Transcriptome Analysis Reveals the Molecular Mechanism of Potentilla anserina L. Polysaccharides in Mitigating Zearalenone-Induced Oxidative Stress in Porcine Sertoli Cells
Abstract
1. Introduction
2. Results
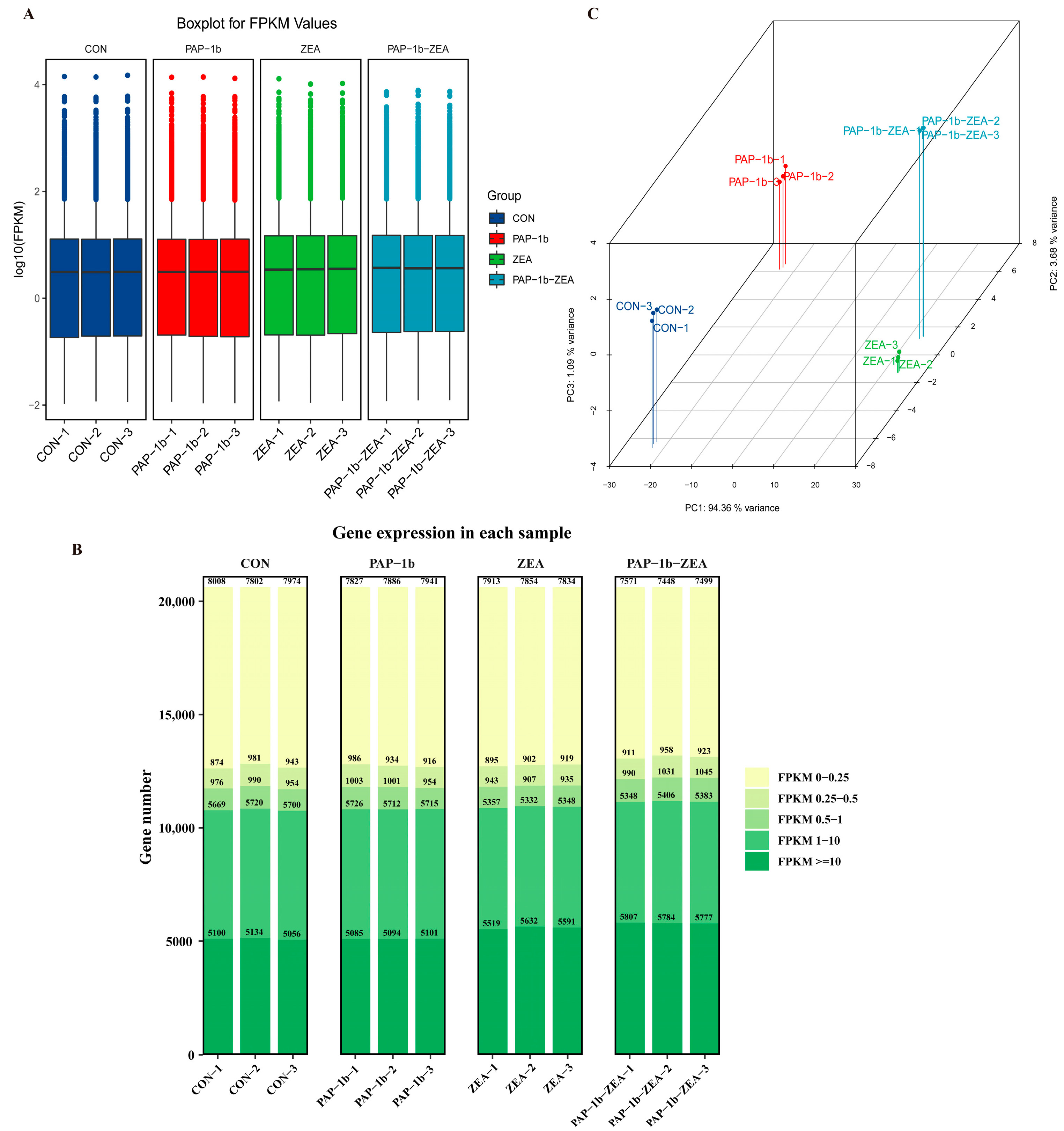
2.1. Data Filtering and Quality Assessment
2.2. Analysis of Overall Gene Expression Levels
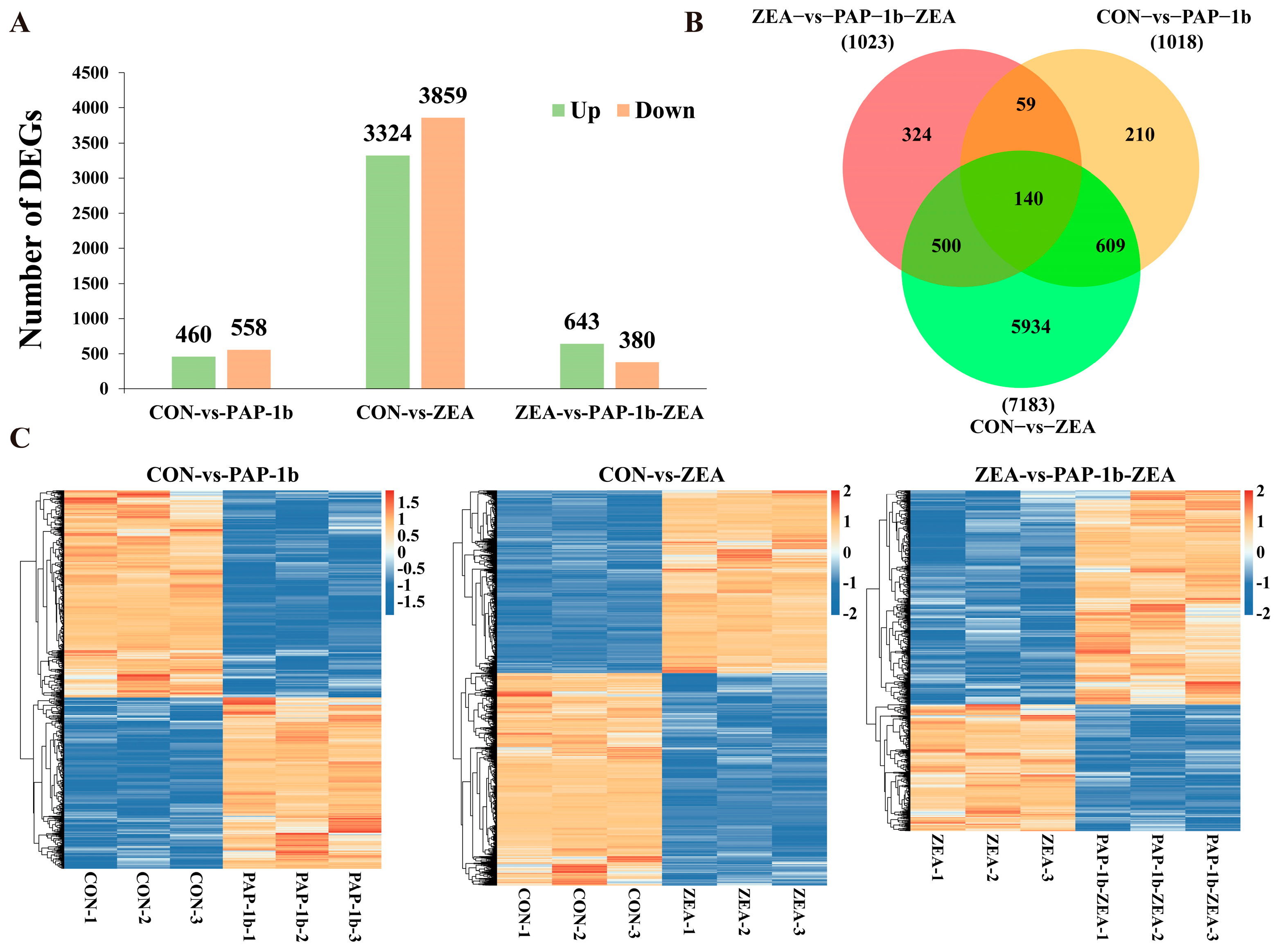
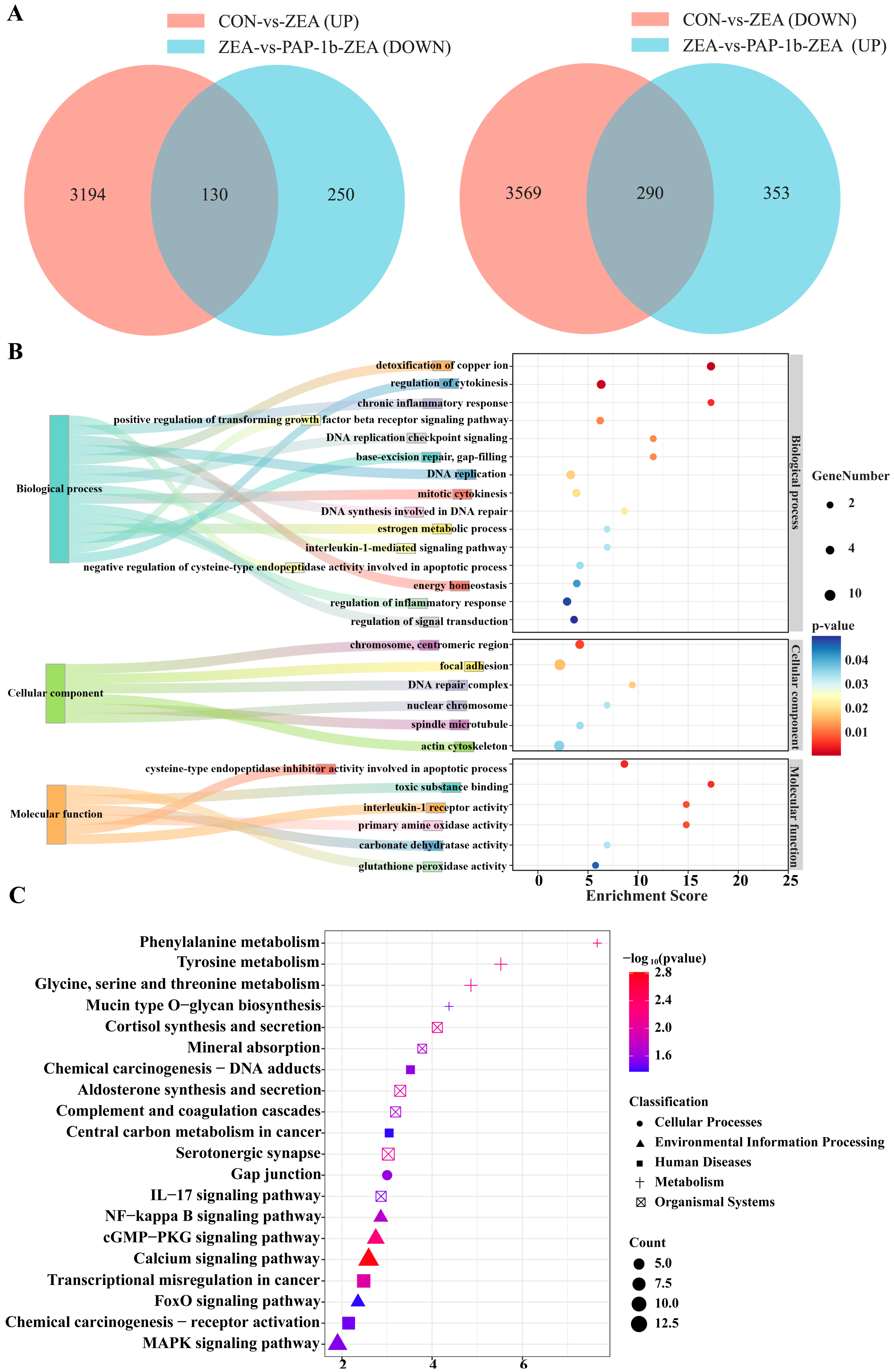
2.3. Identification of DEGs
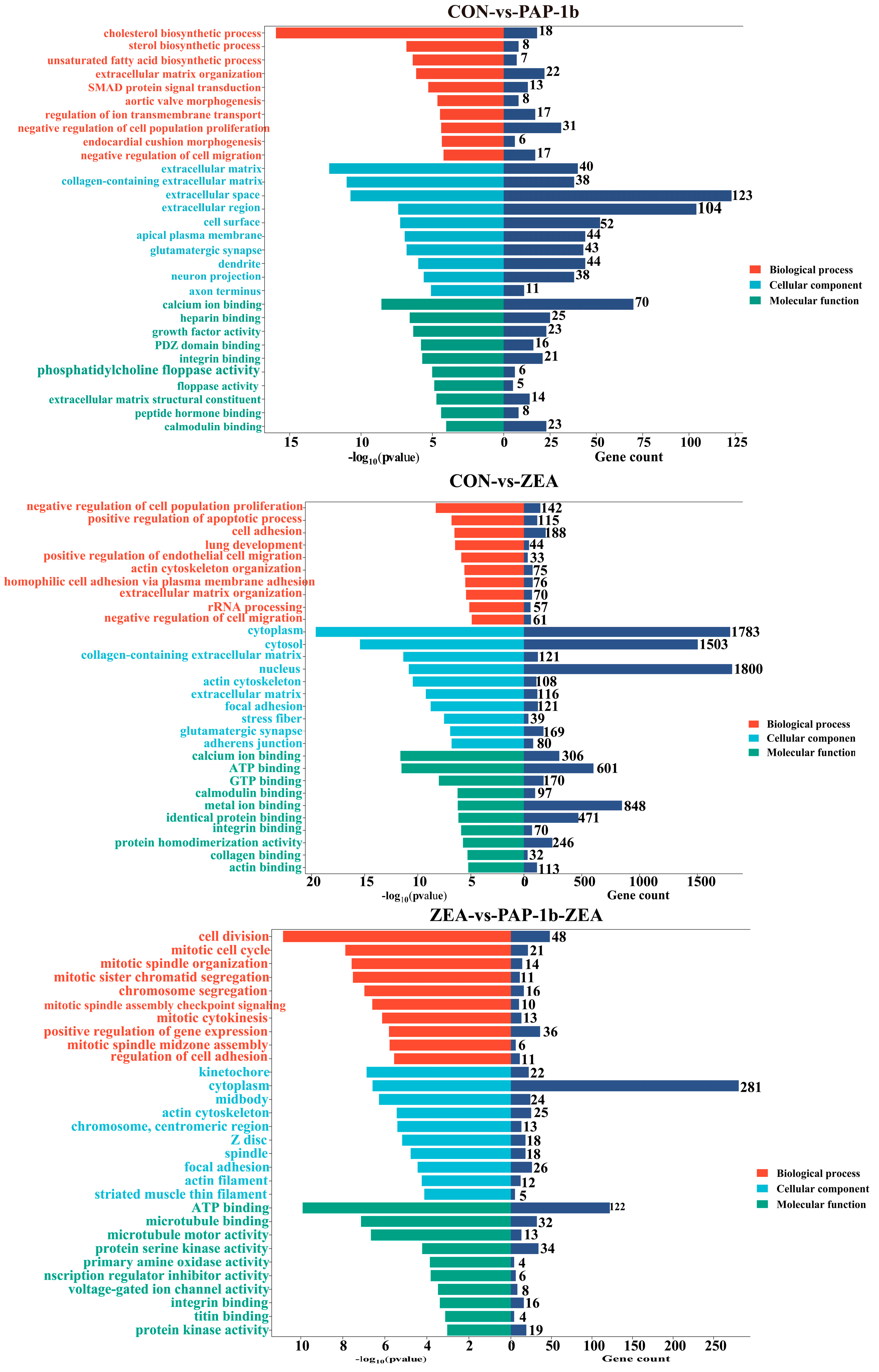
2.4. GO Function Annotation
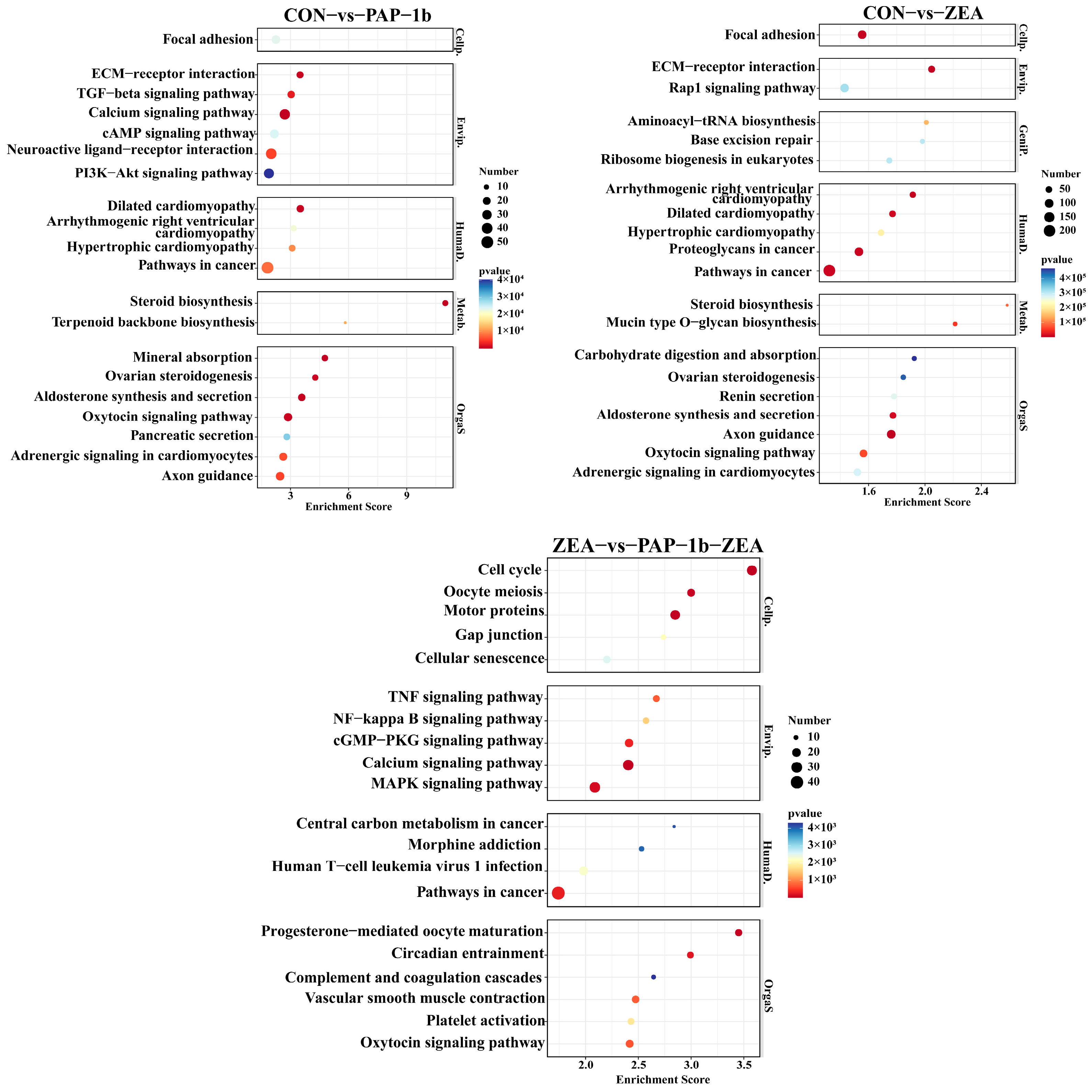
2.5. KEGG Pathway Enrichment Analysis
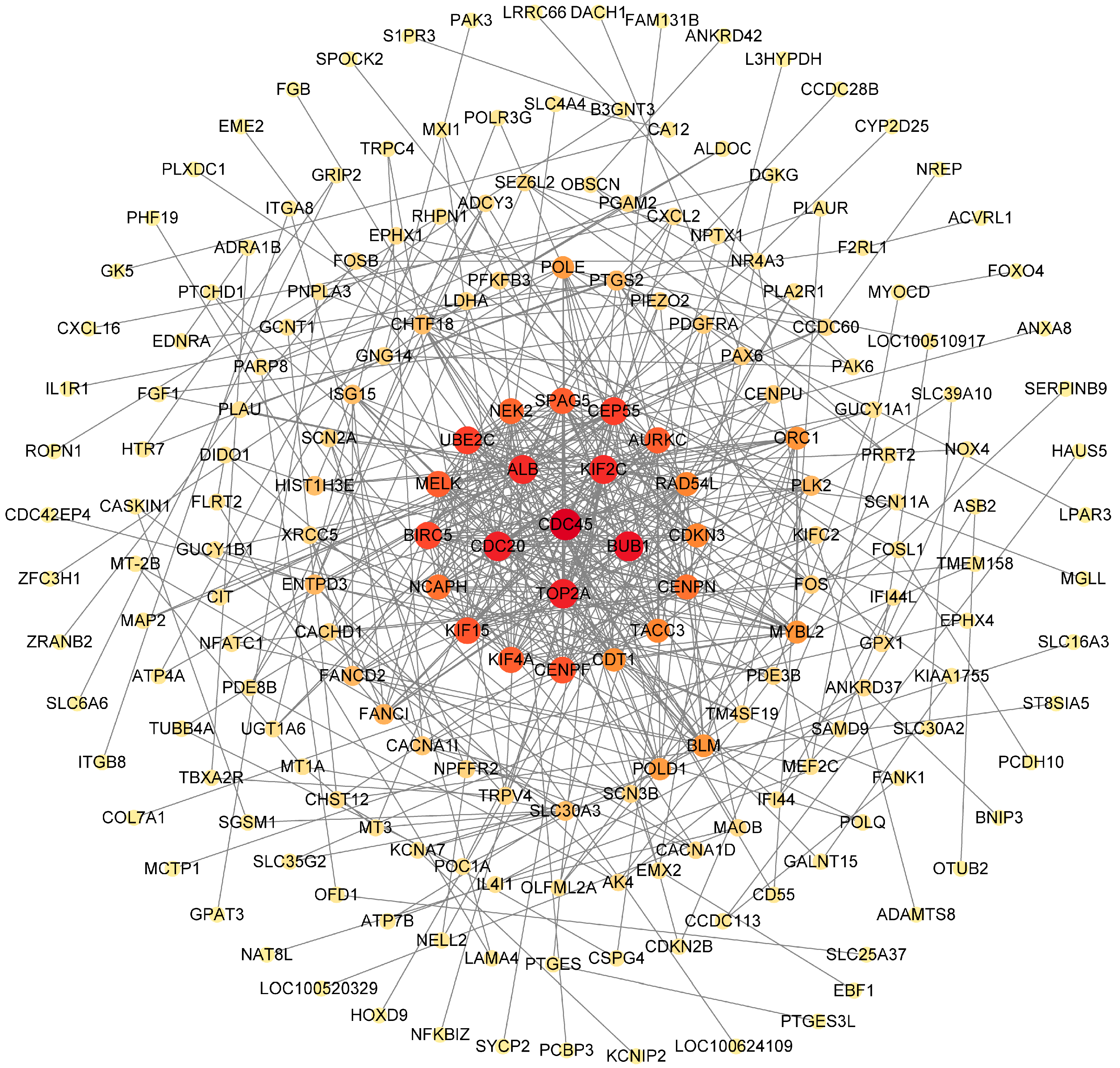
2.6. Further Screening of Key DEGs
2.7. PPI Network
2.8. Validation of Sequencing Results
3. Discussion
4. Materials and Methods
4.1. PAP-1b Preparation and SCs Culture
4.2. Experimental Design
4.3. RNA Extraction and Quality Assessment
4.4. Library Construction and Sequencing
4.5. Raw Data Quality Control and Sequence Comparison Analysis
4.6. Identification of Differentially Expressed Genes (DEGs)
4.7. Functional Enrichment Analysis of DEGs
4.8. Quantitative Real-Time Reverse Transcription–Polymerase Chain Reaction (RT-qPCR)
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Ouyang, B.; Zhang, W.; Guang, C.; Xu, W.; Mu, W. An overview of chemical, physical and biological methods for zearalenone elimination: Recent advances and future prospective. Food Control 2023, 154, 110011. [Google Scholar] [CrossRef]
- Savard, C.; Gawhary, S.; Boyer, A.; Chorfi, Y. Assessment of Zearalenone-Induced Cell Survival and of Global Gene Regulation in Mouse TM4 Sertoli Cells. Toxins 2022, 14, 98. [Google Scholar] [CrossRef]
- Han, X.; Huangfu, B.; Xu, T.; Xu, W.; Asakiya, C.; Huang, K.; He, X. Research Progress of Safety of Zearalenone: A Review. Toxins 2022, 14, 386. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Feng, N.; Wang, Y.; Noll, L.; Xu, S.; Liu, X.; Lu, N.; Zou, H.; Gu, J.; Yuan, Y.; et al. Effects of zearalenone and its derivatives on the synthesis and secretion of mammalian sex steroid hormones: A review. Food Chem. Toxicol. 2019, 126, 262–276. [Google Scholar] [CrossRef]
- Wentzel, J.F.; Lombard, M.J.; Du Plessis, L.H.; Zandberg, L. Evaluation of the cytotoxic properties, gene expression profiles and secondary signalling responses of cultured cells exposed to fumonisin B1, deoxynivalenol and zearalenone mycotoxins. Arch. Toxicol. 2017, 91, 2265–2282. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zeng, Z.; Chen, Y.; Li, R.; Tang, X.; Zhu, X.; Huo, J.; Liu, Y.; Zhang, L.; Chen, J. Genotoxicity of three mycotoxin contaminants of rice: 28-day multi-endpoint assessment in rats. Mutat. Res. Genet. Toxicol. Envorn. Mutagen. 2021, 867, 503369. [Google Scholar] [CrossRef]
- Gao, X.; Xiao, Z.H.; Liu, M.; Zhang, N.Y.; Khalil, M.M.; Gu, C.Q.; Qi, D.S.; Sun, L.H. Dietary Silymarin Supplementation Alleviates Zearalenone-Induced Hepatotoxicity and Reproductive Toxicity in Rats. J. Nutr. 2018, 148, 1209–1216. [Google Scholar] [CrossRef]
- Ben Salah-Abbes, J.; Belgacem, H.; Ezzdini, K.; Abdel-Wahhab, M.A.; Abbes, S. Zearalenone nephrotoxicity: DNA fragmentation, apoptotic gene expression and oxidative stress protected by Lactobacillus plantarum MON03. Toxicon 2020, 175, 28–35. [Google Scholar] [CrossRef]
- Cheng, Q.; Jiang, S.; Huang, L.; Wang, Y.; Yang, W. Zearalenone Exposure Affects the Keap1-Nrf2 Signaling Pathway and Glucose Nutrient Absorption Related Genes of Porcine Jejunal Epithelial Cells. Toxins 2022, 14, 793. [Google Scholar] [CrossRef]
- Yan, R.; Wang, H.; Zhu, J.; Wang, T.; Nepovimova, E.; Long, M.; Li, P.; Kuca, K.; Wu, W. Procyanidins inhibit zearalenone-induced apoptosis and oxidative stress of porcine testis cells through activation of Nrf2 signaling pathway. Food Chem. Toxicol. 2022, 165, 113061. [Google Scholar] [CrossRef]
- Liu, T.; Liu, G.; Xu, Y.; Huang, Y.; Zhang, Y.; Wu, Y.; Xu, Y. Zearalenone Induces Blood-Testis Barrier Damage through Endoplasmic Reticulum Stress-Mediated Paraptosis of Sertoli Cells in Goats. Int. J. Mol. Sci. 2023, 25, 553. [Google Scholar] [CrossRef]
- Cai, P.; Liu, S.; Tu, Y.; Shan, T. Toxicity, biodegradation, and nutritional intervention mechanism of zearalenone. Sci. Total Envorn. 2024, 911, 168648. [Google Scholar] [CrossRef]
- Shen, T.; Miao, Y.; Ding, C.; Fan, W.; Liu, S.; Lv, Y.; Gao, X.; De Boevre, M.; Yan, L.; Okoth, S.; et al. Activation of the p38/MAPK pathway regulates autophagy in response to the CYPOR-dependent oxidative stress induced by zearalenone in porcine intestinal epithelial cells. Food Chem. Toxicol. 2019, 131, 110527. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.L.; Wang, B.J.; Wang, L.; Shan, Y.P.; Zou, H.; Song, R.L.; Wang, T.; Gu, J.H.; Yuan, Y.; Liu, X.Z.; et al. ROS-Mediated Cell Cycle Arrest and Apoptosis Induced by Zearalenone in Mouse Sertoli Cells via ER Stress and the ATP/AMPK Pathway. Toxins 2018, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Lv, L.; Wang, D.; Guo, B.; Lv, J.; Luo, L.; Wen, B.; Kang, Y. Biochemical and molecular responses of maize (Zea mays L.) to 1,2-dibromo-4-(1,2 dibromoethyl) cyclohexane (TBECH) diastereomers: Oxidative stress, DNA damage, antioxidant enzyme gene expression and diversity of root exudates. Sci. Total Envorn. 2021, 753, 141872. [Google Scholar] [CrossRef]
- Cao, L.; Zhao, J.; Ma, L.; Chen, J.; Xu, J.; Rahman, S.U.; Feng, S.; Li, Y.; Wu, J.; Wang, X. Lycopene attenuates zearalenone-induced oxidative damage of piglet sertoli cells through the nuclear factor erythroid-2 related factor 2 signaling pathway. Ecotoxicol. Envorn. Saf. 2021, 225, 112737. [Google Scholar] [CrossRef]
- Zhu, W.; Ge, M.; Li, X.; Wang, J.; Wang, P.; Tai, T.; Wang, Y.; Sun, J.; Shi, G. Hyperoside Attenuates Zearalenone-induced spleen injury by suppressing oxidative stress and inhibiting apoptosis in mice. Int. Immunopharmacol. 2022, 102, 108408. [Google Scholar] [CrossRef]
- Liu, X.L.; Wu, R.Y.; Sun, X.F.; Cheng, S.F.; Zhang, R.Q.; Zhang, T.Y.; Zhang, X.F.; Zhao, Y.; Shen, W.; Li, L. Mycotoxin zearalenone exposure impairs genomic stability of swine follicular granulosa cells in vitro. Int. J. Biol. Sci. 2018, 14, 294–305. [Google Scholar] [CrossRef]
- Lin, X.; Zhu, L.; Gao, X.; Kong, L.; Huang, Y.; Zhao, H.; Chen, Y.; Wen, L.; Li, R.; Wu, J.; et al. Ameliorative effect of betulinic acid against zearalenone exposure triggers testicular dysfunction and oxidative stress in mice via p38/ERK MAPK inhibition and Nrf2-mediated antioxidant defense activation. Ecotoxicol. Envorn. Saf. 2022, 238, 113561. [Google Scholar] [CrossRef]
- Long, M.; Yang, S.H.; Shi, W.; Li, P.; Guo, Y.; Guo, J.; He, J.B.; Zhang, Y. Protective effect of proanthocyanidin on mice Sertoli cell apoptosis induced by zearalenone via the Nrf2/ARE signalling pathway. Envorn. Sci. Pollut. Res. Int. 2017, 24, 26724–26733. [Google Scholar] [CrossRef]
- Cheng, J.; Zhao, L.; Liu, D.; Shen, R.; Bai, D. Potentilla anserine L. polysaccharide protects against cadmium-induced neurotoxicity. Env. Toxicol. Pharmacol. 2022, 90, 103816. [Google Scholar] [CrossRef]
- Shi, J.; Wang, J.; Zhang, J.; Li, X.; Tian, X.; Wang, W.; Wang, P.; Li, M. Polysaccharide extracted from Potentilla anserina L ameliorate acute hypobaric hypoxia-induced brain impairment in rats. Phytother. Res. 2020, 34, 2397–2407. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Cheng, J.; Liu, D.; Gong, H.; Bai, D.; Sun, W. Potentilla anserina polysaccharide alleviates cadmium-induced oxidative stress and apoptosis of H9c2 cells by regulating the MG53-mediated RISK pathway. Chin. J. Nat. Med. 2023, 21, 279–291. [Google Scholar] [CrossRef]
- Chen, J.R.; Yang, Z.Q.; Hu, T.J.; Yan, Z.T.; Niu, T.X.; Wang, L.; Cui, D.A.; Wang, M. Immunomodulatory activity in vitro and in vivo of polysaccharide from Potentilla anserina. Fitoterapia 2010, 81, 1117–1124. [Google Scholar] [CrossRef]
- Han, G.; Hong, S.H.; Lee, S.J.; Hong, S.P.; Cho, C. Transcriptome Analysis of Testicular Aging in Mice. Cells 2021, 10, 2895. [Google Scholar] [CrossRef]
- Pintus, E.; Kadlec, M.; Karlasova, B.; Popelka, M.; Ros-Santaella, J.L. Spermatogenic Activity and Sperm Traits in Post-Pubertal and Adult Tomcats (Felis catus): Implication of Intra-Male Variation in Sperm Size. Cells 2021, 10, 624. [Google Scholar] [CrossRef] [PubMed]
- Petricca, S.; Celenza, G.; Luzi, C.; Cinque, B.; Lizzi, A.R.; Franceschini, N.; Festuccia, C.; Iorio, R. Synergistic Activity of Ketoconazole and Miconazole with Prochloraz in Inducing Oxidative Stress, GSH Depletion, Mitochondrial Dysfunction, and Apoptosis in Mouse Sertoli TM4 Cells. Int. J. Mol. Sci. 2022, 23, 5429. [Google Scholar] [CrossRef]
- Lu, R.; Wang, X.; Zhao, W.; Wang, P.; Zhao, S.; Zhao, X.; Wang, D. Comparative transcriptome and proteome profiles reveal the regulation mechanism of low temperature on garlic greening. Food Res. Int. 2022, 161, 111823. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, H.; Bing, J.; Zhang, G. Integrative transcriptomic and TMT-based proteomic analysis reveals the mechanism by which AtENO2 affects seed germination under salt stress. Front. Plant Sci. 2022, 13, 1035750. [Google Scholar] [CrossRef]
- Wang, H.; Yu, Q.; Ding, X.; Hu, X.; Hou, K.; Liu, X.; Nie, S.; Xie, M. RNA-seq based elucidation of mechanism underlying Ganoderma atrum polysaccharide induced immune activation of murine myeloid-derived dendritic cells. J. Funct. Foods 2019, 55, 104–116. [Google Scholar] [CrossRef]
- Shen, M.; Cai, R.; Li, Z.; Chen, X.; Xie, J. The Molecular Mechanism of Yam Polysaccharide Protected H2O2-Induced Oxidative Damage in IEC-6 Cells. Foods 2023, 12, 262. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Yan, Z.; Du, H.; Song, K.; Gun, S. Structural characteristics of polysaccharide isolated from Potentilla anserina L. and its mitigating effect on Zearalenone-induced oxidative stress in Sertoli cells. Int. J. Biol. Macromol. 2025, 297, 139752. [Google Scholar] [CrossRef]
- Zhao, M.; Hou, J.; Zheng, S.; Ma, X.; Fu, X.; Hu, S.; Zhao, K.; Xu, W. Peucedanum praeruptorum Dunn polysaccharides regulate macrophage inflammatory response through TLR2/TLR4-mediated MAPK and NF-kappaB pathways. Biomed. Pharmacother. 2022, 152, 113258. [Google Scholar] [CrossRef] [PubMed]
- Bing, D.; Wen-Jun, S.; Dachuan, D.; Salman, A.; Fredric, B.K. Plasma membrane cholesterol trafficking in steroidogenesis. FASEB J. 2018, 33, 1389–1400. [Google Scholar] [CrossRef]
- Bellenghi, M.; Puglisi, R.; Pedini, F.; De Feo, A.; Felicetti, F.; Bottero, L.; Sangaletti, S.; Errico, M.C.; Petrini, M.; Gesumundo, C.; et al. SCD5-induced oleic acid production reduces melanoma malignancy by intracellular retention of SPARC and cathepsin B. J. Pathol. 2015, 236, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Hassan, F.; Khan, A.U.; Zaidi, S.; Niazi, M.K.; Ismail, M.A. In Vitro Antioxidant and Inhibitory Study of Picrorhiza kurroa (Kutki), Syzygium aromaticum (Loung), Lawsonia inermis (Henna), Rheum emodi (Revand Chini), Curcuma longa (Haldi) Against Lipid Per-Oxidation in Mice Brain and Liver. Dose Response 2023, 21, 15593258231210431. [Google Scholar] [CrossRef]
- Arjunan, P.; Lin, X.; Tang, Z.; Du, Y.; Kumar, A.; Liu, L.; Yin, X.; Huang, L.; Chen, W.; Chen, Q.; et al. VEGF-B is a potent antioxidant. Proc. Natl. Acad. Sci. USA 2018, 115, 10351–10356. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wang, H.; Li, Y.; Xie, Y.; Huang, C.; Zhao, H.; Miyagishi, M.; Kasim, V. Transcription Factor YY1 Promotes Cell Proliferation by Directly Activating the Pentose Phosphate Pathway. Cancer Res. 2018, 78, 4549–4562. [Google Scholar] [CrossRef]
- Gostimskaya, I.; Grant, C.M. Yeast mitochondrial glutathione is an essential antioxidant with mitochondrial thioredoxin providing a back-up system. Free Radic. Biol. Med. 2016, 94, 55–65. [Google Scholar] [CrossRef]
- Gray, J.P.; Karandrea, S.; Burgos, D.Z.; Jaiswal, A.A.; Heart, E.A. NAD(P)H-dependent quinone oxidoreductase 1 (NQO1) and cytochrome P450 oxidoreductase (CYP450OR) differentially regulate menadione-mediated alterations in redox status, survival and metabolism in pancreatic beta-cells. Toxicol. Lett. 2016, 262, 1–11. [Google Scholar] [CrossRef]
- Palma, F.R.; He, C.; Danes, J.M.; Paviani, V.; Coelho, D.R.; Gantner, B.N.; Bonini, M.G. Mitochondrial Superoxide Dismutase: What the Established, the Intriguing, and the Novel Reveal About a Key Cellular Redox Switch. Antioxid. Redox Signal 2020, 32, 701–714. [Google Scholar] [CrossRef]
- Cheng, B.; Wang, Y.; Ayanlaja, A.A.; Zhu, J.; Kambey, P.A.; Qiu, Z.; Zhang, C.; Hu, W. Glutathione S-Transferases S1, Z1 and A1 Serve as Prognostic Factors in Glioblastoma and Promote Drug Resistance through Antioxidant Pathways. Cells 2022, 11, 3232. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, M.; Loser, A.; Cheng, Q.; Wichmann-Costaganna, M.; Schadel, P.; Werz, O.; Arner, E.S.; Kipp, A.P. Side-by-side comparison of recombinant human glutathione peroxidases identifies overlapping substrate specificities for soluble hydroperoxides. Redox Biol. 2023, 59, 102593. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhu, L.; Yi, X.; Huang, Y.; Zhao, H.; Chen, Y.; Yuan, Z.; Wen, L.; Wu, J.; Yi, J. Betulinic Acid Alleviates Spleen Oxidative Damage Induced by Acute Intraperitoneal Exposure to T-2 Toxin by Activating Nrf2 and Inhibiting MAPK Signaling Pathways. Antioxid. 2021, 10, 158. [Google Scholar] [CrossRef]
- Zhu, L.; Yi, X.; Ma, C.; Luo, C.; Kong, L.; Lin, X.; Gao, X.; Yuan, Z.; Wen, L.; Li, R.; et al. Betulinic Acid Attenuates Oxidative Stress in the Thymus Induced by Acute Exposure to T-2 Toxin via Regulation of the MAPK/Nrf2 Signaling Pathway. Toxins 2020, 12, 540. [Google Scholar] [CrossRef]
- Chen, X.; Wang, X.; Shen, M.; Chen, Y.; Yu, Q.; Yang, J.; Xie, J. Combined RNA-seq and molecular biology technology revealed the protective effect of Cyclocarya paliurus polysaccharide on H2O2-induced oxidative damage in L02 cells thought regulating mitochondrial function, oxidative stress and PI3K/Akt and MAPK signaling pathways. Food Res. Int. 2022, 155, 111080. [Google Scholar] [CrossRef]
- Kwon, D.H.; Cha, H.J.; Lee, H.; Hong, S.H.; Park, C.; Park, S.H.; Kim, G.Y.; Kim, S.; Kim, H.S.; Hwang, H.J.; et al. Protective Effect of Glutathione against Oxidative Stress-induced Cytotoxicity in RAW 264.7 Macrophages through Activating the Nuclear Factor Erythroid 2-Related Factor-2/Heme Oxygenase-1 Pathway. Antioxidants 2019, 8, 82. [Google Scholar] [CrossRef]
- Semenovich, D.S.; Plotnikov, E.Y.; Titko, O.V.; Lukiyenko, E.P.; Kanunnikova, N.P. Effects of Panthenol and N-Acetylcysteine on Changes in the Redox State of Brain Mitochondria under Oxidative Stress In Vitro. Antioxidants 2021, 10, 1699. [Google Scholar] [CrossRef]
- Liu, D.; Xu, J.; Qian, G.; Hamid, M.; Gan, F.; Chen, X.; Huang, K. Selenizing astragalus polysaccharide attenuates PCV2 replication promotion caused by oxidative stress through autophagy inhibition via PI3K/AKT activation. Int. J. Biol. Macromol. 2018, 108, 350–359. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.; Trapnell, C.; Donaghey, J.; Rinn, J.L.; Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011, 12, R22. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, H.; Yan, Z.; Du, H.; Gun, S. Transcriptome Analysis Reveals the Molecular Mechanism of Potentilla anserina L. Polysaccharides in Mitigating Zearalenone-Induced Oxidative Stress in Porcine Sertoli Cells. Antioxidants 2025, 14, 439. https://doi.org/10.3390/antiox14040439
Shi H, Yan Z, Du H, Gun S. Transcriptome Analysis Reveals the Molecular Mechanism of Potentilla anserina L. Polysaccharides in Mitigating Zearalenone-Induced Oxidative Stress in Porcine Sertoli Cells. Antioxidants. 2025; 14(4):439. https://doi.org/10.3390/antiox14040439
Chicago/Turabian StyleShi, Haixia, Zunqiang Yan, Hong Du, and Shuangbao Gun. 2025. "Transcriptome Analysis Reveals the Molecular Mechanism of Potentilla anserina L. Polysaccharides in Mitigating Zearalenone-Induced Oxidative Stress in Porcine Sertoli Cells" Antioxidants 14, no. 4: 439. https://doi.org/10.3390/antiox14040439
APA StyleShi, H., Yan, Z., Du, H., & Gun, S. (2025). Transcriptome Analysis Reveals the Molecular Mechanism of Potentilla anserina L. Polysaccharides in Mitigating Zearalenone-Induced Oxidative Stress in Porcine Sertoli Cells. Antioxidants, 14(4), 439. https://doi.org/10.3390/antiox14040439






