Building a Human Ovarian Antioxidant ceRNA Network “OvAnOx”: A Bioinformatic Perspective for Research on Redox-Related Ovarian Functions and Dysfunctions
Abstract
:1. Introduction
2. Materials and Methods
2.1. PICO Statement
2.2. Gene Ontology and Pathway Analysis, Localization, and Expression of the 21 Antioxidant Enzymes
2.3. Construction and Analysis of LncRNA-miRNA-mRNA Competing Endogenous RNA Networks
2.4. Cellular Localization of miRNAs and lncRNAs Regulating the Antioxidant Genes
3. Results
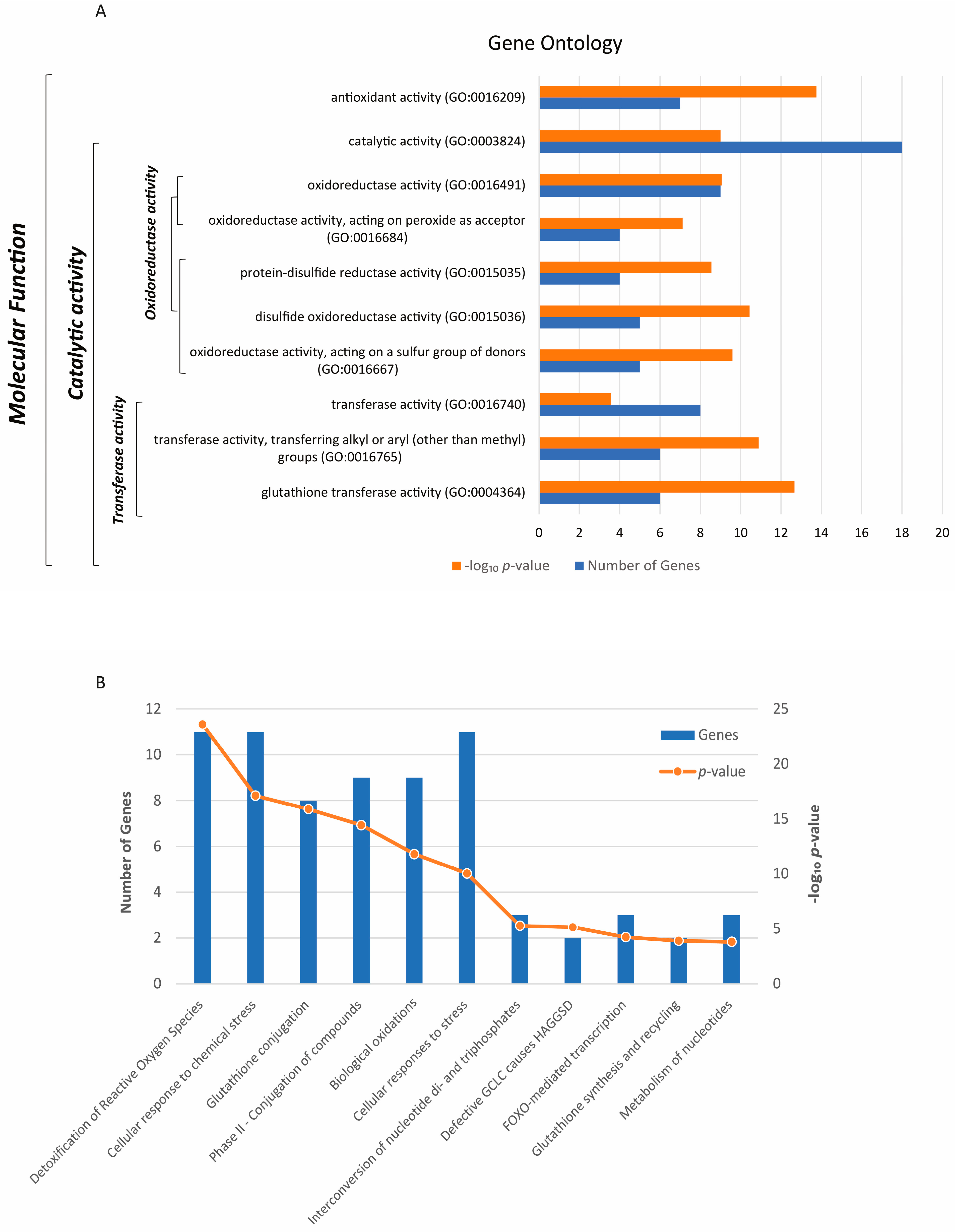
3.1. Antioxidant Genes Control Significant Molecular Functions and Biological Pathways
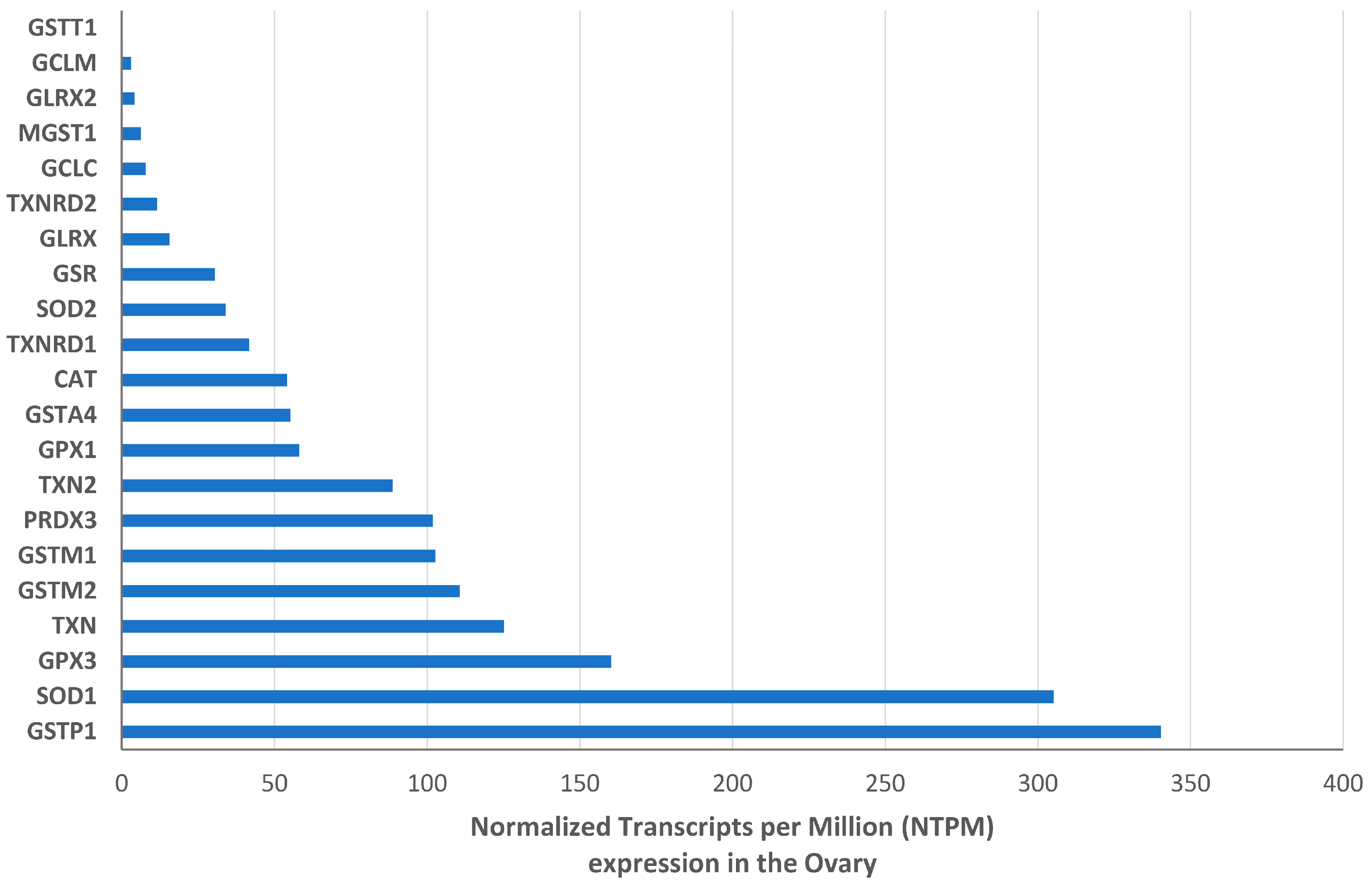
3.2. Expression and Intraovarian Localization of the Antioxidant Enzymes
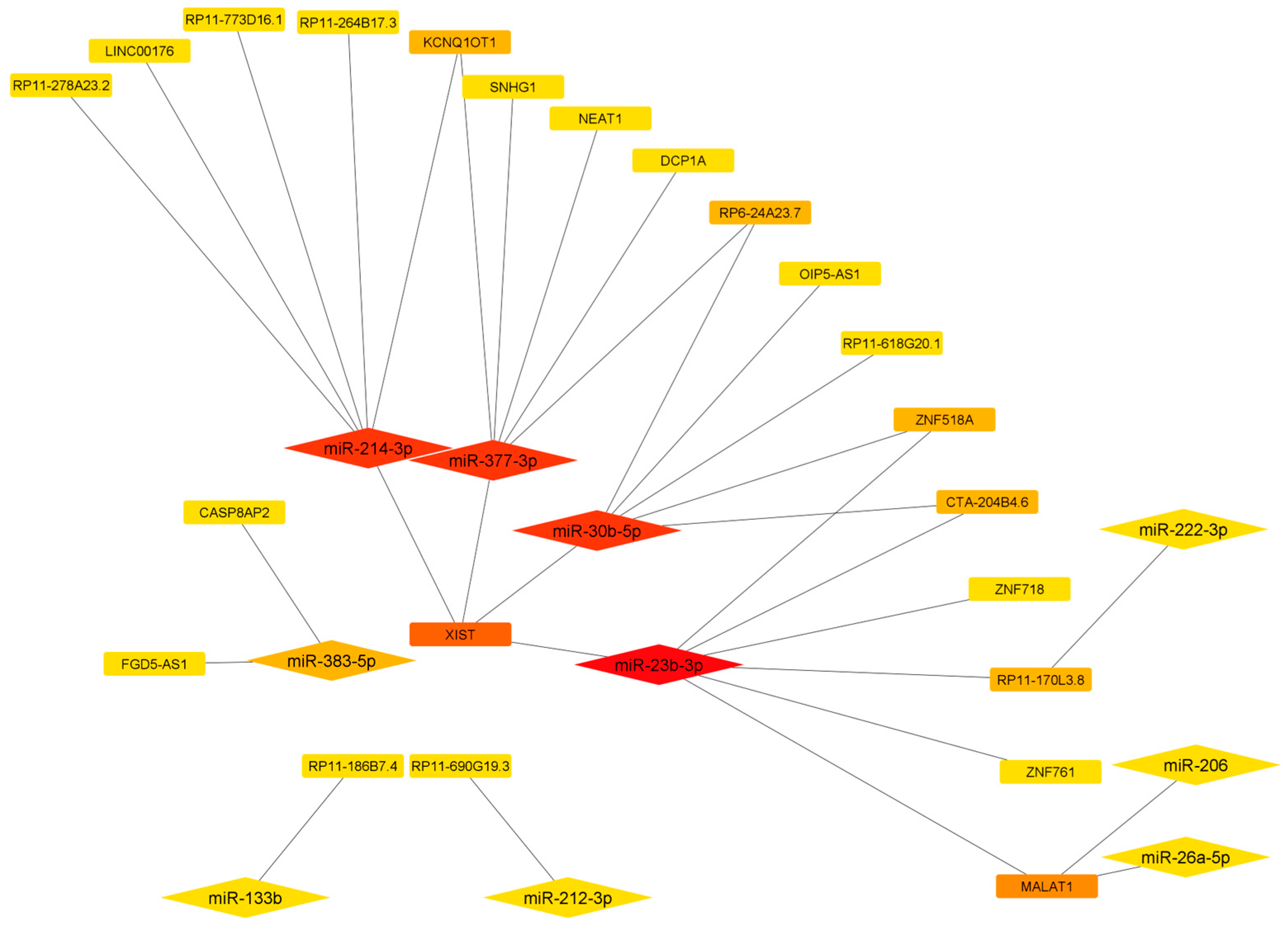
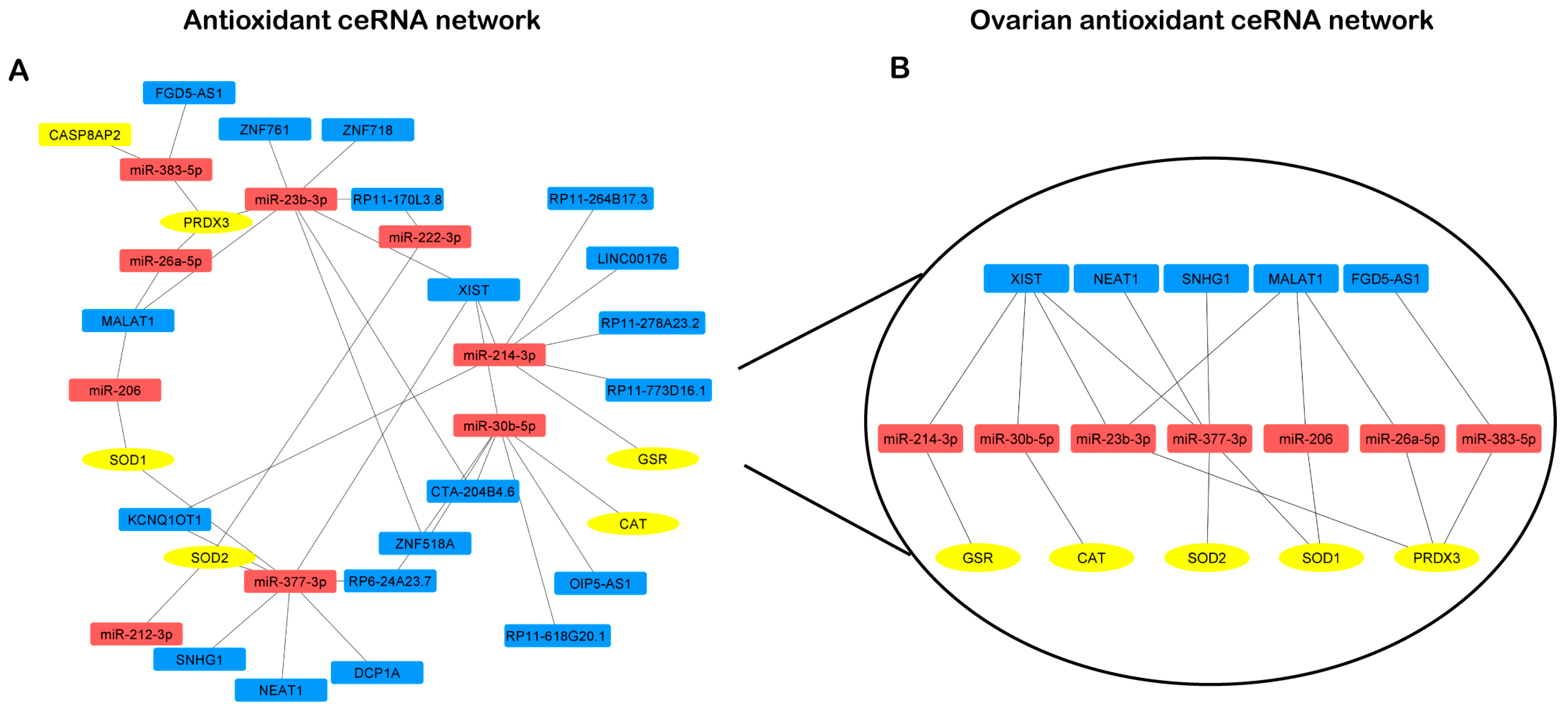
3.3. LncRNA-miRNA-mRNA Competing Endogenous RNA Networks
4. Discussion
4.1. Antioxidant Genes in the Human Ovary
4.2. The OvAnOx ceRNA Network
4.2.1. mRNA Components
4.2.2. lncRNAs Components
4.2.3. The miRNAs Components
4.3. Clinical Implications
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gershon, E.; Dekel, N. Newly Identified Regulators of Ovarian Folliculogenesis and Ovulation. Int. J. Mol. Sci. 2020, 21, 4565. [Google Scholar] [CrossRef] [PubMed]
- Tatone, C.; Amicarelli, F.; Carbone, M.C.; Monteleone, P.; Caserta, D.; Marci, R.; Artini, P.G.; Piomboni, P.; Focarelli, R. Cellular and molecular aspects of ovarian follicle ageing. Hum. Reprod. Update 2008, 14, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Tatone, C.; Di Emidio, G.; Placidi, M.; Rossi, G.; Ruggieri, S.; Taccaliti, C.; D’Alfonso, A.; Amicarelli, F.; Guido, M. AGEs-related dysfunctions in PCOS: Evidence from animal and clinical research. J. Endocrinol. 2021, 251, R1–R9. [Google Scholar] [CrossRef] [PubMed]
- Ruder, E.H.; Hartman, T.J.; Goldman, M.B. Impact of oxidative stress on female fertility. Curr. Opin. Obstet. Gynecol. 2009, 21, 219–222. [Google Scholar] [CrossRef]
- Yan, F.; Zhao, Q.; Li, Y.; Zheng, Z.; Kong, X.; Shu, C.; Liu, Y.; Shi, Y. The role of oxidative stress in ovarian aging: A review. J. Ovarian. Res. 2022, 15, 100. [Google Scholar] [CrossRef]
- Liang, J.; Gao, Y.; Feng, Z.; Zhang, B.; Na, Z.; Li, D. Reactive oxygen species and ovarian diseases: Antioxidant strategies. Redox Biol. 2023, 62, 102659. [Google Scholar] [CrossRef]
- Showell, M.G.; Mackenzie-Proctor, R.; Jordan, V.; Hart, R.J. Antioxidants for female subfertility. Cochrane Database Syst. Rev. 2020, 8, CD007807. [Google Scholar] [CrossRef]
- de Almeida, A.; de Oliveira, J.; da Silva Pontes, L.V.; de Souza Junior, J.F.; Goncalves, T.A.F.; Dantas, S.H.; de Almeida Feitosa, M.S.; Silva, A.O.; de Medeiros, I.A. ROS: Basic Concepts, Sources, Cellular Signaling, and its Implications in Aging Pathways. Oxid. Med. Cell. Longev. 2022, 2022, 1225578. [Google Scholar] [CrossRef]
- Sies, H.; Mailloux, R.J.; Jakob, U. Fundamentals of redox regulation in biology. Nat. Rev. Mol. Cell Biol. 2024, 25, 701–719. [Google Scholar] [CrossRef]
- Lei, X.G.; Zhu, J.H.; Cheng, W.H.; Bao, Y.; Ho, Y.S.; Reddi, A.R.; Holmgren, A.; Arner, E.S. Paradoxical Roles of Antioxidant Enzymes: Basic Mechanisms and Health Implications. Physiol. Rev. 2016, 96, 307–364. [Google Scholar] [CrossRef]
- Nemeth, K.; Bayraktar, R.; Ferracin, M.; Calin, G.A. Non-coding RNAs in disease: From mechanisms to therapeutics. Nat. Rev. Genet. 2024, 25, 211–232. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Gou, L.T.; Zhu, Q.; Liu, M.F. Small RNAs: An expanding world with therapeutic promises. Fundam. Res. 2023, 3, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef]
- Caponnetto, A.; Ferrara, C.; Fazzio, A.; Agosta, N.; Scribano, M.; Vento, M.E.; Borzì, P.; Barbagallo, C.; Stella, M.; Ragusa, M.; et al. A Circular RNA Derived from the Pumilio 1 Gene Could Regulate PTEN in Human Cumulus Cells. Genes 2024, 15, 124. [Google Scholar] [CrossRef]
- Caponnetto, A.; Battaglia, R.; Ferrara, C.; Vento, M.E.; Borzì, P.; Paradiso, M.; Scollo, P.; Purrello, M.; Longobardi, S.; D’Hooghe, T.; et al. Down-regulation of long non-coding RNAs in reproductive aging and analysis of the lncRNA-miRNA-mRNA networks in human cumulus cells. J. Assist. Reprod. Genet. 2022, 39, 919–931. [Google Scholar] [CrossRef]
- Barbagallo, D.; Palermo, C.I.; Barbagallo, C.; Battaglia, R.; Caponnetto, A.; Spina, V.; Ragusa, M.; Di Pietro, C.; Scalia, G.; Purrello, M. Competing endogenous RNA network mediated by circ_3205 in SARS-CoV-2 infected cells. Cell Mol. Life Sci. 2022, 79, 75. [Google Scholar] [CrossRef]
- Nejadi Orang, F.; Abdoli Shadbad, M. Competing endogenous RNA networks and ferroptosis in cancer: Novel therapeutic targets. Cell Death Dis. 2024, 15, 357. [Google Scholar] [CrossRef]
- Ciesielska, S.; Slezak-Prochazka, I.; Bil, P.; Rzeszowska-Wolny, J. Micro RNAs in Regulation of Cellular Redox Homeostasis. Int. J. Mol. Sci. 2021, 22, 6022. [Google Scholar] [CrossRef]
- Kinoshita, C.; Aoyama, K. The Role of Non-Coding RNAs in the Neuroprotective Effects of Glutathione. Int. J. Mol. Sci. 2021, 22, 4245. [Google Scholar] [CrossRef]
- Lim, J.; Luderer, U. Oxidative damage increases and antioxidant gene expression decreases with aging in the mouse ovary. Biol. Reprod. 2011, 84, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Shkolnik, K.; Tadmor, A.; Ben-Dor, S.; Nevo, N.; Galiani, D.; Dekel, N. Reactive oxygen species are indispensable in ovulation. Proc. Natl. Acad. Sci. USA 2011, 108, 1462–1467. [Google Scholar] [CrossRef] [PubMed]
- Kala, M.; Shaikh, M.V.; Nivsarkar, M. Equilibrium between anti-oxidants and reactive oxygen species: A requisite for oocyte development and maturation. Reprod. Med. Biol. 2017, 16, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Jamil, M.; Debbarh, H.; Aboulmaouahib, S.; Aniq Filali, O.; Mounaji, K.; Zarqaoui, M.; Saadani, B.; Louanjli, N.; Cadi, R. Reactive oxygen species in reproduction: Harmful, essential or both? Zygote 2020, 28, 255–269. [Google Scholar] [CrossRef]
- Tian, Y.; Liu, X.; Pei, X.; Gao, H.; Pan, P.; Yang, Y. Mechanism of Mitochondrial Homeostasis Controlling Ovarian Physiology. Endocrinology 2022, 164, bqac189. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tang, J.; Wang, L.; Tan, F.; Song, H.; Zhou, J.; Li, F. Oxidative stress in oocyte aging and female reproduction. J. Cell. Physiol. 2021, 236, 7966–7983. [Google Scholar] [CrossRef]
- Devine, P.J.; Perreault, S.D.; Luderer, U. Roles of reactive oxygen species and antioxidants in ovarian toxicity. Biol. Reprod. 2012, 86, 27. [Google Scholar] [CrossRef]
- Klotz, L.O.; Sanchez-Ramos, C.; Prieto-Arroyo, I.; Urbanek, P.; Steinbrenner, H.; Monsalve, M. Redox regulation of FoxO transcription factors. Redox Biol. 2015, 6, 51–72. [Google Scholar] [CrossRef]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef]
- Saeed-Zidane, M.; Linden, L.; Salilew-Wondim, D.; Held, E.; Neuhoff, C.; Tholen, E.; Hoelker, M.; Schellander, K.; Tesfaye, D. Cellular and exosome mediated molecular defense mechanism in bovine granulosa cells exposed to oxidative stress. PLoS ONE 2017, 12, e0187569. [Google Scholar] [CrossRef] [PubMed]
- Case, A.J. On the Origin of Superoxide Dismutase: An Evolutionary Perspective of Superoxide-Mediated Redox Signaling. Antioxidants 2017, 6, 82. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Bhella, D.; Lindsay, J.G. Reconstitution of the mitochondrial PrxIII antioxidant defence pathway: General properties and factors affecting PrxIII activity and oligomeric state. J. Mol. Biol. 2007, 372, 1022–1033. [Google Scholar] [CrossRef] [PubMed]
- Yewdall, N.A.; Peskin, A.V.; Hampton, M.B.; Goldstone, D.C.; Pearce, F.G.; Gerrard, J.A. Quaternary structure influences the peroxidase activity of peroxiredoxin 3. Biochem. Biophys. Res. Commun. 2018, 497, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Anwar, S.; Alrumaihi, F.; Sarwar, T.; Babiker, A.Y.; Khan, A.A.; Prabhu, S.V.; Rahmani, A.H. Exploring Therapeutic Potential of Catalase: Strategies in Disease Prevention and Management. Biomolecules 2024, 14, 697. [Google Scholar] [CrossRef]
- Trenz, T.S.; Delaix, C.L.; Turchetto-Zolet, A.C.; Zamocky, M.; Lazzarotto, F.; Margis-Pinheiro, M. Going Forward and Back: The Complex Evolutionary History of the GPx. Biology 2021, 10, 1165. [Google Scholar] [CrossRef]
- Chen, T.H.; Wang, H.C.; Chang, C.J.; Lee, S.Y. Mitochondrial Glutathione in Cellular Redox Homeostasis and Disease Manifestation. Int. J. Mol. Sci. 2024, 25, 1314. [Google Scholar] [CrossRef]
- Couto, N.; Wood, J.; Barber, J. The role of glutathione reductase and related enzymes on cellular redox homoeostasis network. Free Radic. Biol. Med. 2016, 95, 27–42. [Google Scholar] [CrossRef]
- Hayes, J.D.; McLellan, L.I. Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Radic. Res. 1999, 31, 273–300. [Google Scholar] [CrossRef]
- Mazari, A.M.A.; Zhang, L.; Ye, Z.W.; Zhang, J.; Tew, K.D.; Townsend, D.M. The Multifaceted Role of Glutathione S-Transferases in Health and Disease. Biomolecules 2023, 13, 688. [Google Scholar] [CrossRef]
- Zhang, H.; Forman, H.J. Glutathione synthesis and its role in redox signaling. Semin. Cell. Dev. Biol. 2012, 23, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.M.; Mieyal, J.J. Protein-thiol oxidation and cell death: Regulatory role of glutaredoxins. Antioxid. Redox Signal. 2012, 17, 1748–1763. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Fan, C.; Zhao, J.; Wang, L.; Duan, D.; Shen, T.; Li, X. Fluorescent Probes for Mammalian Thioredoxin Reductase: Mechanistic Analysis, Construction Strategies, and Future Perspectives. Biosensors 2023, 13, 811. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Lin, Y.; Huang, Y.; Shen, Y.Q.; Chen, Q. Thioredoxin (Trx): A redox target and modulator of cellular senescence and aging-related diseases. Redox Biol. 2024, 70, 103032. [Google Scholar] [CrossRef]
- Matzuk, M.M.; Dionne, L.; Guo, Q.; Kumar, T.R.; Lebovitz, R.M. Ovarian function in superoxide dismutase 1 and 2 knockout mice. Endocrinology 1998, 139, 4008–4011. [Google Scholar] [CrossRef]
- Pretsch, W. Glutathione reductase activity deficiency in homozygous Gr1a1Neu mice does not cause haemolytic anaemia. Genet. Res. 1999, 73, 1–5. [Google Scholar] [CrossRef]
- Rogers, L.K.; Bates, C.M.; Welty, S.E.; Smith, C.V. Diquat induces renal proximal tubule injury in glutathione reductase-deficient mice. Toxicol. Appl. Pharmacol. 2006, 217, 289–298. [Google Scholar] [CrossRef]
- Ho, Y.S.; Xiong, Y.; Ma, W.; Spector, A.; Ho, D.S. Mice lacking catalase develop normally but show differential sensitivity to oxidant tissue injury. J. Biol. Chem. 2004, 279, 32804–32812. [Google Scholar] [CrossRef]
- Wang, S.; He, G.; Chen, M.; Zuo, T.; Xu, W.; Liu, X. The Role of Antioxidant Enzymes in the Ovaries. Oxid. Med. Cell. Longev. 2017, 2017, 4371714. [Google Scholar] [CrossRef]
- Vu, H.V.; Lee, S.; Acosta, T.J.; Yoshioka, S.; Abe, H.; Okuda, K. Roles of prostaglandin F2alpha and hydrogen peroxide in the regulation of Copper/Zinc superoxide dismutase in bovine corpus luteum and luteal endothelial cells. Reprod. Biol. Endocrinol. 2012, 10, 87. [Google Scholar] [CrossRef]
- Tatone, C.; Carbone, M.C.; Falone, S.; Aimola, P.; Giardinelli, A.; Caserta, D.; Marci, R.; Pandolfi, A.; Ragnelli, A.M.; Amicarelli, F. Age-dependent changes in the expression of superoxide dismutases and catalase are associated with ultrastructural modifications in human granulosa cells. Mol. Hum. Reprod. 2006, 12, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Bizon, A.; Tchorz, A.; Madej, P.; Lesniewski, M.; Wojtowicz, M.; Piwowar, A.; Franik, G. The Activity of Superoxide Dismutase, Its Relationship with the Concentration of Zinc and Copper and the Prevalence of rs2070424 Superoxide Dismutase Gene in Women with Polycystic Ovary Syndrome-Preliminary Study. J. Clin. Med. 2022, 11, 2548. [Google Scholar] [CrossRef] [PubMed]
- Seleem, A.K.; El Refaeey, A.A.; Shaalan, D.; Sherbiny, Y.; Badawy, A. Superoxide dismutase in polycystic ovary syndrome patients undergoing intracytoplasmic sperm injection. J. Assist. Reprod. Genet. 2014, 31, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.N.; Chaube, S.K. A moderate increase of hydrogen peroxide level is beneficial for spontaneous resumption of meiosis from diplotene arrest in rat oocytes cultured in vitro. Biores. Open Access 2014, 3, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Matos, L.; Stevenson, D.; Gomes, F.; Silva-Carvalho, J.L.; Almeida, H. Superoxide dismutase expression in human cumulus oophorus cells. Mol. Hum. Reprod. 2009, 15, 411–419. [Google Scholar] [CrossRef]
- Cetica, P.D.; Pintos, L.N.; Dalvit, G.C.; Beconi, M.T. Antioxidant enzyme activity and oxidative stress in bovine oocyte in vitro maturation. IUBMB Life 2001, 51, 57–64. [Google Scholar] [CrossRef]
- Perkins, A.T.; Greig, M.M.; Sontakke, A.A.; Peloquin, A.S.; McPeek, M.A.; Bickel, S.E. Increased levels of superoxide dismutase suppress meiotic segregation errors in aging oocytes. Chromosoma 2019, 128, 215–222. [Google Scholar] [CrossRef]
- Behl, R.; Pandey, R.S. FSH induced stimulation of catalase activity in goat granulosa cells in vitro. Anim. Reprod. Sci. 2002, 70, 215–221. [Google Scholar] [CrossRef]
- Serke, H.; Bausenwein, J.; Hirrlinger, J.; Nowicki, M.; Vilser, C.; Jogschies, P.; Hmeidan, F.A.; Blumenauer, V.; Spanel-Borowski, K. Granulosa cell subtypes vary in response to oxidized low-density lipoprotein as regards specific lipoprotein receptors and antioxidant enzyme activity. J. Clin. Endocrinol. Metab. 2010, 95, 3480–3490. [Google Scholar] [CrossRef]
- Barros, F.D.A.; Adona, P.R.; Guemra, S.; Damiao, B.C.M. Oxidative homeostasis in oocyte competence for in vitro embryo development. Anim. Sci. J. 2019, 90, 1343–1349. [Google Scholar] [CrossRef]
- Luciano, A.M.; Goudet, G.; Perazzoli, F.; Lahuec, C.; Gerard, N. Glutathione content and glutathione peroxidase expression in in vivo and in vitro matured equine oocytes. Mol. Reprod. Dev. 2006, 73, 658–666. [Google Scholar] [CrossRef] [PubMed]
- Katz-Jaffe, M.G.; Lane, S.L.; Parks, J.C.; McCallie, B.R.; Makloski, R.; Schoolcraft, W.B. Antioxidant Intervention Attenuates Aging-Related Changes in the Murine Ovary and Oocyte. Life 2020, 10, 250. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zheng, Y.; Li, J.; Yu, Y.; Zhang, W.; Song, M.; Liu, Z.; Min, Z.; Hu, H.; Jing, Y.; et al. Single-Cell Transcriptomic Atlas of Primate Ovarian Aging. Cell 2020, 180, 585–600.e19. [Google Scholar] [CrossRef] [PubMed]
- Dumollard, R.; Ward, Z.; Carroll, J.; Duchen, M.R. Regulation of redox metabolism in the mouse oocyte and embryo. Development 2007, 134, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Iuchi, Y.; Kawachiya, S.; Fujii, T.; Saito, H.; Kurachi, H.; Fujii, J. Alteration of glutathione reductase expression in the female reproductive organs during the estrous cycle. Biol. Reprod. 2001, 65, 1410–1416. [Google Scholar] [CrossRef] [PubMed]
- Tsai-Turton, M.; Luderer, U. Opposing effects of glutathione depletion and follicle-stimulating hormone on reactive oxygen species and apoptosis in cultured preovulatory rat follicles. Endocrinology 2006, 147, 1224–1236. [Google Scholar] [CrossRef] [PubMed]
- Hoang, Y.D.; Nakamura, B.N.; Luderer, U. Follicle-stimulating hormone and estradiol interact to stimulate glutathione synthesis in rat ovarian follicles and granulosa cells. Biol. Reprod. 2009, 81, 636–646. [Google Scholar] [CrossRef]
- Park, J.I.; Jeon, H.J.; Jung, N.K.; Jang, Y.J.; Kim, J.S.; Seo, Y.W.; Jeong, M.; Chae, H.Z.; Chun, S.Y. Periovulatory expression of hydrogen peroxide-induced sulfiredoxin and peroxiredoxin 2 in the rat ovary: Gonadotropin regulation and potential modification. Endocrinology 2012, 153, 5512–5521. [Google Scholar] [CrossRef]
- Hernández-Cruz, E.Y.; Arancibia-Hernández, Y.L.; Loyola-Mondragón, D.Y.; Pedraza-Chaverri, J. Oxidative Stress and Its Role in Cd-Induced Epigenetic Modifications: Use of Antioxidants as a Possible Preventive Strategy. Oxygen 2022, 2, 177–212. [Google Scholar] [CrossRef]
- Infante-Menendez, J.; Gonzalez-Lopez, P.; Huertas-Larez, R.; Gomez-Hernandez, A.; Escribano, O. Oxidative Stress Modulation by ncRNAs and Their Emerging Role as Therapeutic Targets in Atherosclerosis and Non-Alcoholic Fatty Liver Disease. Antioxidants 2023, 12, 262. [Google Scholar] [CrossRef]
- Fan, H.; Zhou, D.; Zhang, X.; Jiang, M.; Kong, X.; Xue, T.; Gao, L.; Lu, D.; Tao, C.; Wang, L. hsa_circRNA_BECN1 acts as a ceRNA to promote polycystic ovary syndrome progression by sponging the miR-619-5p/Rab5b axis. Mol. Hum. Reprod. 2023, 29, gaad036. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Gong, J.; Guo, Y.; Li, Y.; Huang, H.; Liu, X. Construction of a ceRNA network in polycystic ovary syndrome (PCOS) driven by exosomal lncRNA. Front. Genet. 2022, 13, 979924. [Google Scholar] [CrossRef] [PubMed]
- ElMonier, A.A.; El-Boghdady, N.A.; Fahim, S.A.; Sabry, D.; Elsetohy, K.A.; Shaheen, A.A. LncRNA NEAT1 and MALAT1 are involved in polycystic ovary syndrome pathogenesis by functioning as competing endogenous RNAs to control the expression of PCOS-related target genes. Noncoding RNA Res. 2023, 8, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Ma, L.; Cao, Y.; Zhai, J. Construction of a ceRNA-based lncRNA-mRNA network to identify functional lncRNAs in polycystic ovarian syndrome. Aging 2021, 13, 8481–8496. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhou, Z.; Wang, P.; He, X.; Liu, Y.; Chu, M. The SLC19A1-AS/miR-1343/WNT11 axis is a novel positive regulatory ceRNA network governing goat granulosa cell proliferation. Int. J. Biol. Macromol. 2024, 264, 130658. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Y.; Yang, L.; Du, X.; Li, Q. Nuclear lncRNA NORSF reduces E2 release in granulosa cells by sponging the endogenous small activating RNA miR-339. BMC Biol. 2023, 21, 221. [Google Scholar] [CrossRef]
- Liu, B.; Liu, L.; Sulaiman, Z.; Wang, C.; Wang, L.; Zhu, J.; Liu, S.; Cheng, Z. Comprehensive analysis of lncRNA-miRNA-mRNA ceRNA network and key genes in granulosa cells of patients with biochemical primary ovarian insufficiency. J. Assist. Reprod. Genet. 2024, 41, 15–29. [Google Scholar] [CrossRef]
- Jain, N.; Gupta, P.; Sahoo, S.; Mallick, B. Non-coding RNAs and their cross-talks impacting reproductive health of women. Wiley Interdiscip. Rev. RNA 2022, 13, e1695. [Google Scholar] [CrossRef]
- Lin, N.; Lin, J.Z. Identification of long non-coding RNA biomarkers and signature scoring, with competing endogenous RNA networks- targeted drug candidates for recurrent implantation failure. Hum. Fertil. 2022, 25, 983–992. [Google Scholar] [CrossRef]
- Hu, H.; Jia, Q.; Xi, J.; Zhou, B.; Li, Z. Integrated analysis of lncRNA, miRNA and mRNA reveals novel insights into the fertility regulation of large white sows. BMC Genomics 2020, 21, 636. [Google Scholar] [CrossRef]
- Alfeghaly, C.; Castel, G.; Cazottes, E.; Moscatelli, M.; Moinard, E.; Casanova, M.; Boni, J.; Mahadik, K.; Lammers, J.; Freour, T.; et al. XIST dampens X chromosome activity in a SPEN-dependent manner during early human development. Nat. Struct. Mol. Biol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Liu, X.; Qiukai, E.; Shang, Y.; Zhang, X.; Liu, S.; Zhang, X. Long non-coding RNA Xist regulates oocyte loss via suppressing miR-23b-3p/miR-29a-3p maturation and upregulating STX17 in perinatal mouse ovaries. Cell Death Dis. 2021, 12, 540. [Google Scholar] [CrossRef] [PubMed]
- Avner, R.; Wahrman, J.; Richler, C.; Ayoub, N.; Friedmann, A.; Laufer, N.; Mitrani-Rosenbaum, S. X inactivation-specific transcript expression in mouse oocytes and zygotes. Mol. Hum. Reprod. 2000, 6, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Li, J.; Li, J.; Zhang, N.; Zhou, W.; Ren, L.; Chen, Q.; Li, Y. Construction of Competing Endogenous RNA Networks Incorporating Transcription Factors to Reveal Differences in Granulosa Cells from Patients with Endometriosis. Genet. Test. Mol. Biomarkers 2021, 25, 453–462. [Google Scholar] [CrossRef]
- Liu, M.; Zhu, H.; Li, Y.; Zhuang, J.; Cao, T.; Wang, Y. Expression of serum lncRNA-Xist in patients with polycystic ovary syndrome and its relationship with pregnancy outcome. Taiwan J. Obstet. Gynecol. 2020, 59, 372–376. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Kowluru, R.A. Long Noncoding RNA MALAT1 and Regulation of the Antioxidant Defense System in Diabetic Retinopathy. Diabetes 2021, 70, 227–239. [Google Scholar] [CrossRef]
- Zeng, R.; Zhang, R.; Song, X.; Ni, L.; Lai, Z.; Liu, C.; Ye, W. The long non-coding RNA MALAT1 activates Nrf2 signaling to protect human umbilical vein endothelial cells from hydrogen peroxide. Biochem. Biophys. Res. Commun. 2018, 495, 2532–2538. [Google Scholar] [CrossRef]
- Li, Y.; Xiang, Y.; Song, Y.; Zhang, D.; Tan, L. MALAT1 downregulation is associated with polycystic ovary syndrome via binding with MDM2 and repressing P53 degradation. Mol. Cell. Endocrinol. 2022, 543, 111528. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, P.; Lu, W. lncRNA MALAT1 Regulates Mouse Granulosa Cell Apoptosis and 17beta-Estradiol Synthesis via Regulating miR-205/CREB1 Axis. Biomed. Res. Int. 2021, 2021, 6671814. [Google Scholar] [CrossRef]
- Tu, M.; Wu, Y.; Wang, F.; Huang, Y.; Qian, Y.; Li, J.; Lv, P.; Ying, Y.; Liu, J.; Liu, Y.; et al. Effect of lncRNA MALAT1 on the Granulosa Cell Proliferation and Pregnancy Outcome in Patients with PCOS. Front. Endocrinol. 2022, 13, 825431. [Google Scholar] [CrossRef]
- Wu, L.; Tu, Z.; Bao, Y.; Zhai, Q.; Jin, L. Long noncoding RNA NEAT1 decreases polycystic ovary syndrome progression via the modulation of the microRNA-324-3p and BRD3 axis. Cell Biol. Int. 2022, 46, 2075–2084. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Shimada, M.; Yanaka, K.; Mito, M.; Arai, T.; Takahashi, E.; Fujita, Y.; Fujimori, T.; Standaert, L.; Marine, J.C.; et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 2014, 141, 4618–4627. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.X.; Ke, Y.; Qiu, P.; Gao, J.; Deng, G.P. LncRNA NEAT1 inhibits apoptosis and autophagy of ovarian granulosa cells through miR-654/STC2-mediated MAPK signaling pathway. Exp. Cell Res. 2023, 424, 113473. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, T.; Gomes, A.V. MicroRNAs in the regulation of cellular redox status and its implications in myocardial ischemia-reperfusion injury. Redox Biol. 2020, 36, 101607. [Google Scholar] [CrossRef]
- Bu, H.; Wedel, S.; Cavinato, M.; Jansen-Durr, P. MicroRNA Regulation of Oxidative Stress-Induced Cellular Senescence. Oxid. Med. Cell. Longev. 2017, 2017, 2398696. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Ahmadi, Z.; Samarghandian, S.; Mohammadinejad, R.; Yaribeygi, H.; Sathyapalan, T.; Sahebkar, A. MicroRNA-mediated regulation of Nrf2 signaling pathway: Implications in disease therapy and protection against oxidative stress. Life Sci. 2020, 244, 117329. [Google Scholar] [CrossRef]
- Santonocito, M.; Vento, M.; Guglielmino, M.R.; Battaglia, R.; Wahlgren, J.; Ragusa, M.; Barbagallo, D.; Borzì, P.; Rizzari, S.; Maugeri, M.; et al. Molecular characterization of exosomes and their microRNA cargo in human follicular fluid: Bioinformatic analysis reveals that exosomal microRNAs control pathways involved in follicular maturation. Fertil. Steril. 2014, 102, 1751–1761.e1. [Google Scholar] [CrossRef]
- Battaglia, R.; Vento, M.E.; Ragusa, M.; Barbagallo, D.; La Ferlita, A.; Di Emidio, G.; Borzí, P.; Artini, P.G.; Scollo, P.; Tatone, C.; et al. MicroRNAs Are Stored in Human MII Oocyte and Their Expression Profile Changes in Reproductive Aging. Biol. Reprod. 2016, 95, 131. [Google Scholar] [CrossRef]
- Battaglia, R.; Vento, M.E.; Borzì, P.; Ragusa, M.; Barbagallo, D.; Arena, D.; Purrello, M.; Di Pietro, C. Non-coding RNAs in the Ovarian Follicle. Front. Genet. 2017, 8, 57. [Google Scholar] [CrossRef]
- Battaglia, R.; Musumeci, P.; Ragusa, M.; Barbagallo, D.; Scalia, M.; Zimbone, M.; Lo Faro, J.M.; Borzì, P.; Scollo, P.; Purrello, M.; et al. Ovarian aging increases small extracellular vesicle CD81+ release in human follicular fluid and influences miRNA profiles. Aging 2020, 12, 12324–12341. [Google Scholar] [CrossRef]
- Amin, M.M.J.; Trevelyan, C.J.; Turner, N.A. MicroRNA-214 in Health and Disease. Cells 2021, 10, 3274. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, R.; Caponnetto, A.; Caringella, A.M.; Cortone, A.; Ferrara, C.; Smirni, S.; Iannitti, R.; Purrello, M.; D’Amato, G.; Fioretti, B.; et al. Resveratrol Treatment Induces Mito-miRNome Modification in Follicular Fluid from Aged Women with a Poor Prognosis for In Vitro Fertilization Cycles. Antioxidants 2022, 11, 1019. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Wang, J.; Tao, D.; Zhang, Y.; He, J.; Shi, C. Identification of a novel miRNA from the ovine ovary by a combinatorial approach of bioinformatics and experiments. J. Vet. Med. Sci. 2016, 77, 1617–1624. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dong, C.; Yang, J.; Li, Y.; Feng, J.; Wang, B.; Zhang, J.; Guo, X. The Roles of the miRNAome and Transcriptome in the Ovine Ovary Reveal Poor Efficiency in Juvenile Superovulation. Animals 2021, 11, 239. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Jin, X.; Sheng, Z.; Zhang, Z. miR-206 serves an important role in polycystic ovary syndrome through modulating ovarian granulosa cell proliferation and apoptosis. Exp. Ther. Med. 2021, 21, 179. [Google Scholar] [CrossRef] [PubMed]
- Gad, A.; Sanchez, J.M.; Browne, J.A.; Nemcova, L.; Laurincik, J.; Prochazka, R.; Lonergan, P. Plasma extracellular vesicle miRNAs as potential biomarkers of superstimulatory response in cattle. Sci. Rep. 2020, 10, 19130. [Google Scholar] [CrossRef]
- Zhang, Z.; Sang, M.; Liu, S.; Shao, J.; Cai, Y. Differential expression of long non-coding RNA Regulator of reprogramming and its molecular mechanisms in polycystic ovary syndrome. J. Ovarian. Res. 2021, 14, 79. [Google Scholar] [CrossRef]
- Diaz, M.; Bassols, J.; Lopez-Bermejo, A.; de Zegher, F.; Ibanez, L. Low Circulating Levels of miR-451a in Girls with Polycystic Ovary Syndrome: Different Effects of Randomized Treatments. J. Clin. Endocrinol. Metab. 2020, 105, dgz204. [Google Scholar] [CrossRef]
- Lu, T.; Zou, X.; Liu, G.; Deng, M.; Sun, B.; Guo, Y.; Liu, D.; Li, Y. A Preliminary Study on the Characteristics of microRNAs in Ovarian Stroma and Follicles of Chuanzhong Black Goat during Estrus. Genes 2020, 11, 970. [Google Scholar] [CrossRef]
- De Nardo Maffazioli, G.; Baracat, E.C.; Soares, J.M.; Carvalho, K.C.; Maciel, G.A.R. Evaluation of circulating microRNA profiles in Brazilian women with polycystic ovary syndrome: A preliminary study. PLoS ONE 2022, 17, e0275031. [Google Scholar] [CrossRef]
- Ibrahim, S.; Taqi, M.O.; Sosa, A.S.A.; El-Naby, A.A.H.; Mahmoud, K.G.M.; Darwish, H.R.H.; Abd El Hameed, A.R.; Nawito, M.F. Spatiotemporal expression pattern of miR-205, miR-26a-5p, miR-17-5p, let-7b-5p, and their target genes during different stages of corpus luteum in Egyptian buffaloes. J. Genet. Eng. Biotechnol. 2022, 20, 37. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.Y.; Kim, K.S.; Kim, Y.J.; Kim, S.W.; Kim, H.; Ku, S.Y. Transcriptome Analyses Identify Potential Key microRNAs and Their Target Genes Contributing to Ovarian Reserve. Int. J. Mol. Sci. 2021, 22, 10819. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Fan, F.; Liang, C.; Zhou, Y.; Qiao, X.; Sun, Y.; Jiang, Y.; Kang, L. Variants of pri-miR-26a-5p polymorphisms are associated with values for chicken egg production variables and affects abundance of mature miRNA. Anim. Reprod. Sci. 2019, 201, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Yang, C.; Wu, H.; Chen, Q.; Huang, L.; Li, X.; Tang, H.; Jiang, Y. miR-26a-5p Regulates TNRC6A Expression and Facilitates Theca Cell Proliferation in Chicken Ovarian Follicles. DNA Cell Biol. 2017, 36, 922–929. [Google Scholar] [CrossRef]
- Li, Y.; Wu, X.; Miao, S.; Cao, Q. MiR-383-5p promotes apoptosis of ovarian granulosa cells by targeting CIRP through the PI3K/AKT signaling pathway. Arch. Gynecol. Obstet. 2022, 306, 501–512. [Google Scholar] [CrossRef]

| Cellular Localization | Extracellular Localization | ||
|---|---|---|---|
| Gene Name | Cytoplasm | Mitochondria | Exosomes |
| CAT | |||
| GCLC | |||
| GCLM | |||
| GLRX | |||
| GLRX2 | |||
| GPX1 | |||
| GPX3 | |||
| GSR | |||
| GSTA4 | |||
| GSTM1 | |||
| GSTM2 | |||
| GSTP1 | |||
| GSTT1 | |||
| MGST1 | |||
| PRDX3 | |||
| SOD1 | |||
| SOD2 | |||
| TXN | |||
| TXN2 | |||
| TXNRD1 | |||
| TXNRD2 | |||
| Gene Name | FF | O | CC | GC | TC | LC | SC | ND |
|---|---|---|---|---|---|---|---|---|
| CAT | ||||||||
| GCLC | ||||||||
| GCLM | ||||||||
| GLRX | ||||||||
| GLRX2 | ||||||||
| GPX1 | ||||||||
| GPX3 | ||||||||
| GSR | ||||||||
| GSTA4 | ||||||||
| GSTM1 | ||||||||
| GSTM2 | ||||||||
| GSTP1 | ||||||||
| GSTT1 | ||||||||
| MGST1 | ||||||||
| PRDX3 | ||||||||
| SOD1 | ||||||||
| SOD2 | ||||||||
| TXN | ||||||||
| TXN2 | ||||||||
| TXNRD1 | ||||||||
| TXNRD2 |
| CAT | GCLC | GCLM | GSR | GSTP1 | PRDX3 | SOD1 | SOD2 | TXN2 | TXNRD2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| miR-16-5p | ||||||||||
| miR-17-3p | ||||||||||
| miR-23b-3p | ||||||||||
| miR-26a-5p | ||||||||||
| miR-27a-5p | ||||||||||
| miR-30b-5p | ||||||||||
| miR-106b | ||||||||||
| miR-133a | ||||||||||
| mir-146a | ||||||||||
| miR-186-5p | ||||||||||
| miR-206 | ||||||||||
| miR-212-3p | ||||||||||
| miR-214-3p | ||||||||||
| miR-222-3p | ||||||||||
| miR-377-3p | ||||||||||
| miR-383-3p | ||||||||||
| miR-425-5p | ||||||||||
| miR-433-5p | ||||||||||
| miR-513a-3p | ||||||||||
| miR-3929 | ||||||||||
| miR-5191 | ||||||||||
| miR-6823-5p |
| miR-23b-3p | miR-26a-5p | miR-30b-5p | miR-133b | miR-206 | miR-212-3p | miR-214-3p | miR-222-3p | miR-377-3p | miR-383-5p | |
|---|---|---|---|---|---|---|---|---|---|---|
| CASP8AP2 | ||||||||||
| CTA-204B4.6 | ||||||||||
| DCP1A | ||||||||||
| FGD5-AS1 | ||||||||||
| KCNQ1OT1 | ||||||||||
| LINC00176 | ||||||||||
| MALAT1 | ||||||||||
| NEAT1 | ||||||||||
| OIP5-AS1 | ||||||||||
| RP11-170L3.8 | ||||||||||
| RP11-186B7.4 | ||||||||||
| RP11-264B17.3 | ||||||||||
| RP11-278A23.2 | ||||||||||
| RP11-618G20.1 | ||||||||||
| RP11-690G19.3 | ||||||||||
| RP11-773D16.1 | ||||||||||
| RP6-24A23.7 | ||||||||||
| SNHG1 | ||||||||||
| XIST | ||||||||||
| ZNF518A | ||||||||||
| ZNF718 | ||||||||||
| ZNF761 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tatone, C.; Di Emidio, G.; Battaglia, R.; Di Pietro, C. Building a Human Ovarian Antioxidant ceRNA Network “OvAnOx”: A Bioinformatic Perspective for Research on Redox-Related Ovarian Functions and Dysfunctions. Antioxidants 2024, 13, 1101. https://doi.org/10.3390/antiox13091101
Tatone C, Di Emidio G, Battaglia R, Di Pietro C. Building a Human Ovarian Antioxidant ceRNA Network “OvAnOx”: A Bioinformatic Perspective for Research on Redox-Related Ovarian Functions and Dysfunctions. Antioxidants. 2024; 13(9):1101. https://doi.org/10.3390/antiox13091101
Chicago/Turabian StyleTatone, Carla, Giovanna Di Emidio, Rosalia Battaglia, and Cinzia Di Pietro. 2024. "Building a Human Ovarian Antioxidant ceRNA Network “OvAnOx”: A Bioinformatic Perspective for Research on Redox-Related Ovarian Functions and Dysfunctions" Antioxidants 13, no. 9: 1101. https://doi.org/10.3390/antiox13091101









