Identification and Analysis of the Superoxide Dismutase (SOD) Gene Family and Potential Roles in High-Temperature Stress Response of Herbaceous Peony (Paeonia lactiflora Pall.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of PlSOD Genes in P. lactiflora
2.2. Physicochemical Characteristics and Subcellular Localization
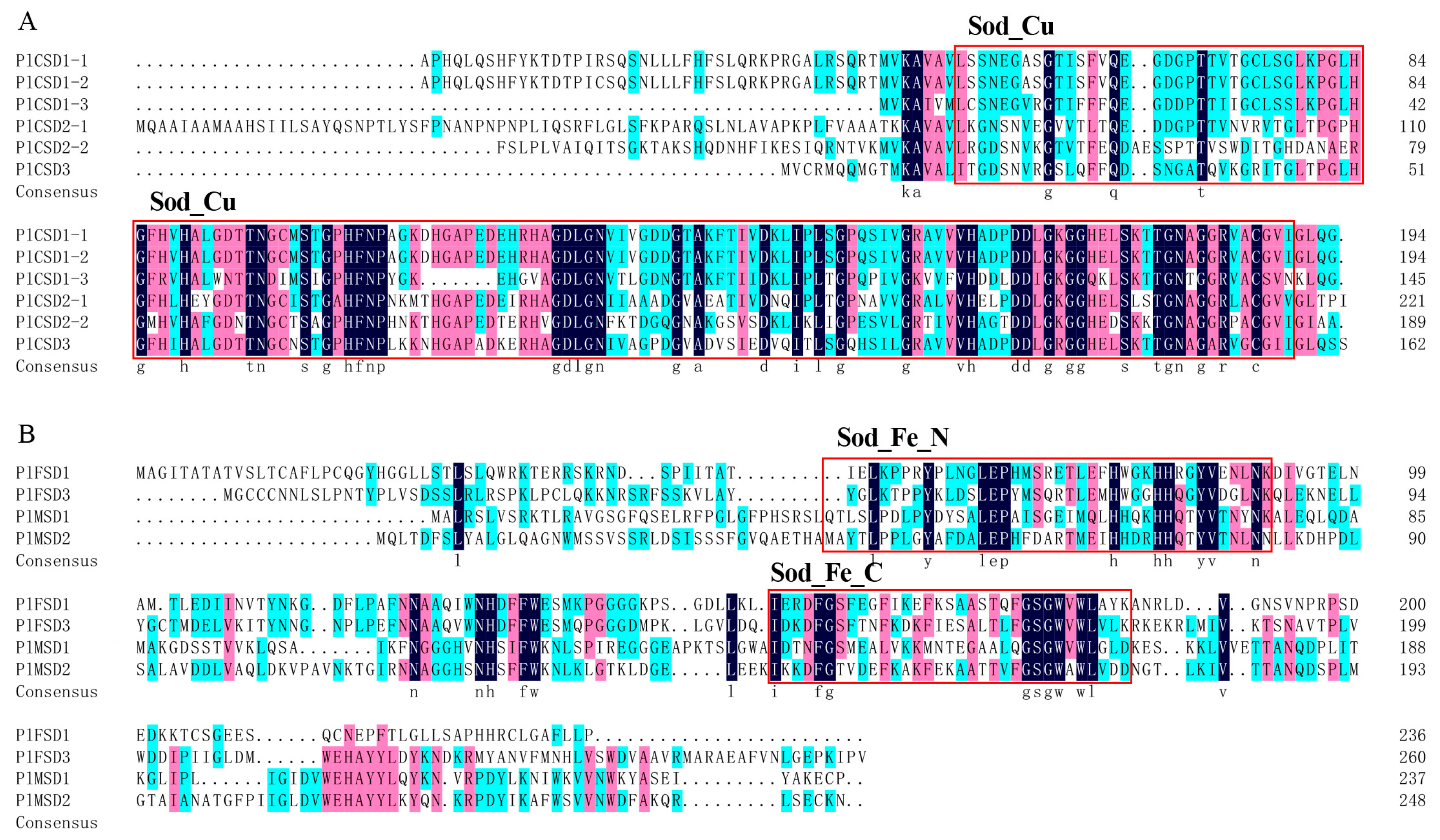
2.3. Phylogenetic Analysis, Motif, and Conserved Domain Analysis
2.4. Expression Patterns of PlSOD Genes Based on RNA-Seq Data
2.5. Plant Materials
2.6. Treatments and Sampling
2.7. Determination of Chl Fluorescence
2.8. Measurements of ROS-Related Physiological and Biochemical Indices
2.9. RNA Extraction and qRT-PCR Analysis
2.10. Statistical Analysis
3. Results
3.1. Identification of SOD Gene Family in P. lactiflora
3.2. Phylogenetic Analysis of PlSODs
3.3. Phylogeny and Conserved Motifs of PlSOD Genes
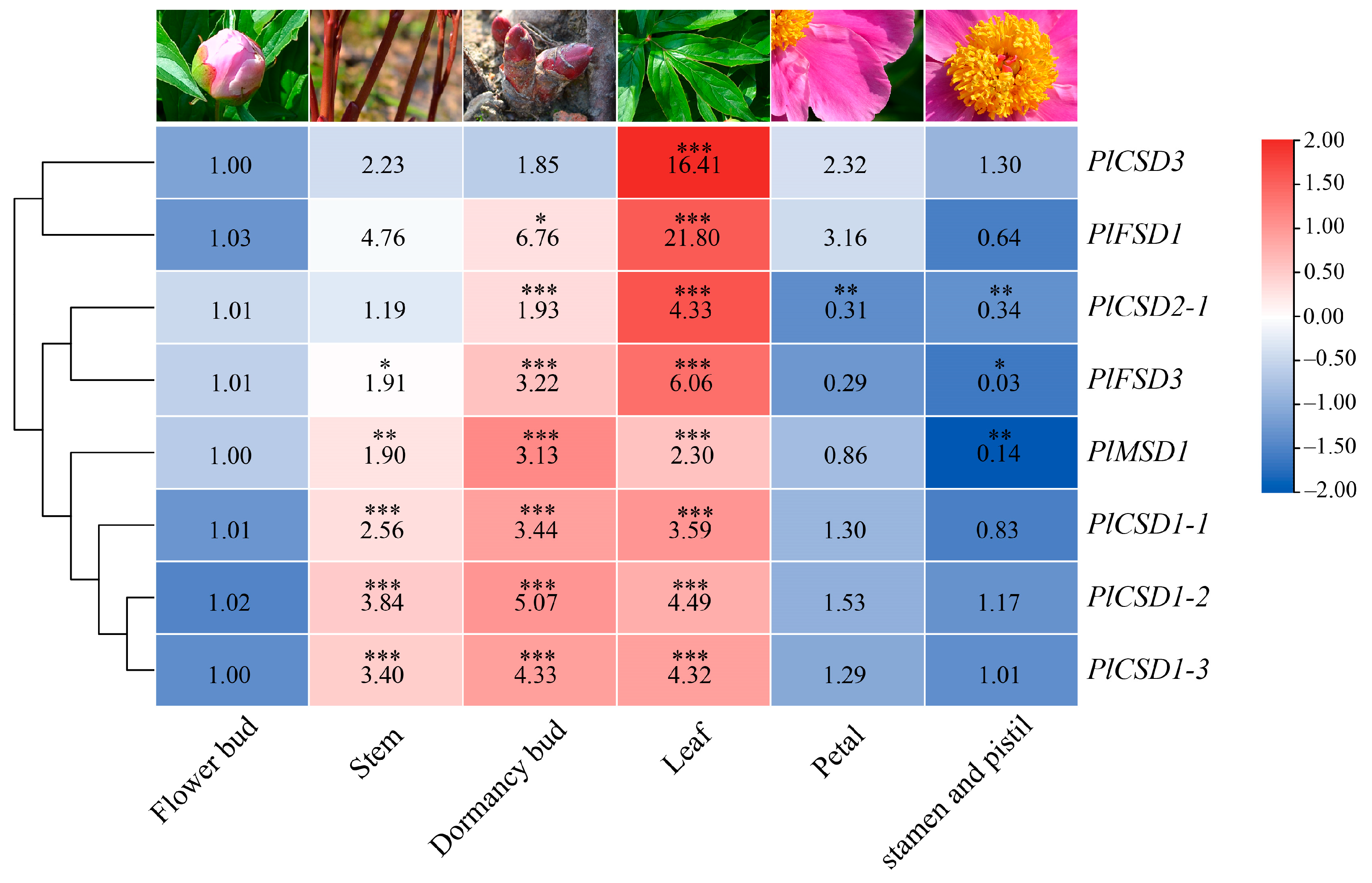
3.4. Expression Analysis of PlSOD Genes in Different Tissues
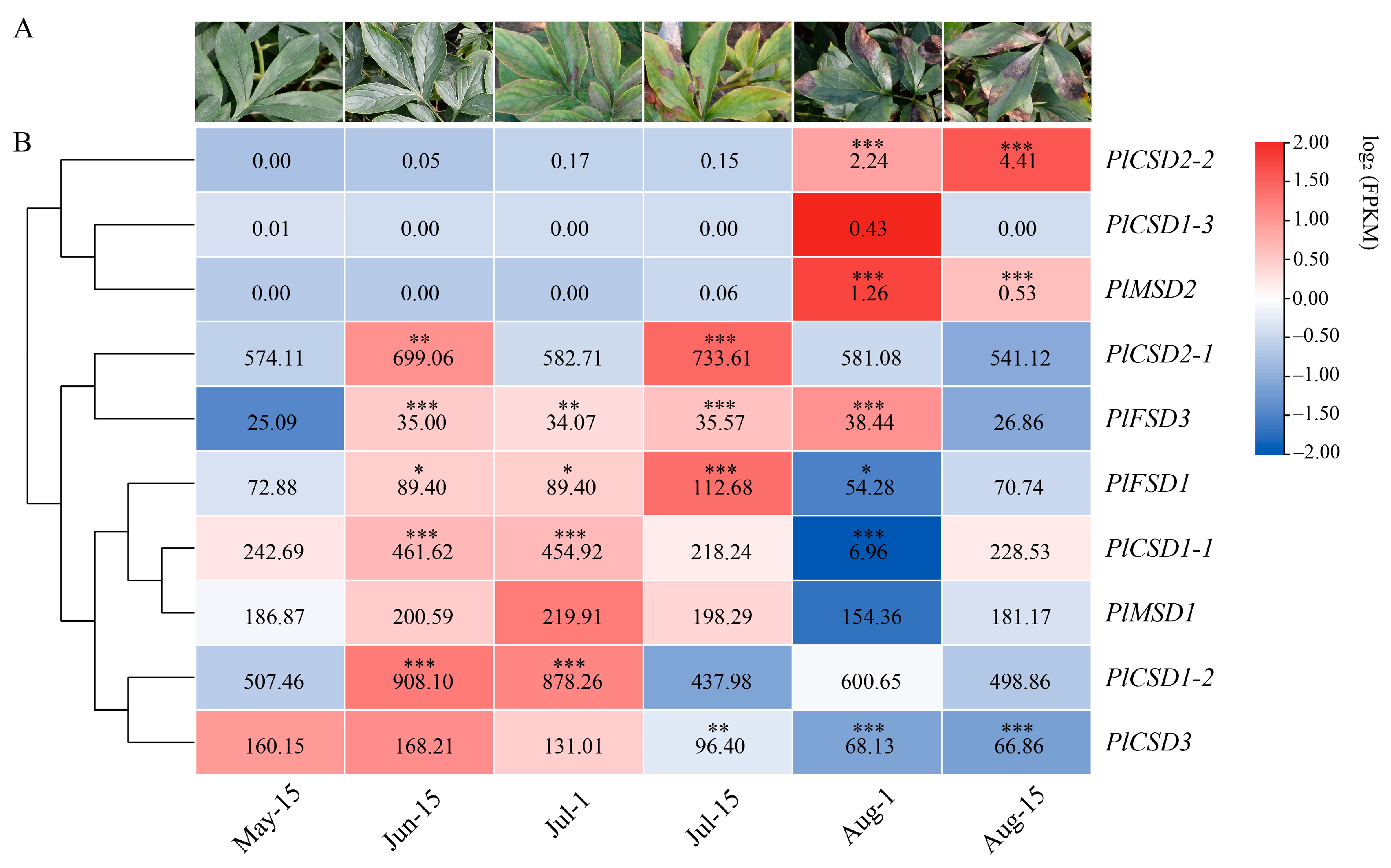
3.5. Expression Analysis of PlSODs under Natural High-Temperature Stress
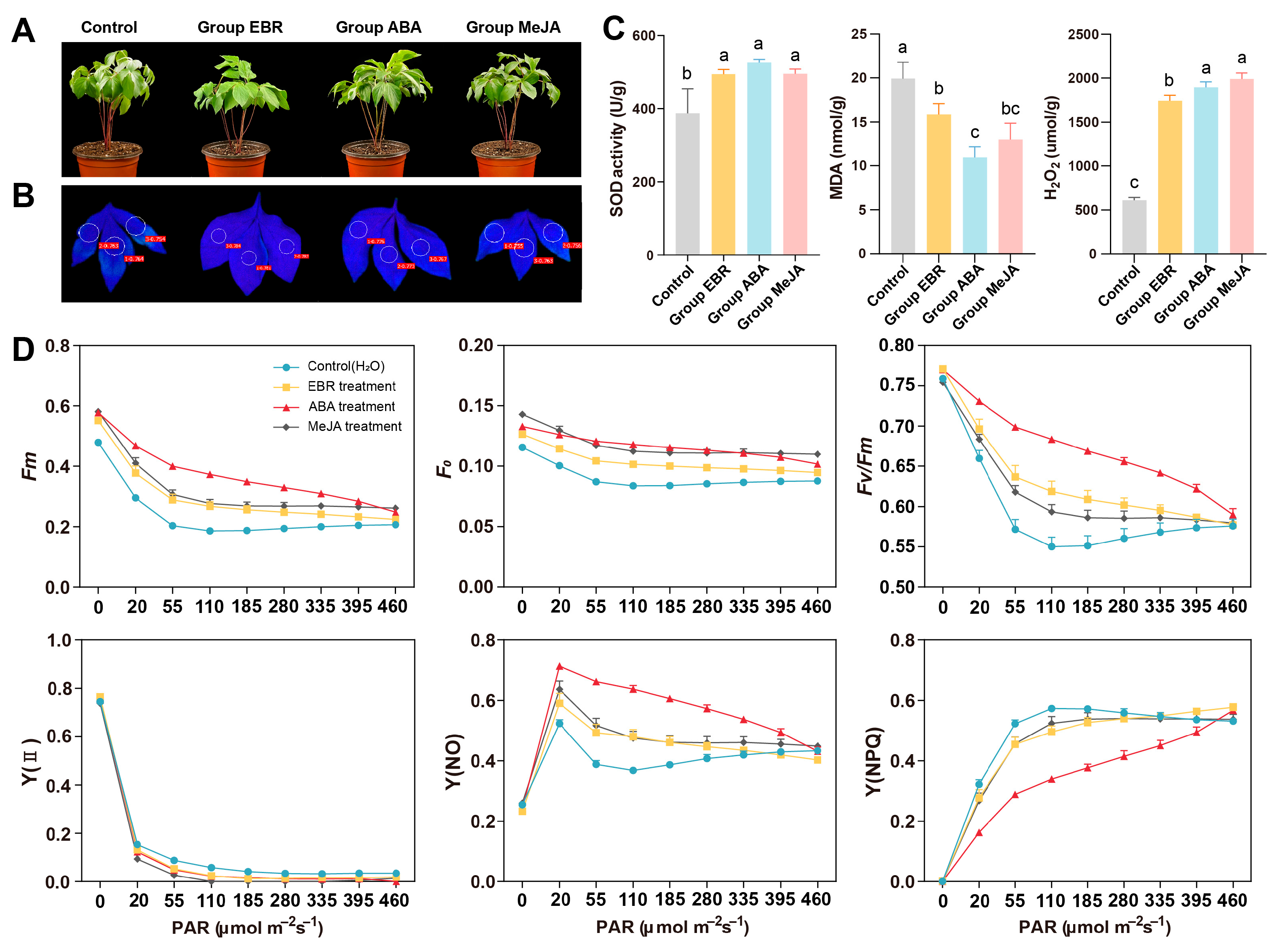
3.6. Phenotypic and Physiological Responses to High-Temperature Stress
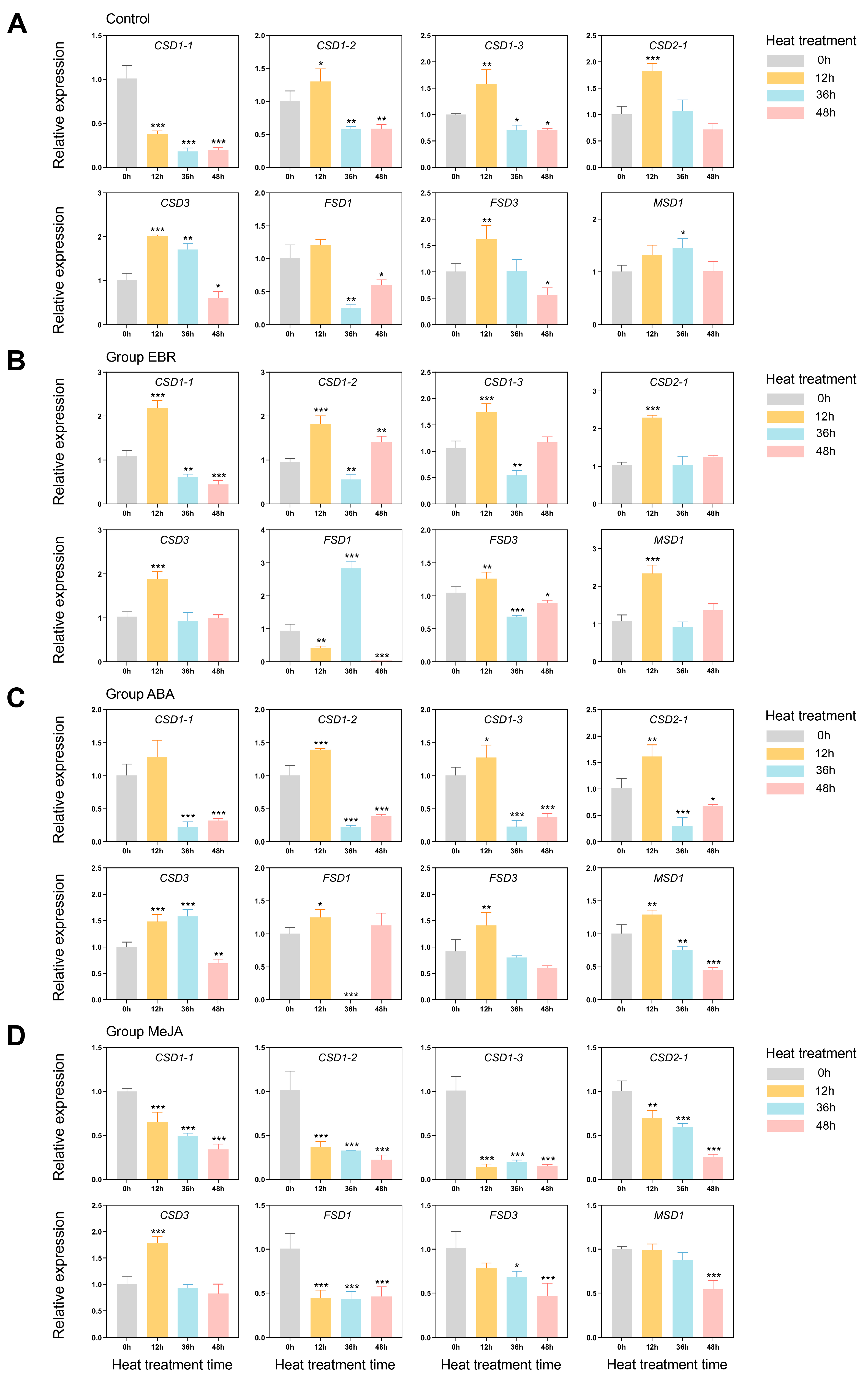
3.7. Exogenous Hormones and High-Temperature Stress-Induced Expression Profiles of PlSOD Family Genes
4. Discussion
4.1. Characteristics and Evolutionary Analysis of PlSODs in P. lactiflora
4.2. PlSODs Are Widely Involved in High-Temperature Stress Response
4.3. EBR and ABA Treatments Can Enhance the Tolerance of P. lactiflora under High-Temperature Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gupta, S.; Dong, Y.; Dijkwel, P.P.; Mueller-Roeber, B.; Gechev, T.S. Genome-Wide Analysis of ROS Antioxidant Genes in Resurrection Species Suggest an Involvement of Distinct ROS Detoxification Systems during Desiccation. Int. J. Mol. Sci. 2019, 20, 3101. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Oxidative Stress, Antioxidants and Stress Tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, K.K. ROS Homeostasis in Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Parvin, K.; Bhuiyan, T.F.; Anee, T.I.; Nahar, K.; Hossen, M.S.; Zulfiqar, F.; Alam, M.M.; Fujita, M. Regulation of ROS Metabolism in Plants under Environmental Stress: A Review of Recent Experimental Evidence. Int. J. Mol. Sci. 2020, 21, 8695. [Google Scholar] [CrossRef]
- Qi, S.; Wang, X.; Wu, Q.; Xing, S.; Li, X.; Zhao, F.; Yu, Y.; Qi, S.; Wang, X.; Wu, Q.; et al. Morphological, Physiological and Transcriptomic Analyses Reveal Potential Candidate Genes Responsible for Salt Stress in Rosa rugosa. Ornam. Plant Res. 2023, 3, 21. [Google Scholar] [CrossRef]
- Rajput, V.D.; Harish; Singh, R.K.; Verma, K.K.; Sharma, L.; Quiroz-Figueroa, F.R.; Meena, M.; Gour, V.S.; Minkina, T.; Sushkova, S.; et al. Recent Developments in Enzymatic Antioxidant Defence Mechanism in Plants with Special Reference to Abiotic Stress. Biology 2021, 10, 267. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive Oxygen Species and Antioxidant Machinery in Abiotic Stress Tolerance in Crop Plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Ali, S.; Tyagi, A.; Bae, H. ROS Interplay between Plant Growth and Stress Biology: Challenges and Future Perspectives. Plant Physiol. Biochem. 2023, 203, 108032. [Google Scholar] [CrossRef]
- Fink, R.C.; Scandalios, J.G. Molecular Evolution and Structure–Function Relationships of the Superoxide Dismutase Gene Families in Angiosperms and Their Relationship to Other Eukaryotic and Prokaryotic Superoxide Dismutases. Arch. Biochem. Biophys. 2002, 399, 19–36. [Google Scholar] [CrossRef]
- Schmidt, A.; Gube, M.; Schmidt, A.; Kothe, E. In Silico Analysis of Nickel Containing Superoxide Dismutase Evolution and Regulation. J. Basic Microbiol. 2009, 49, 109–118. [Google Scholar] [CrossRef]
- Dupont, C.L.; Neupane, K.; Shearer, J.; Palenik, B. Diversity, Function and Evolution of Genes Coding for Putative Ni-containing Superoxide Dismutases. Environ. Microbiol. 2008, 10, 1831–1843. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, Q.; Zhong, S.; Yang, H.; Ren, T.; Chen, C.; Tan, F.; Cao, G.; Liu, J.; Luo, P. Genome-Wide Identification of Superoxide Dismutase and Expression in Response to Fruit Development and Biological Stress in Akebia Trifoliata: A Bioinformatics Study. Antioxidants 2023, 12, 726. [Google Scholar] [CrossRef] [PubMed]
- Stephenie, S.; Chang, Y.P.; Gnanasekaran, A.; Esa, N.M.; Gnanaraj, C. An Insight on Superoxide Dismutase (SOD) from Plants for Mammalian Health Enhancement. J. Funct. Foods 2020, 68, 103917. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Chen, Y.; Wang, S.; Fang, Y.; Zhang, X.; Wu, Y.; Xue, D. Genome-wide Identification of the SOD Gene Family and Expression Analysis under Drought and Salt Stress in Barley. Plant Growth Regul. 2021, 94, 49–60. [Google Scholar] [CrossRef]
- Kliebenstein, D.J.; Monde, R.-A.; Last, R.L. Superoxide Dismutase in Arabidopsis: An Eclectic Enzyme Family with Disparate Regulation and Protein Localization. Plant Physiol. 1998, 118, 637–650. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, X.; Deng, F.; Yuan, R.; Shen, F. Genome-Wide Characterization and Expression Analyses of Superoxide Dismutase (SOD) Genes in Gossypium Hirsutum. BMC Genom. 2017, 18, 376. [Google Scholar] [CrossRef]
- Feng, K.; Yu, J.; Cheng, Y.; Ruan, M.; Wang, R.; Ye, Q.; Zhou, G.; Li, Z.; Yao, Z.; Yang, Y.; et al. The SOD Gene Family in Tomato: Identification, Phylogenetic Relationships, and Expression Patterns. Front. Plant Sci. 2016, 7, 1279. [Google Scholar] [CrossRef]
- Huo, C.; He, L.; Yu, T.; Ji, X.; Li, R.; Zhu, S.; Zhang, F.; Xie, H.; Liu, W. The Superoxide Dismutase Gene Family in Nicotiana Tabacum: Genome-Wide Identification, Characterization, Expression Profiling and Functional Analysis in Response to Heavy Metal Stress. Front. Plant Sci. 2022, 13, 904105. [Google Scholar] [CrossRef]
- Lu, W.; Duanmu, H.; Qiao, Y.; Jin, X.; Yu, Y.; Yu, L.; Chen, C. Genome-Wide Identification and Characterization of the Soybean SOD Family during Alkaline Stress. PeerJ 2020, 8, e8457. [Google Scholar] [CrossRef]
- Feng, X.; Lai, Z.; Lin, Y.; Lai, G.; Lian, C. Genome-Wide Identification and Characterization of the Superoxide Dismutase Gene Family in Musa Acuminata Cv. Tianbaojiao (AAA Group). BMC Genom. 2015, 16, 823. [Google Scholar] [CrossRef]
- Su, W.; Raza, A.; Gao, A.; Jia, Z.; Zhang, Y.; Hussain, M.A.; Mehmood, S.S.; Cheng, Y.; Lv, Y.; Zou, X. Genome-Wide Analysis and Expression Profile of Superoxide Dismutase (SOD) Gene Family in Rapeseed (Brassica napus L.) under Different Hormones and Abiotic Stress Conditions. Antioxidants 2021, 10, 1182. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Wu, L.J.; Yu, Z.L. Effect of Salt and Drought Stress on Antioxidant Enzymes Activities and SOD Isoenzymes of Liquorice (Glycyrrhiza Uralensis Fisch). Plant Growth Regul. 2006, 49, 157–165. [Google Scholar] [CrossRef]
- Samota, M.K.; Awana, M.; Krishnan, V.; Kumar, S.; Tyagi, A.; Pandey, R.; Mithra, S.V.A.; Singh, A. A Novel Micronutrients and Methyl Jasmonate Cocktail of Elicitors via Seed Priming Improves Drought Tolerance by Mitigating Oxidative Stress in Rice (Oryza Sativa L.). Protoplasma 2023, 261, 553–570. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.S.; Bang, S.G.; Ahn, M.-A.; Kim, G.; Kim, E.; Eom, S.H.; Hyun, T.K. Molecular Cloning and Functional Characterization of Heat Stress-Responsive Superoxide Dismutases in Garlic (Allium Sativum L.). Antioxidants 2021, 10, 815. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Liao, X.; He, M.; Li, X.; Wang, Z.; Ma, H.; Yu, S.; Liu, S. Tolerance Analysis of Chloroplast OsCu/Zn-SOD Overexpressing Rice under NaCl and NaHCO3 Stress. PLoS ONE 2017, 12, e0186052. [Google Scholar] [CrossRef]
- Shiraya, T.; Mori, T.; Maruyama, T.; Sasaki, M.; Takamatsu, T.; Oikawa, K.; Itoh, K.; Kaneko, K.; Ichikawa, H.; Mitsui, T. Golgi/Plastid-type Manganese Superoxide Dismutase Involved in Heat-stress Tolerance during Grain Filling of Rice. Plant Biotechnol. J. 2015, 13, 1251–1263. [Google Scholar] [CrossRef]
- Yan, H.; Li, Q.; Park, S.-C.; Wang, X.; Liu, Y.; Zhang, Y.; Tang, W.; Kou, M.; Ma, D. Overexpression of CuZnSOD and APX Enhance Salt Stress Tolerance in Sweet Potato. Plant Physiol. Biochem. 2016, 109, 20–27. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J. Plant Hormone Regulation of Abiotic Stress Responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Devireddy, A.R.; Zandalinas, S.; Fichman, Y.; Mittler, R. Integration of Reactive Oxygen Species and Hormone Signaling during Abiotic Stress. Plant J. 2021, 105, 459–476. [Google Scholar] [CrossRef]
- Nie, W.-F.; Wang, M.-M.; Xia, X.-J.; Zhou, Y.-H.; Shi, K.; Chen, Z.; Yu, J.Q. Silencing of Tomato RBOH1 and MPK2 Abolishes Brassinosteroid-Induced H₂O₂ Generation and Stress Tolerance. Plant Cell Environ. 2013, 36, 789–803. [Google Scholar] [CrossRef]
- Larkindale, J.; Knight, M.R. Protection against Heat Stress-Induced Oxidative Damage in Arabidopsis Involves Calcium, Abscisic Acid, Ethylene, and Salicylic Acid. Plant Physiol. 2002, 128, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Huang, Y.; Dong, X.; Wang, R.; Tang, M.; Cai, J.; Chen, J.; Zhang, X.; Nie, G. Exogenous Methyl Jasmonate Improves Heat Tolerance of Perennial Ryegrass Through Alteration of Osmotic Adjustment, Antioxidant Defense, and Expression of Jasmonic Acid-Responsive Genes. Front. Plant Sci. 2021, 12, 664519. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shi, X.; Zhang, R.; Zhang, K.; Shao, L.; Xu, T.; Li, D.; Zhang, D.; Zhang, J.; Xia, Y. Impact of Summer Heat Stress Inducing Physiological and Biochemical Responses in Herbaceous Peony Cultivars (Paeonia Lactiflora Pall.) from Different Latitudes. Ind. Crops Prod. 2022, 184, 115000. [Google Scholar] [CrossRef]
- Xin, Z.; Yang, W.; Duan, Y.; Wang, W.; Niu, L.; Sun, D.; Zhang, Y. Bioactive Components Antibacterial Activities of Hydrolate Extracts by Optimization Conditions from Paeonia Ostii, T. Hong & J. X. Zhang. Ind. Crops Prod. 2022, 188, 115737. [Google Scholar] [CrossRef]
- Zhao, D.; Xia, X.; Su, J.; Wei, M.; Wu, Y.; Tao, J. Overexpression of Herbaceous Peony HSP70 Confers High Temperature Tolerance. BMC Genom. 2019, 20, 70. [Google Scholar] [CrossRef]
- Zhang, T.; Tang, Y.; Luan, Y.; Cheng, Z.; Wang, X.; Tao, J.; Zhao, D. Herbaceous Peony AP2/ERF Transcription Factor Binds the Promoter of the Tryptophan Decarboxylase Gene to Enhance High-temperature Stress Tolerance. Plant Cell Environ. 2022, 45, 2729–2743. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, R.; Zhang, K.; Shao, L.; Xu, T.; Shi, X.; Li, D.; Zhang, J.; Xia, Y. Development of a Multi-Criteria Decision-Making Approach for Evaluating the Comprehensive Application of Herbaceous Peony at Low Latitudes. Int. J. Mol. Sci. 2022, 23, 14342. [Google Scholar] [CrossRef]
- Wu, Y.-Q.; Zhao, D.-Q.; Han, C.-X.; Tao, J. Biochemical and Molecular Responses of Herbaceous Peony to High Temperature Stress. Can. J. Plant Sci. 2016, 96, 474–484. [Google Scholar] [CrossRef]
- Zhao, D.; Han, C.; Zhou, C.; Tao, J. Shade Ameliorates High Temperature-Induced Inhibition of Growth in Herbaceous Peony (Paeonia Lactiflora). Int. J. Agric. Biol. 2015, 17, 911–919. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as Designed by Its Users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Klee, E.W.; Ellis, L.B.M. Evaluating Eukaryotic Secreted Protein Prediction. BMC Bioinform. 2005, 6, 256. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc: A Package of Web Servers for Predicting Subcellular Localization of Proteins in Various Organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Aiyar, A. The Use of CLUSTAL W and CLUSTAL X for Multiple Sequence Alignment. Methods Mol. Biol. 2000, 132, 221–241. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Cheng, P.; Bi, D.; Chen, J.; Zhao, M.; Wang, Y.; Wang, H.; Cao, P.; Huang, C. Genome-Wide Identification and Analysis of TCP Transcription Factor Genes in Rosa Chinensis in Response to Abiotic Stress and Fungal Diseases. Ornam. Plant Res. 2023, 3, 3. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, X.; Chen, X.; Zhang, R.; Guo, J.; Wang, Q.; Li, D.; Shao, L.; Shi, X.; Han, J.; et al. Establishment of a Homologous Silencing System with Intact-Plant Infiltration and Minimized Operation for Studying Gene Function in Herbaceous Peonies. Int. J. Mol. Sci. 2024, 25, 4412. [Google Scholar] [CrossRef]
- Shao, L.; Xu, T.; Wang, X.; Zhang, R.; Wang, X.; Ren, Z.; Zhang, J.; Xia, Y.; Li, D. Integrative Comparative Assessment of Cold Acclimation in Evergreen and Deciduous Iris Species. Antioxidants 2022, 11, 977. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, R.; Huang, Q.; Shi, X.; Li, D.; Shao, L.; Xu, T.; Horvath, D.P.; Xia, Y.; Zhang, J. Comparative Study on Physiological Responses and Gene Expression of Bud Endodormancy Release Between Two Herbaceous Peony Cultivars (Paeonia Lactiflora Pall.) with Contrasting Chilling Requirements. Front. Plant Sci. 2022, 12, 772285. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 4, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Guruprasad, K.; Reddy, B.V.; Pandit, M.W. Correlation between Stability of a Protein and Its Dipeptide Composition: A Novel Approach for Predicting in Vivo Stability of a Protein from Its Primary Sequence. Protein Eng. 1990, 4, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Fernández-Ocaña, A.; Carreras, A.; Valderrama, R.; Luque, F.; Esteban, F.J.; Rodríguez-Serrano, M.; Chaki, M.; Pedrajas, J.R.; Sandalio, L.M.; et al. The Expression of Different Superoxide Dismutase Forms Is Cell-Type Dependent in Olive (Olea Europaea L.) Leaves. Plant Cell Physiol. 2006, 47, 984–994. [Google Scholar] [CrossRef] [PubMed]
- Altenhoff, A.M.; Dessimoz, C. Phylogenetic and Functional Assessment of Orthologs Inference Projects and Methods. PLoS Comput. Biol. 2009, 5, e1000262. [Google Scholar] [CrossRef] [PubMed]
- Ganko, E.W.; Meyers, B.C.; Vision, T.J. Divergence in Expression between Duplicated Genes in Arabidopsis. Mol. Biol. Evol. 2007, 24, 2298–2309. [Google Scholar] [CrossRef]
- Guo, Y.; Xue, J.; Song, G.; Su, X.; Yang, K.; Zhou, L. Diurnal variation characteristics of photosynthesis and chlorophyll fluorescence of six grape varieties in greenhouse in Hotan area. Acta Bot. Boreali-Occident. Sin. 2024, 44, 1208–1217. [Google Scholar]
- Zhu, L.; Scafaro, A.P.; Vierling, E.; Ball, M.C.; Posch, B.C.; Stock, F.; Atkin, O.K. Heat Tolerance of a Tropical–Subtropical Rainforest Tree Species Polyscias Elegans: Time-dependent Dynamic Responses of Physiological Thermostability and Biochemistry. New Phytol. 2023, 241, 715–731. [Google Scholar] [CrossRef]
- Khan, W.U.; Khan, L.U.; Chen, D.; Chen, F. Comparative Analyses of Superoxide Dismutase (SOD) Gene Family and Expression Profiling under Multiple Abiotic Stresses in Water Lilies. Horticulturae 2023, 9, 781. [Google Scholar] [CrossRef]
- Hu, X.; Hao, C.; Cheng, Z.-M.; Zhong, Y. Genome-Wide Identification, Characterization, and Expression Analysis of the Grapevine Superoxide Dismutase (SOD) Family. Int. J. Genom. 2019, 2019, 7350414. [Google Scholar] [CrossRef]
- Jiang, W.; Yang, L.; He, Y.; Zhang, H.; Li, W.; Chen, H.; Ma, D.; Yin, J. Genome-Wide Identification and Transcriptional Expression Analysis of Superoxide Dismutase (SOD) Family in Wheat (Triticum Aestivum). PeerJ 2019, 7, e8062. [Google Scholar] [CrossRef] [PubMed]
- Zelko, I.N.; Mariani, T.J.; Folz, R.J. Superoxide Dismutase Multigene Family: A Comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) Gene Structures, Evolution, and Expression. Free Radic. Biol. Med. 2002, 33, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.; Jiang, C.; Chen, L.; Paul, A.; Chatterjee, A.; Shen, G. Achieving Abiotic Stress Tolerance in Plants through Antioxidative Defense Mechanisms. Front. Plant Sci. 2023, 14, 1110622. [Google Scholar] [CrossRef] [PubMed]
- Han, L.-M.; Hua, W.-P.; Cao, X.-Y.; Yan, J.-A.; Chen, C.; Wang, Z.-Z. Genome-Wide Identification and Expression Analysis of the Superoxide Dismutase (SOD) Gene Family in Salvia Miltiorrhiza. Gene 2020, 742, 144603. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ouyang, L.; Zhou, D.; Cai, Y.; He, H. Superoxide Dismutase Family Genes in Watermelon and Their Responses to Different Abiotic Stresses. Front. Agric. Sci. Eng. 2021, 8, 645. [Google Scholar] [CrossRef]
- Yu, S.; Wang, C.; Wang, Q.; Sun, Q.; Zhang, Y.; Dong, J.; Yin, Y.; Zhang, S.; Yu, G. Identification and Analysis of SOD Family Genes in Peanut (Arachis Hypogaea L.) and Their Potential Roles in Stress Responses. Agronomy 2023, 13, 1959. [Google Scholar] [CrossRef]
- Yu, W.; Kong, G.; Chao, J.; Yin, T.; Tian, H.; Ya, H.; He, L.; Zhang, H. Genome-Wide Identification of the Rubber Tree Superoxide Dismutase (SOD) Gene Family and Analysis of Its Expression under Abiotic Stress. PeerJ 2022, 10, e14251. [Google Scholar] [CrossRef]
- Rehman, S.; Rashid, A.; Manzoor, M.A.; Li, L.; Sun, W.; Riaz, M.W.; Li, D.; Zhuge, Q. Genome-Wide Evolution and Comparative Analysis of Superoxide Dismutase Gene Family in Cucurbitaceae and Expression Analysis of Lagenaria Siceraria Under Multiple Abiotic Stresses. Front. Genet. 2022, 12, 784878. [Google Scholar] [CrossRef]
- Elmongy, M.S.; Abd El-Baset, M.M. Melatonin Application Induced Physiological and Molecular Changes in Carnation (Dianthus Caryophyllus L.) under Heat Stress. Horticulturae 2024, 10, 122. [Google Scholar] [CrossRef]
- Alam, M.N.; Zhang, L.; Yang, L.; Islam, M.R.; Liu, Y.; Luo, H.; Yang, P.; Wang, Q.; Chan, Z. Transcriptomic Profiling of Tall Fescue in Response to Heat Stress and Improved Thermotolerance by Melatonin and 24-Epibrassinolide. BMC Genom. 2018, 19, 224. [Google Scholar] [CrossRef]
- Zhou, J.; Jiang, X.; Agathokleous, E.; Lu, X.; Yang, Z.; Li, R. High Temperature Inhibits Photosynthesis of Chrysanthemum (Chrysanthemum Morifolium Ramat.) Seedlings More than Relative Humidity. Front. Plant Sci. 2023, 14, 1272013. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhou, Y.; Yang, X.; Zhang, B.; Xu, F.; Wang, Y.; Song, C.; Yi, M.; Ma, N.; Zhou, X.; et al. The Heat Stress Transcription Factor LlHsfA4 Enhanced Basic Thermotolerance through Regulating ROS Metabolism in Lilies (Lilium Longiflorum). Int. J. Mol. Sci. 2022, 23, 572. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cai, J.; Liu, F.; Dai, T.; Cao, W.; Wollenweber, B.; Jiang, D. Multiple Heat Priming Enhances Thermo-Tolerance to a Later High Temperature Stress via Improving Subcellular Antioxidant Activities in Wheat Seedlings. Plant Physiol. Biochem. 2014, 74, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Euring, D.; Cha, J.Y.; Lin, Z.; Lu, M.; Huang, L.-J.; Kim, W.Y. Plant Hormone-Mediated Regulation of Heat Tolerance in Response to Global Climate Change. Front. Plant Sci. 2021, 11, 627969. [Google Scholar] [CrossRef]
- Suzuki, N.; Bassil, E.; Hamilton, J.S.; Inupakutika, M.A.; Zandalinas, S.I.; Tripathy, D.; Luo, Y.; Dion, E.; Fukui, G.; Kumazaki, A.; et al. ABA Is Required for Plant Acclimation to a Combination of Salt and Heat Stress. PLoS ONE 2016, 11, e0147625. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z. Molecular Basis for Jasmonate and Ethylene Signal Interactions in Arabidopsis. J. Exp. Bot. 2014, 65, 5743–5748. [Google Scholar] [CrossRef]
- Su, Q.; Zheng, X.; Tian, Y.; Wang, C. Exogenous Brassinolide Alleviates Salt Stress in Malus Hupehensis Rehd. by Regulating the Transcription of NHX-Type Na+(K+)/H+ Antiporters. Front. Plant Sci. 2020, 11, 38. [Google Scholar] [CrossRef]
- Goraya, G.K.; Kaur, B.; Asthir, B.; Bala, S.; Kaur, G.; Farooq, M. Rapid Injuries of High Temperature in Plants. J. Plant Biol. 2017, 60, 298–305. [Google Scholar] [CrossRef]
- Lippmann, R.; Babben, S.; Menger, A.; Delker, C.; Quint, M. Development of Wild and Cultivated Plants under Global Warming Conditions. Curr. Biol. 2019, 29, R1326–R1338. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, H.; Lin, W.; Jahan, M.S.; Wang, J.; Sun, J.; Jiang, J.; Gu, W.; Zou, J.; Shu, S.; et al. Foliar Application of a Mixture of Putrescine, Melatonin, Proline, and Potassium Fulvic Acid Alleviates High Temperature Stress of Cucumber Plants Grown in the Greenhouse. Technol. Hortic. 2022, 2, 6. [Google Scholar] [CrossRef]
- Mitsui, T.; Yamakawa, H.; Kobata, T. Molecular Physiological Aspects of Chalking Mechanism in Rice Grains under High-Temperature Stress. Plant Prod. Sci. 2016, 19, 22–29. [Google Scholar] [CrossRef]
- Wang, X.; Xu, C.; Cai, X.; Wang, Q.; Dai, S. Heat-Responsive Photosynthetic and Signaling Pathways in Plants: Insight from Proteomics. Int. J. Mol. Sci. 2017, 18, 2191. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Ding, Y.; Zhu, C. Sensitivity and Responses of Chloroplasts to Heat Stress in Plants. Front. Plant Sci. 2020, 11, 375. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.K.; Andersen, S.B.; Ottosen, C.-O.; Rosenqvist, E. Wheat Cultivars Selected for High Fv/Fm under Heat Stress Maintain High Photosynthesis, Total Chlorophyll, Stomatal Conductance, Transpiration and Dry Matter. Physiol. Plant. 2015, 153, 284–298. [Google Scholar] [CrossRef]
- Murata, N.; Takahashi, S.; Nishiyama, Y.; Allakhverdiev, S.I. Photoinhibition of Photosystem II under Environmental Stress. Biochim. Biophys. Acta BBA Bioenerg. 2007, 1767, 414–421. [Google Scholar] [CrossRef]


| Distilled Water Treatment | 1 µM EBR Treatment | 100 µM ABA Treatment | 100 µM MeJA Treatment | |
|---|---|---|---|---|
| High-temperature treatment (42 °C) | Control | Group EBR | Group ABA | Group MeJA |
| Gene Name | Gene ID | Amino Acid | Molecular Weight | PI | Instability Index | GRAVY | Subcellular Location | Pfam Domain |
|---|---|---|---|---|---|---|---|---|
| PlCSD1-1 | Peony_Unigene_344154 | 194 | 20,123.57 | 7.38 | 28.85 | −0.28 | Chl | CZ |
| PlCSD1-2 | Peony_Unigene_468541 | 194 | 20,070.52 | 6.94 | 26.58 | −0.244 | Chl | CZ |
| PlCSD1-3 | Peony_Unigene_151283 | 145 | 15,318.57 | 6.95 | 10.21 | −0.028 | Chl, Cyt. | CZ |
| PlCSD2-1 | Peony_Unigene_483257 | 221 | 22,713.7 | 6.1 | 20.01 | 0.011 | Chl | CZ |
| PlCSD2-2 | Peony_Unigene_424839 | 189 | 19,737.87 | 6.9 | 28.47 | −0.514 | Chl, Cyt. | CZ |
| PlCSD3 | Peony_Unigene_299453 | 163 | 16,766.82 | 6.74 | 18.39 | −0.209 | Chl, Cyt. | CZ |
| PlFSD1 | Peony_Unigene_433554 | 236 | 26,301.77 | 6.71 | 37.04 | −0.43 | Chl | IMA, IMC |
| PlFSD3 | Peony_Unigene_453151 | 261 | 29,972.54 | 8.17 | 41.09 | −0.35 | Chl | IMA, IMC |
| PlMSD1 | Peony_Unigene_259976 | 237 | 26,472.27 | 8.98 | 48.54 | −0.346 | Mit | IMA, IMC |
| PlMSD2 | Peony_Unigene_061444 | 248 | 27,771.29 | 6.04 | 26.19 | −0.344 | Mit | IMA, IMC |
| Gene | Alpha Helix (%) | Beta Turn (%) | Extended Strand (%) | Random Coil (%) |
|---|---|---|---|---|
| PlCSD1-1 | 10.31 | 5.15 | 30.41 | 54.12 |
| PlCSD1-2 | 13.40 | 6.70 | 28.87 | 51.03 |
| PlCSD1-3 | 8.11 | 7.43 | 34.46 | 50.00 |
| PlCSD2-1 | 19.46 | 8.14 | 26.24 | 46.15 |
| PlCSD2-2 | 12.17 | 6.35 | 32.28 | 49.21 |
| PlCSD3 | 9.20 | 5.52 | 30.67 | 54.60 |
| PlFSD1 | 32.63 | 3.81 | 16.53 | 47.03 |
| PlFSD3 | 37.55 | 3.07 | 15.33 | 44.06 |
| PlMSD1 | 52.74 | 4.22 | 12.66 | 30.38 |
| PlMSD2 | 53.63 | 3.63 | 14.11 | 28.63 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Li, D.; Guo, J.; Wang, Q.; Zhang, K.; Wang, X.; Shao, L.; Luo, C.; Xia, Y.; Zhang, J. Identification and Analysis of the Superoxide Dismutase (SOD) Gene Family and Potential Roles in High-Temperature Stress Response of Herbaceous Peony (Paeonia lactiflora Pall.). Antioxidants 2024, 13, 1128. https://doi.org/10.3390/antiox13091128
Chen X, Li D, Guo J, Wang Q, Zhang K, Wang X, Shao L, Luo C, Xia Y, Zhang J. Identification and Analysis of the Superoxide Dismutase (SOD) Gene Family and Potential Roles in High-Temperature Stress Response of Herbaceous Peony (Paeonia lactiflora Pall.). Antioxidants. 2024; 13(9):1128. https://doi.org/10.3390/antiox13091128
Chicago/Turabian StyleChen, Xiaoxuan, Danqing Li, Junhong Guo, Qiyao Wang, Kaijing Zhang, Xiaobin Wang, Lingmei Shao, Cheng Luo, Yiping Xia, and Jiaping Zhang. 2024. "Identification and Analysis of the Superoxide Dismutase (SOD) Gene Family and Potential Roles in High-Temperature Stress Response of Herbaceous Peony (Paeonia lactiflora Pall.)" Antioxidants 13, no. 9: 1128. https://doi.org/10.3390/antiox13091128
APA StyleChen, X., Li, D., Guo, J., Wang, Q., Zhang, K., Wang, X., Shao, L., Luo, C., Xia, Y., & Zhang, J. (2024). Identification and Analysis of the Superoxide Dismutase (SOD) Gene Family and Potential Roles in High-Temperature Stress Response of Herbaceous Peony (Paeonia lactiflora Pall.). Antioxidants, 13(9), 1128. https://doi.org/10.3390/antiox13091128








