of randomized clinical trials. Front. Endocrinol. 2024, 14, 1282699. [Google Scholar] [CrossRef] [PubMed]
- Amini-Salehi, E.; Nayak, S.S.; Maddineni, G.; Mahapatro, A.; Keivanlou, M.-H.; Moghadam, S.S.; Vakilpour, A.; Aleali, M.S.; Joukar, F.; Hashemi, M. Can modulation of gut microbiota affect anthropometric indices in patients with non-alcoholic fatty liver disease? An umbrella meta-analysis of randomized controlled trials. Ann. Med. Surg. 2024, 86, 2900–2910. [Google Scholar] [CrossRef]
- Rasaei, N.; Heidari, M.; Esmaeili, F.; Khosravi, S.; Baeeri, M.; Tabatabaei-Malazy, O.; Emamgholipour, S. The effects of prebiotic, probiotic or synbiotic supplementation on overweight/obesity indicators: An umbrella review of the trials’ meta-analyses. Front. Endocrinol. 2024, 15, 1277921. [Google Scholar] [CrossRef]
- Moravejolahkami, A.R.; Shakibaei, M.; Fairley, A.M.; Sharma, M. Probiotics, prebiotics, and synbiotics in type 1 diabetes mellitus: A systematic review and meta-analysis of clinical trials. Diabetes/Metab. Res. Rev. 2024, 40, e3655. [Google Scholar] [CrossRef]
- Vandenbempt, V.; Eski, S.E.; Brahma, M.K.; Li, A.; Negueruela, J.; Bruggeman, Y.; Demine, S.; Xiao, P.; Cardozo, A.K.; Baeyens, N. HAMSAB diet ameliorates dysfunctional signaling in pancreatic islets in autoimmune diabetes. iScience 2024, 27, 108694. [Google Scholar] [CrossRef]
- Krautkramer, K.A.; Kreznar, J.H.; Romano, K.A.; Vivas, E.I.; Barrett-Wilt, G.A.; Rabaglia, M.E.; Keller, M.P.; Attie, A.D.; Rey, F.E.; Denu, J.M. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol. Cell 2016, 64, 982–992. [Google Scholar] [CrossRef]
- Ge, Y.; Zadeh, M.; Mohamadzadeh, M. Vitamin B12 coordinates ileal epithelial cell and microbiota functions to resist Salmonella infection in mice. J. Exp. Med. 2022, 219, e20220057. [Google Scholar] [CrossRef]
- Mei, X.; Li, Y.; Zhang, X.; Zhai, X.; Yang, Y.; Li, Z.; Li, L. Maternal Phlorizin Intake Protects Offspring from Maternal Obesity-Induced Metabolic Disorders in Mice via Targeting Gut Microbiota to Activate the SCFA-GPR43 Pathway. J. Agric. Food Chem. 2024, 72, 4703–4725. [Google Scholar] [CrossRef]
- Chen, M.; Li, S.; Arora, I.; Yi, N.; Sharma, M.; Li, Z.; Tollefsbol, T.O.; Li, Y. Maternal soybean diet on prevention of obesity-related breast cancer through early-life gut microbiome and epigenetic regulation. J. Nutr. Biochem. 2022, 110, 109119. [Google Scholar] [CrossRef]
- Lu, X.; Chen, B.; Xu, D.; Hu, W.; Wang, X.; Dai, Y.; Wang, Q.; Peng, Y.; Chen, K.; Zhao, D. Epigenetic programming mediates abnormal gut microbiota and disease susceptibility in offspring with prenatal dexamethasone exposure. Cell Rep. Med. 2024, 5, 101398. [Google Scholar] [CrossRef]
- Li, W.; Han, Z.; Yin, X.; Zhou, R.; Liu, H. CDX2 alleviates hypoxia-induced apoptosis and oxidative stress in spermatogenic cells through suppression of reactive oxygen species-mediated Wnt/β-catenin pathway. J. Appl. Toxicol. 2024, 44, 853–862. [Google Scholar] [CrossRef]
- Hill, E.M.; Howard, C.D.; Bale, T.L.; Jašarević, E. Perinatal exposure to tetracycline contributes to lasting developmental effects on offspring. Anim. Microbiome 2021, 3, 37. [Google Scholar] [CrossRef]
- Yaskolka Meir, A.; Keller, M.; Hoffmann, A.; Rinott, E.; Tsaban, G.; Kaplan, A.; Zelicha, H.; Hagemann, T.; Ceglarek, U.; Isermann, B. The effect of polyphenols on DNA methylation-assessed biological age attenuation: The DIRECT PLUS randomized controlled trial. BMC Med. 2023, 21, 364. [Google Scholar] [CrossRef]
- Salas-Perez, F.; Assmann, T.S.; Ramos-Lopez, O.; Martínez, J.A.; Riezu-Boj, J.I.; Milagro, F.I. Crosstalk between gut microbiota and epigenetic markers in obesity development: Relationship between Ruminococcus, BMI, and MACROD2/SEL1L2 methylation. Nutrients 2023, 15, 1550. [Google Scholar] [CrossRef] [PubMed]
- Morales, C.; Arias-Carrasco, R.; Maracaja-Coutinho, V.; Seron, P.; Lanas, F.; Salazar, L.A.; Saavedra, N. Differences in Bacterial Small RNAs in Stool Samples from Hypercholesterolemic and Normocholesterolemic Subjects. Int. J. Mol. Sci. 2023, 24, 7213. [Google Scholar] [CrossRef]
- Tarallo, S.; Ferrero, G.; De Filippis, F.; Francavilla, A.; Pasolli, E.; Panero, V.; Cordero, F.; Segata, N.; Grioni, S.; Pensa, R.G. Stool microRNA profiles reflect different dietary and gut microbiome patterns in healthy individuals. Gut 2022, 71, 1302–1314. [Google Scholar] [CrossRef]
- Noronha, N.Y.; Noma, I.H.Y.; Fernandes Ferreira, R.; Rodrigues, G.d.S.; Martins, L.d.S.; Watanabe, L.M.; Pinhel, M.A.d.S.; Mello Schineider, I.; Diani, L.M.; Carlos, D. Association between the relative abundance of phyla actinobacteria, vitamin C consumption, and DNA methylation of genes linked to immune response pathways. Front. Nutr. 2024, 11, 1373499. [Google Scholar] [CrossRef]
- Kovatcheva, M.; Melendez, E.; Chondronasiou, D.; Pietrocola, F.; Bernad, R.; Caballe, A.; Junza, A.; Capellades, J.; Holguín-Horcajo, A.; Prats, N. Vitamin B12 is a limiting factor for induced cellular plasticity and tissue repair. Nat. Metab. 2023, 5, 1911–1930. [Google Scholar] [CrossRef]
- Lurz, E.; Horne, R.G.; Määttänen, P.; Wu, R.Y.; Botts, S.R.; Li, B.; Rossi, L.; Johnson-Henry, K.C.; Pierro, A.; Surette, M.G. Vitamin B12 deficiency alters the gut microbiota in a murine model of colitis. Front. Nutr. 2020, 7, 83. [Google Scholar] [CrossRef]
- Forgie, A.J.; Pepin, D.M.; Ju, T.; Tollenaar, S.; Sergi, C.M.; Gruenheid, S.; Willing, B.P. Over supplementation with vitamin B12 alters microbe-host interactions in the gut leading to accelerated Citrobacter rodentium colonization and pathogenesis in mice. Microbiome 2023, 11, 21. [Google Scholar] [CrossRef]
- Sun, X.; Wen, J.; Guan, B.; Li, J.; Luo, J.; Li, J.; Wei, M.; Qiu, H. Folic acid and zinc improve hyperuricemia by altering the gut microbiota of rats with high-purine diet-induced hyperuricemia. Front. Microbiol. 2022, 13, 907952. [Google Scholar] [CrossRef]
- Jourova, L.; Anzenbacherova, E.; Dostal, Z.; Anzenbacher, P.; Briolotti, P.; Rigal, E.; Daujat-Chavanieu, M.; Gerbal-Chaloin, S. Butyrate, a typical product of gut microbiome, affects function of the AhR gene, being a possible agent of crosstalk between gut microbiome, and hepatic drug metabolism. J. Nutr. Biochem. 2022, 107, 109042. [Google Scholar] [CrossRef]
- Ge, X.; Zheng, M.; Hu, M.; Fang, X.; Geng, D.; Liu, S.; Wang, L.; Zhang, J.; Guan, L.; Zheng, P. Butyrate ameliorates quinolinic acid–induced cognitive decline in obesity models. J. Clin. Investig. 2023, 133, e154612. [Google Scholar] [CrossRef]
- Caetano-Silva, M.E.; Rund, L.; Hutchinson, N.T.; Woods, J.A.; Steelman, A.J.; Johnson, R.W. Inhibition of inflammatory microglia by dietary fiber and short-chain fatty acids. Sci. Rep. 2023, 13, 2819. [Google Scholar] [CrossRef]
- Deng, Y.; McDonald, O.G.; Means, A.L.; Peek Jr, R.M.; Washington, M.K.; Acra, S.A.; Polk, D.B.; Yan, F. Exposure to p40 in early life prevents intestinal inflammation in adulthood through inducing a long-lasting epigenetic imprint on TGFβ. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 1327–1345. [Google Scholar] [CrossRef]
- Akamine, Y.; Millman, J.F.; Uema, T.; Okamoto, S.; Yonamine, M.; Uehara, M.; Kozuka, C.; Kaname, T.; Shimabukuro, M.; Kinjo, K. Fermented brown rice beverage distinctively modulates the gut microbiota in Okinawans with metabolic syndrome: A randomized controlled trial. Nutr. Res. 2022, 103, 68–81. [Google Scholar] [CrossRef]
- Hutchinson, N.T.; Wang, S.S.; Rund, L.A.; Caetano-Silva, M.E.; Allen, J.M.; Johnson, R.W.; Woods, J.A. Effects of an inulin fiber diet on the gut microbiome, colon, and inflammatory biomarkers in aged mice. Exp. Gerontol. 2023, 176, 112164. [Google Scholar] [CrossRef]
- Gadallah, S.H.; Eissa, S.; Ghanem, H.M.; Ahmed, E.K.; Hasanin, A.H.; El Mahdy, M.M.; Matboli, M. Probiotic-prebiotic-synbiotic modulation of (YAP1, LATS1 and NF2 mRNAs/miR-1205/lncRNA SRD5A3-AS1) panel in NASH animal model. Biomed. Pharmacother. 2021, 140, 111781. [Google Scholar] [CrossRef]
- Stachowska, E.; Maciejewska-Markiewicz, D.; Palma, J.; Mielko, K.A.; Qasem, B.; Kozłowska-Petriczko, K.; Ufnal, M.; Sokolowska, K.E.; Hawryłkowicz, V.; Załęska, P. Precision nutrition in NAFLD: Effects of a high-fiber intervention on the serum metabolome of NAFD patients—A pilot study. Nutrients 2022, 14, 5355. [Google Scholar] [CrossRef]
- AlOlaby, R.R.; Zafarullah, M.; Barboza, M.; Peng, G.; Varian, B.J.; Erdman, S.E.; Lebrilla, C.; Tassone, F. Differential Methylation Profile in Fragile X Syndrome-Prone Offspring Mice after in Utero Exposure to Lactobacillus Reuteri. Genes 2022, 13, 1300. [Google Scholar] [CrossRef]
- Sharma, N.; Navik, U.; Tikoo, K. Unveiling the presence of epigenetic mark by Lactobacillus supplementation in high-fat diet-induced metabolic disorder in Sprague-Dawley rats. J. Nutr. Biochem. 2020, 84, 108442. [Google Scholar] [CrossRef]
- Lilja, S.; Stoll, C.; Krammer, U.; Hippe, B.; Duszka, K.; Debebe, T.; Höfinger, I.; König, J.; Pointner, A.; Haslberger, A. Five days periodic fasting elevates levels of longevity related christensenella and sirtuin expression in humans. Int. J. Mol. Sci. 2021, 22, 2331. [Google Scholar] [CrossRef]
- Wang, R.; Halimulati, M.; Huang, X.; Ma, Y.; Li, L.; Zhang, Z. Sulforaphane-driven reprogramming of gut microbiome and metabolome ameliorates the progression of hyperuricemia. J. Adv. Res. 2023, 52, 19–28. [Google Scholar] [CrossRef]
- Li, B.; Zhang, H.; Shi, L.; Li, R.; Luo, Y.; Deng, Y.; Li, S.; Li, R.; Liu, Z. Saccharomyces boulardii alleviates DSS-induced intestinal barrier dysfunction and inflammation in humanized mice. Food Funct. 2022, 13, 102–112. [Google Scholar] [CrossRef]
- Liu, X.; Hu, G.; Wang, A.; Long, G.; Yang, Y.; Wang, D.; Zhong, N.; Jia, J. Black tea reduces diet-induced obesity in mice via modulation of gut microbiota and gene expression in host tissues. Nutrients 2022, 14, 1635. [Google Scholar] [CrossRef] [PubMed]
- Abdulrahman, A.O.; Alzubaidi, M.Y.; Nadeem, M.S.; Khan, J.A.; Rather, I.A.; Khan, M.I. Effects of urolithins on obesity-associated gut dysbiosis in rats fed on a high-fat diet. Int. J. Food Sci. Nutr. 2021, 72, 923–934. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Wang, R.; Lei, J.; Feng, L.; Zhou, B. Urolithin B protects mice from diet-induced obesity, insulin resistance, and intestinal inflammation by regulating gut microbiota composition. Food Funct. 2024, 15, 7518–7533. [Google Scholar] [CrossRef] [PubMed]
- Greco, C.M.; Garetto, S.; Montellier, E.; Liu, Y.; Chen, S.; Baldi, P.; Sassone-Corsi, P.; Lucci, J. A non-pharmacological therapeutic approach in the gut triggers distal metabolic rewiring capable of ameliorating diet-induced dysfunctions encompassed by metabolic syndrome. Sci. Rep. 2020, 10, 12915. [Google Scholar] [CrossRef] [PubMed]
- Broadfield, L.A.; Saigal, A.; Szamosi, J.C.; Hammill, J.A.; Bezverbnaya, K.; Wang, D.; Gautam, J.; Tsakiridis, E.E.; Di Pastena, F.; McNicol, J. Metformin-induced reductions in tumor growth involves modulation of the gut microbiome. Mol. Metab. 2022, 61, 101498. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, Y.; Liu, J.; Zuo, J.; Yan, L.; Thring, R.W.; Ba, X.; Qi, D.; Wu, M.; Gao, Y. Tauroursodeoxycholic acid functions as a critical effector mediating insulin sensitization of metformin in obese mice. Redox Biol. 2022, 57, 102481. [Google Scholar] [CrossRef]
| Intervention | Study Subjects | Gut Microbiota Alteration | Functional Changes | Mechanism | Ref. | |
|---|---|---|---|---|---|---|
| Vitamins | Vitamin C | Humans | Increases Actinobacteria | Boosts immune functions, glucose homeostasis, and cell metabolism | DNA methylation alterations of 116 genes (13 hypo and 103 hypermethylated) | [134] |
| Vitamin B12 | Stem cells, in vitro | Not applicable | Boosts cell regeneration | Mediates H3K36me3 generation | [135] | |
| Vitamin B12 deficiency (4 weeks) | Mice with DSS (dextran sodium sulphate) challenge | No change in normal mice but altered abundance of 30 genera | Reduces DSS-induced epithelial tissue damage | Unknown | [136] | |
| Vitamin B12, excess amount (1000-fold) | Mice | Decreases α diversity, Clostridia vadin BB60 and Lachnospiraceae NK4A136 groups, but increases Parasutterella | Immune activation, production of IL-17A and the IL-12/23p40 subunit cytokines in colon | Unknown | [137] | |
| Folic acid and zinc | Rats (hyperuricemia, induced by a high-purine diet) | Increase the abundance of probiotic bacteria and reduce pathogenic bacteria | Improve hyperuricemia | Unknown | [138] | |
| Postbiotics | Butyrate | Human primary liver cells | Not applicable | Increases AhR expression and its target genes | Unknown | [139] |
| Butyrate | Humans with obesity and diet-induced obese mice | Unknown | Decreases quinolinic acid- induced BDNF suppression and improves cognition | Increases H3K18ac at BDNF promoter | [140] | |
| Acetate and butyrate | In vitro on microglia | Unknown | Reduces microglia cytokine production | Reduces HDAC activity and NF-κB nuclear translocation | [141] | |
| Lactate | Human cells and mouse | Macrophage exposed to lactate-producing bacteria | Enhances Arg1 (a metabolic gene) and wound healing | Histone lactylation | [73] | |
| p40, a probiotic functional factor | In early life of mice | Modulate gut microbiota | Long-lasting TGFβ production by intestinal epithelial cells, expands Tregs and mitigates gut inflammation | Epigenetic increase of TGFβ expression by H3K4me1/3 persisting into adulthood | [142] | |
| Prebiotics | Fermented brown vs. white rice | Patients with metabolic syndrome | Increases species belonging to the Clostridia class | Reduces inflammation | Increases blood SCFAs | [143] |
| Inulin (a soluble fiber) | Mice and in vitro studies | Unknown | Reduces microglia TNF-α secretion | Increases gut SCFA production and its blood level | [144] | |
| Inulin fiber and multi-strain probiotics | High-fat/sucrose diet-induced steatohepatitis in rats. | Unknown | Improves steatosis, inflammation, liver enzymes, fibrosis, and lipid panel; decreases TGFβ1 (a fibrotic marker) and IL6 | Decreases hepatic Yap1 and miR-1205 expression, and upregulated Lats1, Nf2 and lncRNA SRD5A3-AS1 | [145] | |
| High fiber diet | Humans’ NAFLD | Potentially change gut microbiome | Reduces liver steatosis | Decreases serum SCFAs (unexpectedly) | [146] | |
| Probiotics or fecal microbiota transplatation | Fecal microbial transplantation | Humans | Gut microbiome alterations, including Prevotella ASVs | Modulates plasma metabolome and the epigenome of immune cells | Prevotella ASVs correlated with methylation of AFAP1 involved in mitochondrial function, and insulin sensitivity | [83] |
| Lactobacillus reuteri | Pregnant mice | Gut microbiome alterations | Potential prevention of autism-like symptoms | Altered DNA methylation of genes linked to neuro and synaptogenesis, synaptic transmission, reelin signaling, etc. in offspring | [147] | |
| Lactobacillus suplementation | High fat diet (HFD) induced insulin-resistant rats | Altered gut microbiota composition in favor of Lactobacillus | Reduces hyper-glycemia, hyper-insulinemia, hyper lipidemia, and hepatic- intestinal damage | Mitigates methylation of H3K79me2 and demethylation of H3K27me3 and reduces Foxo1 expression | [148] | |
| Periodic fasting | 5 days of periodic fasting | Humans | Increased gut microbiota diversity, Prevotella, Lactobacillus, and Christensenella abundance | Improves metabolism | Increases mitochondrial DNA, SIRT1, SIRT3, and miRlet7b-p expression in blood cells | [149] |
| Nutritional compounds | Sulforaphane | Rats | Improves gut microbial diversity and functions | Reduces uric acid level | Epigenetic modification of Nrf2 gene | [150] |
| Saccharomyces boulardii | DSS-induced colitis in humanized mice | Increase microbial SCFAs production | Mitigates colon damage and inflammatory responses | Modulates the cytokine profile | [151] | |
| Black tee | HFD feeding mice | Reverses HFD-induced gut dysbiosis | Prevents obesity | DNA methylation alterations, including imprinted genes in the spermatozoa of HFD mice | [152] | |
| Urolithins | HFD obese rats and mice | Modulated gut microbiota and in mice increase population of Akkermansia spp. | Decreases body weight, inflammation, ROS, insulin resistance and restores serum lipid profile | Unknown | [153,154] | |
| Policaptil Gel Retard | HFD feeding mice | Increases gut Bacteroidetes and decreases Firmicutes | Decreases food intake and body weight, improves metabolic state | Modulates expression of metabolic genes and rescues Igfbp2 expression | [155] | |
| Prescribed drugs | Metformin, oral | Mice | Increases SCFA- producing microbes like Lachnospiraceae, Alistipes, and Ruminococcaceae | Decreases colon adenocarcinoma proliferation | Increases circulating propionate and butyrate | [156] |
| Metformin | ob/ob mice (genetically modified obese mice) | Reduces Bifidobacterium and increases Akkermanisia muciniphlia proportion | Increases tauroursodeoxycholic acid, which reduces ROS and intestinal inflammation | Tauroursodeoxycholic acid blocks KEAP1 binding to Nrf2, leading to Nrf2 nuclear translocation, initiating antioxidant gene expression | [157] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mostafavi Abdolmaleky, H.; Zhou, J.-R. Gut Microbiota Dysbiosis, Oxidative Stress, Inflammation, and Epigenetic Alterations in Metabolic Diseases. Antioxidants 2024, 13, 985. https://doi.org/10.3390/antiox13080985
Mostafavi Abdolmaleky H, Zhou J-R. Gut Microbiota Dysbiosis, Oxidative Stress, Inflammation, and Epigenetic Alterations in Metabolic Diseases. Antioxidants. 2024; 13(8):985. https://doi.org/10.3390/antiox13080985
Chicago/Turabian StyleMostafavi Abdolmaleky, Hamid, and Jin-Rong Zhou. 2024. "Gut Microbiota Dysbiosis, Oxidative Stress, Inflammation, and Epigenetic Alterations in Metabolic Diseases" Antioxidants 13, no. 8: 985. https://doi.org/10.3390/antiox13080985
APA StyleMostafavi Abdolmaleky, H., & Zhou, J.-R. (2024). Gut Microbiota Dysbiosis, Oxidative Stress, Inflammation, and Epigenetic Alterations in Metabolic Diseases. Antioxidants, 13(8), 985. https://doi.org/10.3390/antiox13080985







