Multiple Mechanisms of Action of Sulfodyne®, a Natural Antioxidant, against Pathogenic Effects of SARS-CoV-2 Infection
Abstract
:1. Introduction
2. Results
2.1. Increased Susceptibility of Nrf2 KO Mice to SARS-CoV-2 Infection
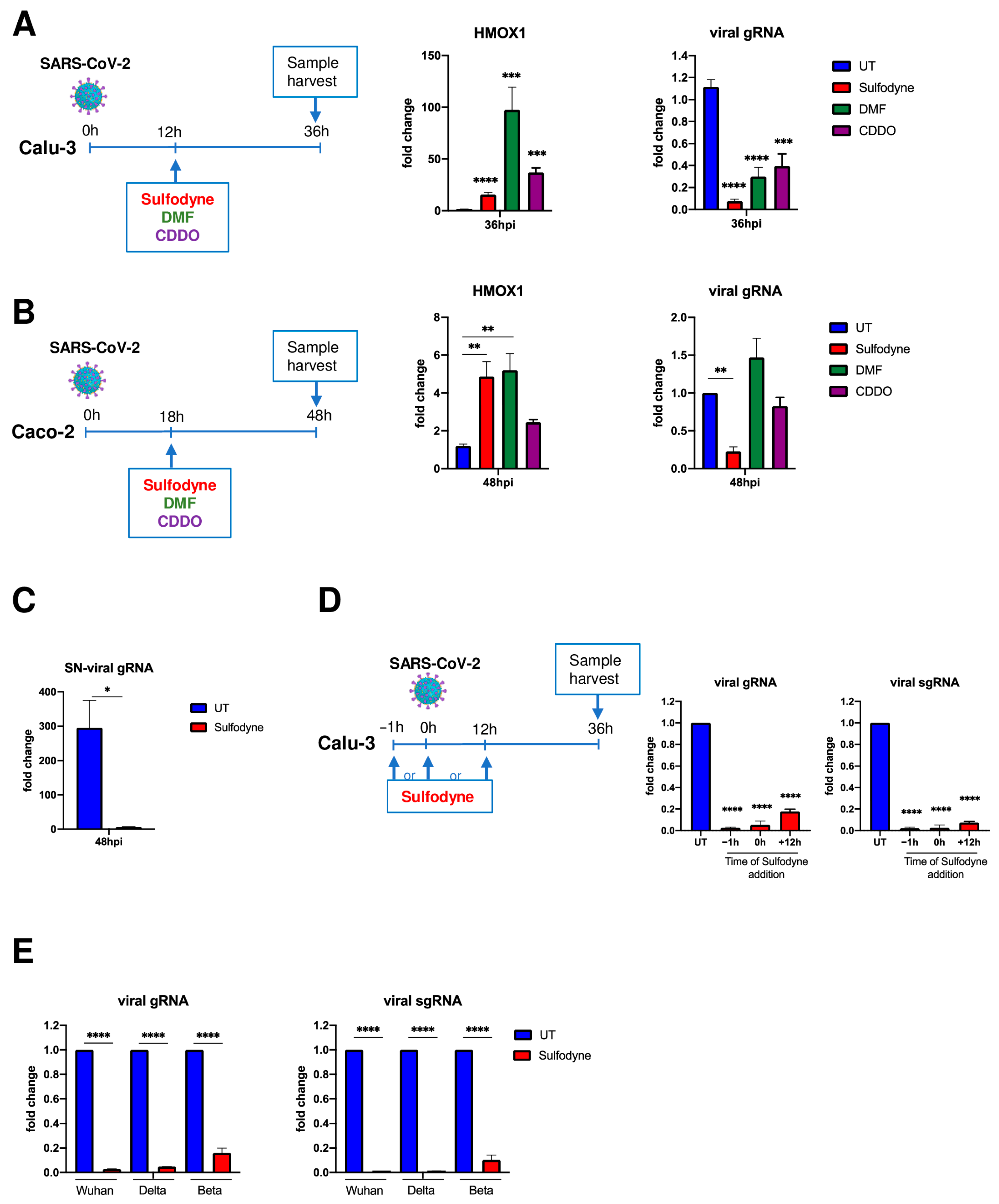
2.2. Sulfodyne® Exhibits Strong Antiviral Activity in Epithelial Cell Lines
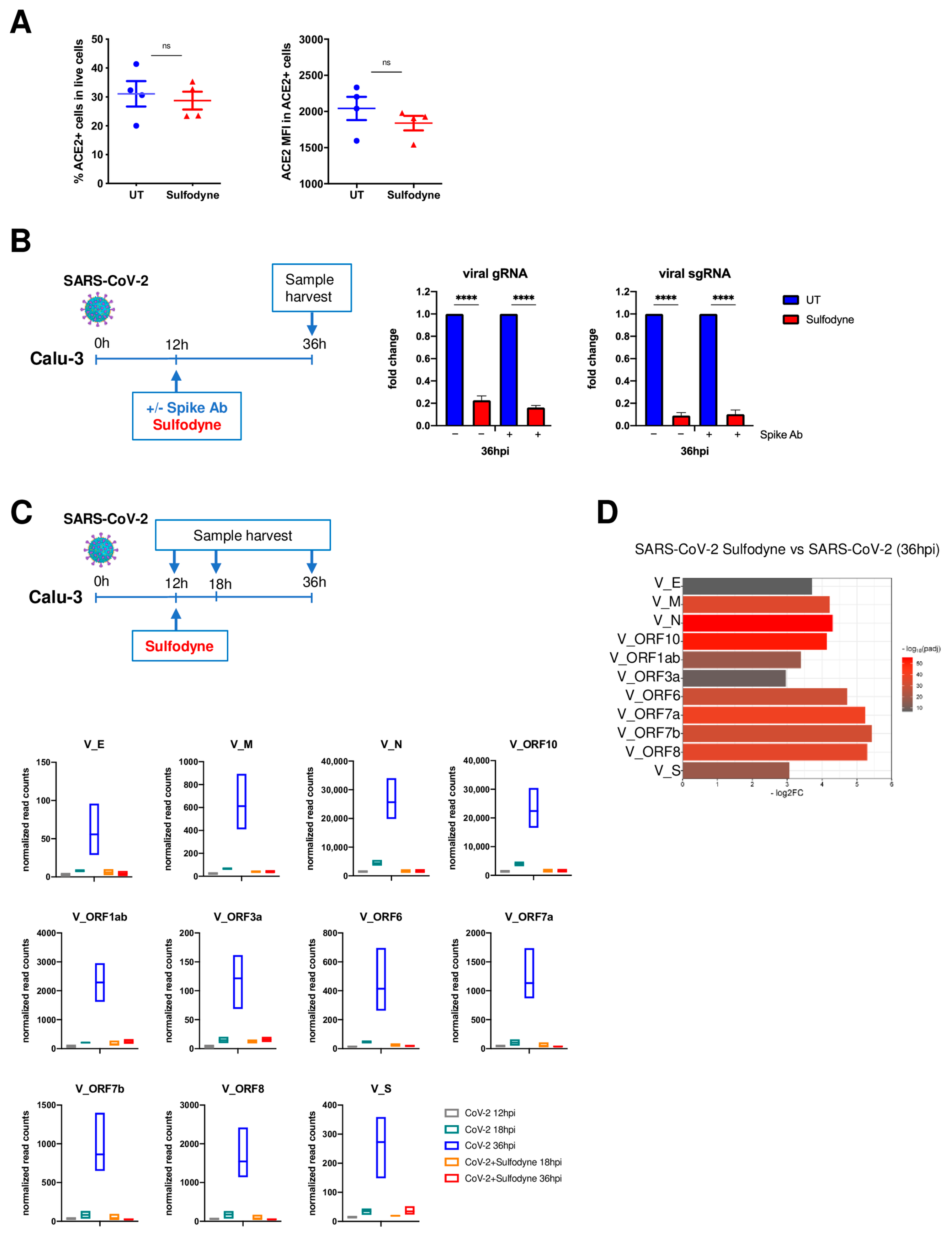
2.3. Sulfodyne® Inhibits SARS-CoV-2 Replication
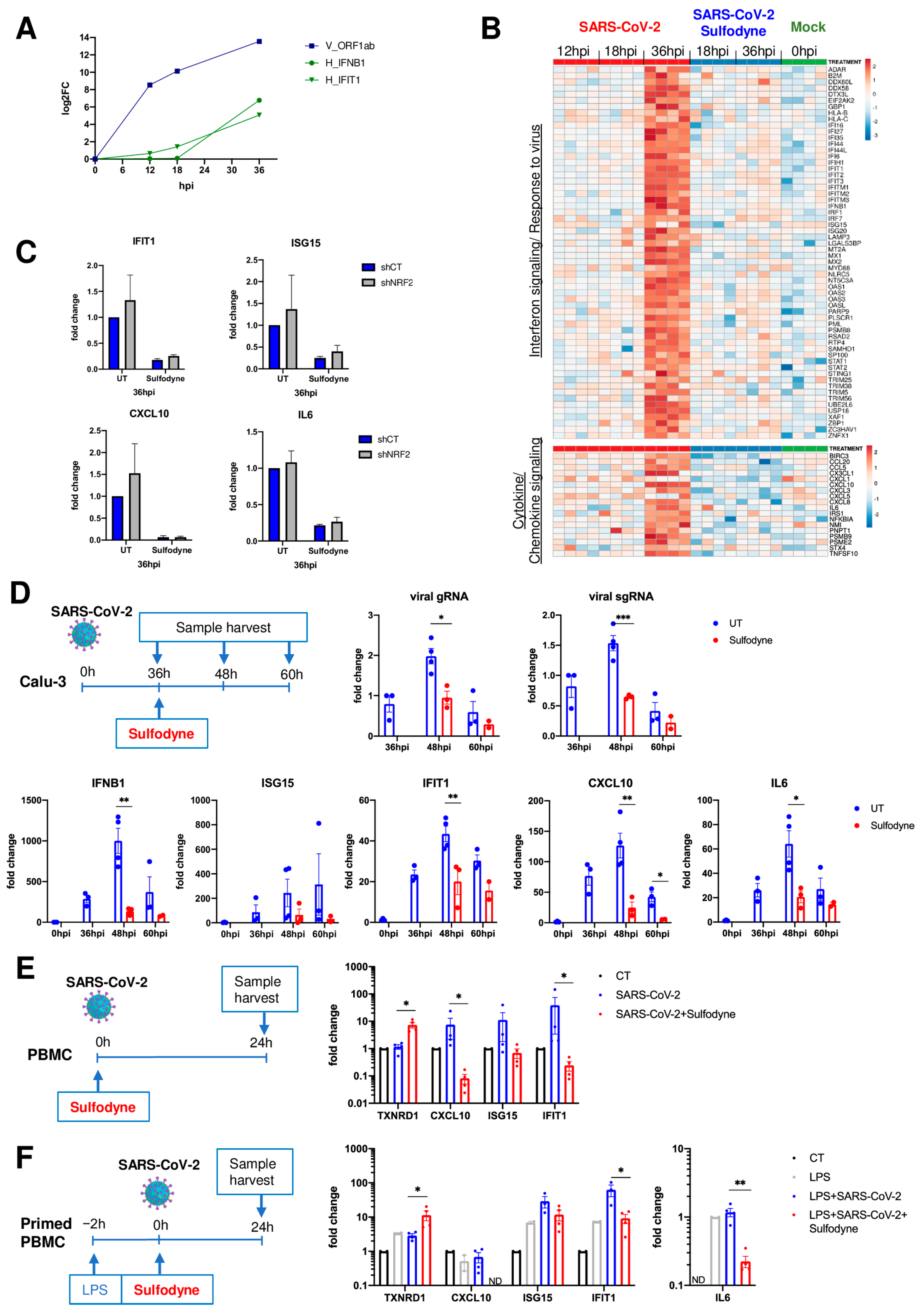
2.4. Sulfodyne® Regulates Host Metabolic Pathways during SARS-CoV-2 Infection
2.5. Anti-Inflammatory Action of Sulfodyne® Is Independent of SARS-CoV-2 Replication
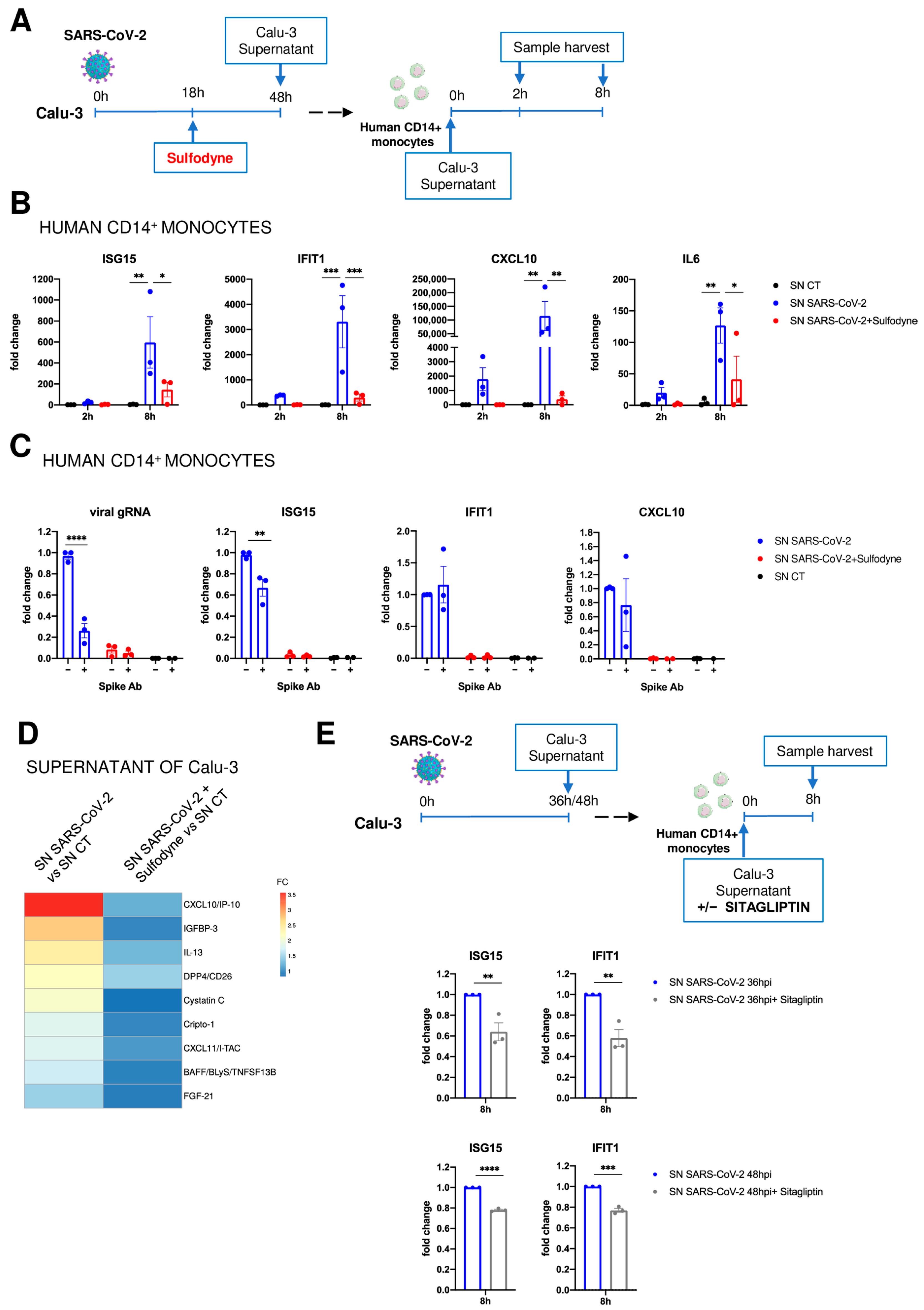
2.6. Immunomodulatory Impact of Sulfodyne® on By-Stander Monocytes
2.7. Sulfodyne® Treatment Confers Protection against SARS-CoV-2 Infection In Vivo
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Treatments and Virus
4.2. RNA Extraction and RT-qPCR
4.3. RNA-Seq
4.4. Protein Extraction and Capillary-Based Immunoblotting (WES Simple Western)
4.5. Flow Cytometry
4.6. Antibodies
4.7. ShRNA
4.8. Cytokine Measurement
4.9. Primary Cells
4.10. Hamsters and In Vivo Infection
4.11. Mice and In Vivo Infection
4.12. Viral Load
4.13. Histopathological Analysis
4.14. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Toussi, S.S.; Hammond, J.L.; Gerstenberger, B.S.; Anderson, A.S. Therapeutics for COVID-19. Nat. Microbiol. 2023, 8, 771–786. [Google Scholar] [CrossRef]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal 2018, 29, 1727–1745. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Kensler, T.W.; Motohashi, H. The KEAP1-NRF2 System: A Thiol-Based Sensor-Effector Apparatus for Maintaining Redox Homeostasis. Physiol. Rev. 2018, 98, 1169–1203. [Google Scholar] [CrossRef]
- Kobayashi, E.H.; Suzuki, T.; Funayama, R.; Nagashima, T.; Hayashi, M.; Sekine, H.; Tanaka, N.; Moriguchi, T.; Motohashi, H.; Nakayama, K.; et al. Nrf2 Suppresses Macrophage Inflammatory Response by Blocking Proinflammatory Cytokine Transcription. Nat. Commun. 2016, 7, 11624. [Google Scholar] [CrossRef]
- Hybertson, B.M.; Gao, B.; Bose, S.K.; McCord, J.M. Oxidative Stress in Health and Disease: The Therapeutic Potential of Nrf2 Activation. Mol. Aspects Med. 2011, 32, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Ramezani, A.; Nahad, M.P.; Faghihloo, E. The Role of Nrf2 Transcription Factor in Viral Infection. J. Cell Biochem. 2018, 119, 6366–6382. [Google Scholar] [CrossRef]
- Alam, M.S.; Czajkowsky, D.M. SARS-CoV-2 Infection and Oxidative Stress: Pathophysiological Insight into Thrombosis and Therapeutic Opportunities. Cytokine Growth Factor. Rev. 2022, 63, 44–57. [Google Scholar] [CrossRef]
- Gain, C.; Song, S.; Angtuaco, T.; Satta, S.; Kelesidis, T. The Role of Oxidative Stress in the Pathogenesis of Infections with Coronaviruses. Front. Microbiol. 2022, 13, 1111930. [Google Scholar] [CrossRef]
- Coronel, P.M.V.; Pereira, I.C.; Basilio, D.C.L.S.; Espinoça, I.T.; de Souza, K.F.S.; Ota, R.S.N.; de Almeida, E.B.; Paredes-Gamero, E.J.; Wilhelm Filho, D.; Perdomo, R.T.; et al. Biomarkers of Oxidative Stress and Inflammation in Subjects with COVID-19: Characterization and Prognosis of the Disease. Microb. Pathog. 2023, 184, 106339. [Google Scholar] [CrossRef]
- Olagnier, D.; Farahani, E.; Thyrsted, J.; Blay-Cadanet, J.; Herengt, A.; Idorn, M.; Hait, A.; Hernaez, B.; Knudsen, A.; Iversen, M.B.; et al. SARS-CoV2-Mediated Suppression of NRF2-Signaling Reveals Potent Antiviral and Anti-Inflammatory Activity of 4-Octyl-Itaconate and Dimethyl Fumarate. Nat. Commun. 2020, 11, 4938. [Google Scholar] [CrossRef] [PubMed]
- Gümüş, H.; Erat, T.; Öztürk, İ.; Demir, A.; Koyuncu, I. Oxidative Stress and Decreased Nrf2 Level in Pediatric Patients with COVID-19. J. Med. Virol. 2022, 94, 2259–2264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, J.; Wang, L.; Aliyari, S.; Cheng, G. SARS-CoV-2 Virus NSP14 Impairs NRF2/HMOX1 Activation by Targeting Sirtuin 1. Cell Mol. Immunol. 2022, 19, 872–882. [Google Scholar] [CrossRef]
- Liu, L.; Du, J.; Yang, S.; Zheng, B.; Shen, J.; Huang, J.; Cao, L.; Huang, S.; Liu, X.; Guo, L.; et al. SARS-CoV-2 ORF3a Sensitizes Cells to Ferroptosis via Keap1-NRF2 Axis. Redox Biol. 2023, 63, 102752. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Haas de Mello, A.; Morris, D.R.; Jones-Hall, Y.L.; Ivanciuc, T.; Sattler, R.A.; Paessler, S.; Menachery, V.D.; Garofalo, R.P.; Casola, A. SARS-CoV-2 Inhibits NRF2-Mediated Antioxidant Responses in Airway Epithelial Cells and in the Lung of a Murine Model of Infection. Microbiol. Spectr. 2023, 11, e0037823. [Google Scholar] [CrossRef]
- McCord, J.M.; Hybertson, B.M.; Cota-Gomez, A.; Geraci, K.P.; Gao, B. Nrf2 Activator PB125® as a Potential Therapeutic Agent against COVID-19. Antioxidants 2020, 9, 518. [Google Scholar] [CrossRef]
- Rothan, H.A.; Zhong, Y.; Sanborn, M.A.; Teoh, T.C.; Ruan, J.; Yusof, R.; Hang, J.; Henderson, M.J.; Fang, S. Small Molecule Grp94 Inhibitors Block Dengue and Zika Virus Replication. Antiviral Res. 2019, 171, 104590. [Google Scholar] [CrossRef]
- Ordonez, A.A.; Bullen, C.K.; Villabona-Rueda, A.F.; Thompson, E.A.; Turner, M.L.; Merino, V.F.; Yan, Y.; Kim, J.; Davis, S.L.; Komm, O.; et al. Sulforaphane Exhibits Antiviral Activity against Pandemic SARS-CoV-2 and Seasonal HCoV-OC43 Coronaviruses in Vitro and in Mice. Commun. Biol. 2022, 5, 242. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Fahey, J.W.; Kostov, R.V.; Kensler, T.W. KEAP1 and Done? Targeting the NRF2 Pathway with Sulforaphane. Trends Food Sci. Technol. 2017, 69, 257–269. [Google Scholar] [CrossRef]
- Habtemariam, S. Anti-Inflammatory Therapeutic Mechanisms of Isothiocyanates: Insights from Sulforaphane. Biomedicines 2024, 12, 1169. [Google Scholar] [CrossRef]
- Hu, C.; Eggler, A.L.; Mesecar, A.D.; van Breemen, R.B. Modification of Keap1 Cysteine Residues by Sulforaphane. Chem. Res. Toxicol. 2011, 24, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Treasure, K.; Harris, J.; Williamson, G. Exploring the Anti-Inflammatory Activity of Sulforaphane. Immunol. Cell Biol. 2023, 101, 805–828. [Google Scholar] [CrossRef] [PubMed]
- Heiss, E.; Herhaus, C.; Klimo, K.; Bartsch, H.; Gerhäuser, C. Nuclear Factor Kappa B Is a Molecular Target for Sulforaphane-Mediated Anti-Inflammatory Mechanisms. J. Biol. Chem. 2001, 276, 32008–32015. [Google Scholar] [CrossRef]
- Múnera-Rodríguez, A.M.; Leiva-Castro, C.; Sobrino, F.; López-Enríquez, S.; Palomares, F. Sulforaphane-Mediated Immune Regulation through Inhibition of NF-kB and MAPK Signaling Pathways in Human Dendritic Cells. Biomed. Pharmacother. 2024, 177, 117056. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-N.; Kim, D.-H.; Kim, E.-H.; Lee, M.-H.; Kundu, J.K.; Na, H.-K.; Cha, Y.-N.; Surh, Y.-J. Sulforaphane Inhibits Phorbol Ester-Stimulated IKK-NF-κB Signaling and COX-2 Expression in Human Mammary Epithelial Cells by Targeting NF-κB Activating Kinase and ERK. Cancer Lett. 2014, 351, 41–49. [Google Scholar] [CrossRef]
- Heiss, E.; Gerhäuser, C. Time-Dependent Modulation of Thioredoxin Reductase Activity Might Contribute to Sulforaphane-Mediated Inhibition of NF-kappaB Binding to DNA. Antioxid. Redox Signal 2005, 7, 1601–1611. [Google Scholar] [CrossRef]
- Ma, C.; Gu, C.; Lian, P.; Wazir, J.; Lu, R.; Ruan, B.; Wei, L.; Li, L.; Pu, W.; Peng, Z.; et al. Sulforaphane Alleviates Psoriasis by Enhancing Antioxidant Defense through KEAP1-NRF2 Pathway Activation and Attenuating Inflammatory Signaling. Cell Death Dis. 2023, 14, 768. [Google Scholar] [CrossRef]
- Cuadrado, A.; Pajares, M.; Benito, C.; Jiménez-Villegas, J.; Escoll, M.; Fernández-Ginés, R.; Garcia Yagüe, A.J.; Lastra, D.; Manda, G.; Rojo, A.I.; et al. Can Activation of NRF2 Be a Strategy against COVID-19? Trends Pharmacol. Sci. 2020, 41, 598–610. [Google Scholar] [CrossRef]
- Chen, Z.; Du, R.; Cooper, L.; Achi, J.G.; Dong, M.; Ran, Y.; Zhang, J.; Zhan, P.; Rong, L.; Cui, Q. Sulforaphane Is a Reversible Covalent Inhibitor of 3-Chymotrypsin-like Protease of SARS-CoV-2. J. Med. Virol. 2023, 95, e28609. [Google Scholar] [CrossRef]
- Gasparello, J.; D’Aversa, E.; Papi, C.; Gambari, L.; Grigolo, B.; Borgatti, M.; Finotti, A.; Gambari, R. Sulforaphane Inhibits the Expression of Interleukin-6 and Interleukin-8 Induced in Bronchial Epithelial IB3-1 Cells by Exposure to the SARS-CoV-2 Spike Protein. Phytomedicine 2021, 87, 153583. [Google Scholar] [CrossRef]
- Kiser, C.; Gonul, C.P.; Olcum, M.; Genc, S. Inhibitory Effects of Sulforaphane on NLRP3 Inflammasome Activation. Mol. Immunol. 2021, 140, 175–185. [Google Scholar] [CrossRef]
- Fahey, J.W.; Wade, K.L.; Wehage, S.L.; Holtzclaw, W.D.; Liu, H.; Talalay, P.; Fuchs, E.; Stephenson, K.K. Stabilized Sulforaphane for Clinical Use: Phytochemical Delivery Efficiency. Mol. Nutr. Food Res. 2017, 61, 1600766. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Ghosh, A.; Lo, C.-S.; Chenier, I.; Scholey, J.W.; Filep, J.G.; Ingelfinger, J.R.; Zhang, S.-L.; Chan, J.S.D. Nrf2 Deficiency Upregulates Intrarenal Angiotensin-Converting Enzyme-2 and Angiotensin 1-7 Receptor Expression and Attenuates Hypertension and Nephropathy in Diabetic Mice. Endocrinology 2018, 159, 836–852. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ma, J.; Jiang, Y. Transcription Factor Nrf2 as a Potential Therapeutic Target for COVID-19. Cell Stress. Chaperones 2023, 28, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Appelberg, S.; Gupta, S.; Svensson Akusjärvi, S.; Ambikan, A.T.; Mikaeloff, F.; Saccon, E.; Végvári, Á.; Benfeitas, R.; Sperk, M.; Ståhlberg, M.; et al. Dysregulation in Akt/mTOR/HIF-1 Signaling Identified by Proteo-Transcriptomics of SARS-CoV-2 Infected Cells. Emerg. Microbes Infect. 2020, 9, 1748–1760. [Google Scholar] [CrossRef]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and Evasion of Type I Interferon Responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef]
- Li, M.; Ferretti, M.; Ying, B.; Descamps, H.; Lee, E.; Dittmar, M.; Lee, J.S.; Whig, K.; Kamalia, B.; Dohnalová, L.; et al. Pharmacological Activation of STING Blocks SARS-CoV-2 Infection. Sci. Immunol. 2021, 6, eabi9007. [Google Scholar] [CrossRef]
- Thorne, L.G.; Reuschl, A.-K.; Zuliani-Alvarez, L.; Whelan, M.V.X.; Turner, J.; Noursadeghi, M.; Jolly, C.; Towers, G.J. SARS-CoV-2 Sensing by RIG-I and MDA5 Links Epithelial Infection to Macrophage Inflammation. EMBO J. 2021, 40, e107826. [Google Scholar] [CrossRef]
- Kazmierski, J.; Friedmann, K.; Postmus, D.; Emanuel, J.; Fischer, C.; Jansen, J.; Richter, A.; Bosquillon de Jarcy, L.; Schüler, C.; Sohn, M.; et al. Nonproductive Exposure of PBMCs to SARS-CoV-2 Induces Cell-Intrinsic Innate Immune Responses. Mol. Syst. Biol. 2022, 18, e10961. [Google Scholar] [CrossRef]
- Gudowska-Sawczuk, M.; Mroczko, B. What Is Currently Known about the Role of CXCL10 in SARS-CoV-2 Infection? Int. J. Mol. Sci. 2022, 23, 3673. [Google Scholar] [CrossRef]
- Sebastián-Martín, A.; Sánchez, B.G.; Mora-Rodríguez, J.M.; Bort, A.; Díaz-Laviada, I. Role of Dipeptidyl Peptidase-4 (DPP4) on COVID-19 Physiopathology. Biomedicines 2022, 10, 2026. [Google Scholar] [CrossRef]
- Mulvihill, E.E.; Drucker, D.J. Pharmacology, Physiology, and Mechanisms of Action of Dipeptidyl Peptidase-4 Inhibitors. Endocr. Rev. 2014, 35, 992–1019. [Google Scholar] [CrossRef]
- Yazbeck, R.; Jaenisch, S.E.; Abbott, C.A. Dipeptidyl Peptidase 4 Inhibitors: Applications in Innate Immunity? Biochem. Pharmacol. 2021, 188, 114517. [Google Scholar] [CrossRef] [PubMed]
- Imai, M.; Iwatsuki-Horimoto, K.; Hatta, M.; Loeber, S.; Halfmann, P.J.; Nakajima, N.; Watanabe, T.; Ujie, M.; Takahashi, K.; Ito, M.; et al. Syrian Hamsters as a Small Animal Model for SARS-CoV-2 Infection and Countermeasure Development. Proc. Natl. Acad. Sci. USA 2020, 117, 16587–16595. [Google Scholar] [CrossRef]
- Golden, J.W.; Cline, C.R.; Zeng, X.; Garrison, A.R.; Carey, B.D.; Mucker, E.M.; White, L.E.; Shamblin, J.D.; Brocato, R.L.; Liu, J.; et al. Human Angiotensin-Converting Enzyme 2 Transgenic Mice Infected with SARS-CoV-2 Develop Severe and Fatal Respiratory Disease. JCI Insight 2020, 5, e142032. [Google Scholar] [CrossRef]
- Bartolini, D.; Stabile, A.M.; Bastianelli, S.; Giustarini, D.; Pierucci, S.; Busti, C.; Vacca, C.; Gidari, A.; Francisci, D.; Castronari, R.; et al. SARS-CoV2 Infection Impairs the Metabolism and Redox Function of Cellular Glutathione. Redox Biol. 2021, 45, 102041. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wang, X.; Sun, Y.; Zhao, H.; Cheng, F.; Wang, J.; Yang, F.; Hu, J.; Zhang, H.; Wang, C.-C.; et al. SARS-CoV-2 ORF8 Reshapes the ER through Forming Mixed Disulfides with ER Oxidoreductases. Redox Biol. 2022, 54, 102388. [Google Scholar] [CrossRef] [PubMed]
- Mullen, P.J.; Garcia, G.; Purkayastha, A.; Matulionis, N.; Schmid, E.W.; Momcilovic, M.; Sen, C.; Langerman, J.; Ramaiah, A.; Shackelford, D.B.; et al. SARS-CoV-2 Infection Rewires Host Cell Metabolism and Is Potentially Susceptible to mTORC1 Inhibition. Nat. Commun. 2021, 12, 1876. [Google Scholar] [CrossRef]
- Angeloni, C.; Turroni, S.; Bianchi, L.; Fabbri, D.; Motori, E.; Malaguti, M.; Leoncini, E.; Maraldi, T.; Bini, L.; Brigidi, P.; et al. Novel Targets of Sulforaphane in Primary Cardiomyocytes Identified by Proteomic Analysis. PLoS ONE 2013, 8, e83283. [Google Scholar] [CrossRef]
- Zhang, Y.; Gilmour, A.; Ahn, Y.-H.; de la Vega, L.; Dinkova-Kostova, A.T. The Isothiocyanate Sulforaphane Inhibits mTOR in an NRF2-Independent Manner. Phytomedicine 2021, 86, 153062. [Google Scholar] [CrossRef]
- Dana, A.-H.; Alejandro, S.-P. Role of Sulforaphane in Endoplasmic Reticulum Homeostasis through Regulation of the Antioxidant Response. Life Sci. 2022, 299, 120554. [Google Scholar] [CrossRef] [PubMed]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.-H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Béziat, V.; et al. Autoantibodies against Type I IFNs in Patients with Life-Threatening COVID-19. Science 2020, 370, eabd4585. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.D.; Hodeib, S.; Korol, C.; et al. Inborn Errors of Type I IFN Immunity in Patients with Life-Threatening COVID-19. Science 2020, 370, eabd4570. [Google Scholar] [CrossRef] [PubMed]
- Bastard, P.; Gervais, A.; Le Voyer, T.; Rosain, J.; Philippot, Q.; Manry, J.; Michailidis, E.; Hoffmann, H.-H.; Eto, S.; Garcia-Prat, M.; et al. Autoantibodies Neutralizing Type I IFNs Are Present in ~4% of Uninfected Individuals over 70 Years Old and Account for ~20% of COVID-19 Deaths. Sci. Immunol. 2021, 6, eabl4340. [Google Scholar] [CrossRef]
- Brunet-Ratnasingham, E.; Morin, S.; Randolph, H.E.; Labrecque, M.; Bélair, J.; Lima-Barbosa, R.; Pagliuzza, A.; Marchitto, L.; Hultström, M.; Niessl, J.; et al. Sustained IFN Signaling Is Associated with Delayed Development of SARS-CoV-2-Specific Immunity. Nat. Commun. 2024, 15, 4177. [Google Scholar] [CrossRef]
- Park, A.; Iwasaki, A. Type I and Type III Interferons—Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe 2020, 27, 870–878. [Google Scholar] [CrossRef]
- Lee, J.S.; Shin, E.-C. The Type I Interferon Response in COVID-19: Implications for Treatment. Nat. Rev. Immunol. 2020, 20, 585–586. [Google Scholar] [CrossRef]
- Zhou, Z.; Ren, L.; Zhang, L.; Zhong, J.; Xiao, Y.; Jia, Z.; Guo, L.; Yang, J.; Wang, C.; Jiang, S.; et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe 2020, 27, 883–890. [Google Scholar] [CrossRef]
- Lee, J.S.; Park, S.; Jeong, H.W.; Ahn, J.Y.; Choi, S.J.; Lee, H.; Choi, B.; Nam, S.K.; Sa, M.; Kwon, J.-S.; et al. Immunophenotyping of COVID-19 and Influenza Highlights the Role of Type I Interferons in Development of Severe COVID-19. Sci. Immunol. 2020, 5, eabd1554. [Google Scholar] [CrossRef]
- Galani, I.-E.; Rovina, N.; Lampropoulou, V.; Triantafyllia, V.; Manioudaki, M.; Pavlos, E.; Koukaki, E.; Fragkou, P.C.; Panou, V.; Rapti, V.; et al. Untuned Antiviral Immunity in COVID-19 Revealed by Temporal Type I/III Interferon Patterns and Flu Comparison. Nat. Immunol. 2021, 22, 32–40. [Google Scholar] [CrossRef]
- Jhuti, D.; Rawat, A.; Guo, C.M.; Wilson, L.A.; Mills, E.J.; Forrest, J.I. Interferon Treatments for SARS-CoV-2: Challenges and Opportunities. Infect. Dis. Ther. 2022, 11, 953–972. [Google Scholar] [CrossRef] [PubMed]
- Phetsouphanh, C.; Darley, D.R.; Wilson, D.B.; Howe, A.; Munier, C.M.L.; Patel, S.K.; Juno, J.A.; Burrell, L.M.; Kent, S.J.; Dore, G.J.; et al. Immunological Dysfunction Persists for 8 Months Following Initial Mild-to-Moderate SARS-CoV-2 Infection. Nat. Immunol. 2022, 23, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Barnett, K.C.; Xie, Y.; Asakura, T.; Song, D.; Liang, K.; Taft-Benz, S.A.; Guo, H.; Yang, S.; Okuda, K.; Gilmore, R.C.; et al. An Epithelial-Immune Circuit Amplifies Inflammasome and IL-6 Responses to SARS-CoV-2. Cell Host Microbe 2023, 31, 243–259. [Google Scholar] [CrossRef]
- Leon, J.; Michelson, D.A.; Olejnik, J.; Chowdhary, K.; Oh, H.S.; Hume, A.J.; Galván-Peña, S.; Zhu, Y.; Chen, F.; Vijaykumar, B.; et al. A Virus-Specific Monocyte Inflammatory Phenotype Is Induced by SARS-CoV-2 at the Immune-Epithelial Interface. Proc. Natl. Acad. Sci. USA 2022, 119, e2116853118. [Google Scholar] [CrossRef] [PubMed]
- Cipolla, B.G.; Mandron, E.; Lefort, J.M.; Coadou, Y.; Della Negra, E.; Corbel, L.; Le Scodan, R.; Azzouzi, A.R.; Mottet, N. Effect of Sulforaphane in Men with Biochemical Recurrence after Radical Prostatectomy. Cancer Prev. Res. 2015, 8, 712–719. [Google Scholar] [CrossRef]
- Hu, R.; Hebbar, V.; Kim, B.-R.; Chen, C.; Winnik, B.; Buckley, B.; Soteropoulos, P.; Tolias, P.; Hart, R.P.; Kong, A.-N.T. In Vivo Pharmacokinetics and Regulation of Gene Expression Profiles by Isothiocyanate Sulforaphane in the Rat. J. Pharmacol. Exp. Ther. 2004, 310, 263–271. [Google Scholar] [CrossRef]
- Yagishita, Y.; Fahey, J.W.; Dinkova-Kostova, A.T.; Kensler, T.W. Broccoli or Sulforaphane: Is It the Source or Dose That Matters? Molecules 2019, 24, 3593. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romeo, P.-H.; Conquet, L.; Messiaen, S.; Pascal, Q.; Moreno, S.G.; Bravard, A.; Bernardino-Sgherri, J.; Dereuddre-Bosquet, N.; Montagutelli, X.; Le Grand, R.; et al. Multiple Mechanisms of Action of Sulfodyne®, a Natural Antioxidant, against Pathogenic Effects of SARS-CoV-2 Infection. Antioxidants 2024, 13, 1083. https://doi.org/10.3390/antiox13091083
Romeo P-H, Conquet L, Messiaen S, Pascal Q, Moreno SG, Bravard A, Bernardino-Sgherri J, Dereuddre-Bosquet N, Montagutelli X, Le Grand R, et al. Multiple Mechanisms of Action of Sulfodyne®, a Natural Antioxidant, against Pathogenic Effects of SARS-CoV-2 Infection. Antioxidants. 2024; 13(9):1083. https://doi.org/10.3390/antiox13091083
Chicago/Turabian StyleRomeo, Paul-Henri, Laurine Conquet, Sébastien Messiaen, Quentin Pascal, Stéphanie G. Moreno, Anne Bravard, Jacqueline Bernardino-Sgherri, Nathalie Dereuddre-Bosquet, Xavier Montagutelli, Roger Le Grand, and et al. 2024. "Multiple Mechanisms of Action of Sulfodyne®, a Natural Antioxidant, against Pathogenic Effects of SARS-CoV-2 Infection" Antioxidants 13, no. 9: 1083. https://doi.org/10.3390/antiox13091083






