Dissecting the Mycobacterium bovis BCG Response to Macrophage Infection to Help Prioritize Targets for Anti-Tuberculosis Drug and Vaccine Discovery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mycobacterial Culture
2.2. Macrophage Culture and Infection
2.3. Mycobacterial RNA Extraction and RNA Sequencing
2.4. Transcriptomic Analyses
3. Results
3.1. Mycobacterial Transcriptional Adaptations to Macrophage Infection
3.2. Fatty Acid Metabolism and Cholesterol Catabolism
3.3. PE/PPE Family
3.4. Cytochrome P450 Family
3.5. Overlap with Gene Essentiality Datasets
3.6. Comparison to the TB Vaccination Pipeline
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2021. World Health Organization. 2021. Available online: https://apps.who.int/iris/handle/10665/346387 (accessed on 28 November 2021).
- Bettencourt, P.J.G. The 100th anniversary of bacille Calmette-Guérin (BCG) and the latest vaccines against COVID-19. Int. J. Tuberc. Lung Dis. 2021, 25, 611–613. [Google Scholar] [CrossRef]
- Colditz, G.A.; Brewer, T.F.; Berkey, C.S.; Wilson, M.E.; Burdick, E.; Fineberg, H.V.; Mosteller, F. Efficacy of BCG Vaccine in the Prevention of Tuberculosis. JAMA J. Am. Med. Assoc. 1994, 271, 698–702. [Google Scholar] [CrossRef]
- Fine, P.E. Variation in protection by BCG: Implications of and for heterologous immunity. Lancet 1995, 346, 1339–1345. [Google Scholar] [CrossRef]
- Kaufmann, S.H.E.; Weiner, J.; Fordham von Reyn, C. Novel approaches to tuberculosis vaccine development. Int. J. Infect. Dis. 2017, 56, 263–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorman, S.E.; Nahid, P.; Kurbatova, E.V.; Phillips, P.P.J.; Bryant, K.; Dooley, K.E.; Engle, M.; Goldberg, S.V.; Phan, H.T.T.; Hakim, J.; et al. Four-Month Rifapentine Regimens with or without Moxifloxacin for Tuberculosis. N. Engl. J. Med. 2021, 384, 1705–1718. [Google Scholar] [CrossRef]
- Oh, S.; Trifonov, L.; Yadav, V.D.; Barry, C.E.I.; Boshoff, H.I. Tuberculosis Drug Discovery: A Decade of Hit Assessment for Defined Targets. Front. Cell. Infect. Microbiol. 2021, 11, 188. [Google Scholar] [CrossRef]
- Ulrichs, T.; Kaufmann, S.H.E. New insights into the function of granulomas in human tuberculosis. J. Pathol. 2005, 208, 261–269. [Google Scholar] [CrossRef]
- Schnappinger, D.; Ehrt, S.; Voskuil, M.I.; Liu, Y.; Mangan, J.A.; Monahan, I.M.; Dolganov, G.; Efron, B.; Butcher, P.; Nathan, C.; et al. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages. J. Exp. Med. 2003, 198, 693–704. [Google Scholar] [CrossRef] [Green Version]
- Rohde, K.H.; Abramovitch, R.B.; Russell, D.G. Mycobacterium tuberculosis Invasion of Macrophages: Linking Bacterial Gene Expression to Environmental Cues. Cell Host Microbe 2007, 2, 352–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tailleux, L.; Waddell, S.J.; Pelizzola, M.; Mortellaro, A.; Withers, M.; Tanne, A.; Castagnoli, P.R.; Gicquel, B.; Stoker, N.G.; Butcher, P.D.; et al. Probing Host Pathogen Cross-Talk by Transcriptional Profiling of Both Mycobacterium tuberculosis and Infected Human Dendritic Cells and Macrophages. PLoS ONE 2008, 3, e1403. [Google Scholar] [CrossRef] [PubMed]
- Rienksma, R.A.; Suarez-Diez, M.; Mollenkopf, H.-J.; Dolganov, G.M.; Dorhoi, A.; Schoolnik, G.K.; Santos, V.A.M.D.; Kaufmann, S.H.E.; Schaap, P.J.; Gengenbacher, M. Comprehensive insights into transcriptional adaptation of intracellular mycobacteria by microbe-enriched dual RNA sequencing. BMC Genom. 2015, 16, 34. [Google Scholar] [CrossRef] [Green Version]
- Peterson, E.J.R.; Bailo, R.; Rothchild, A.C.; Arrieta-Ortiz, M.L.; Kaur, A.; Pan, M.; Mai, D.; Abidi, A.A.; Cooper, C.; Aderem, A.; et al. Path-seq identifies an essential mycolate remodeling program for mycobacterial host adaptation. Mol. Syst. Biol. 2019, 15, e8584. [Google Scholar] [CrossRef] [PubMed]
- Rengarajan, J.; Bloom, B.R.; Rubin, E.J. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. USA 2005, 102, 8327–8332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffin, J.E.; Gawronski, J.D.; DeJesus, M.A.; Ioerger, T.R.; Akerley, B.J.; Sassetti, C.M. High-Resolution Phenotypic Profiling Defines Genes Essential for Mycobacterial Growth and Cholesterol Catabolism. PLoS Pathog. 2011, 7, e1002251. [Google Scholar] [CrossRef] [Green Version]
- Bettencourt, P.; Müller, J.; Nicastri, A.; Cantillon, D.; Madhavan, M.; Charles, P.D.; Fotso, C.B.; Wittenberg, R.; Bull, N.; Pinpathomrat, N.; et al. Identification of antigens presented by MHC for vaccines against tuberculosis. NPJ Vaccines 2020, 5, 2. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Borgers, K.; Ou, J.-Y.; Zheng, P.-X.; Tiels, P.; Van Hecke, A.; Plets, E.; Michielsen, G.; Festjens, N.; Callewaert, N.; Lin, Y.-C. Reference genome and comparative genome analysis for the WHO reference strain for Mycobacterium bovis BCG Danish, the present tuberculosis vaccine. BMC Genom. 2019, 20, 561. [Google Scholar] [CrossRef] [Green Version]
- Caspi, R.; Billington, R.; Ferrer, L.; Foerster, H.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Mueller, L.A.; et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016, 44, D471–D480. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-D.; Guinn, K.M.; Harrell, M.I.; Liao, R.; Voskuil, M.I.; Tompa, M.; Schoolnik, G.K.; Sherman, D.R. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 2003, 48, 833–843. [Google Scholar] [CrossRef] [Green Version]
- Kendall, S.L.; Withers, M.; Soffair, C.N.; Moreland, N.J.; Gurcha, S.; Sidders, B.; Frita, R.; Bokum, A.T.; Besra, G.S.; Lott, J.S.; et al. A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Mol. Microbiol. 2007, 65, 684–699. [Google Scholar] [CrossRef] [Green Version]
- Segal, W.; Bloch, H. Biochemical differentiation of mycobacterium tuberculosis grown in vivo and in vitro. J. Bacteriol. 1956, 72, 132–141. [Google Scholar] [CrossRef] [Green Version]
- Munoz-Elias, E.J.; McKinney, J.D. Carbon metabolism of intracellular bacteria. Cell. Microbiol. 2006, 8, 10–22. [Google Scholar] [CrossRef]
- Wilburn, K.M.; Fieweger, R.A.; Vander Ven, B.C. Cholesterol and fatty acids grease the wheels of Mycobacterium tuberculosis pathogenesis. Pathog. Dis. 2018, 76. [Google Scholar] [CrossRef]
- McGuire, A.M.; Weiner, B.; Park, S.T.; Wapinski, I.; Raman, S.; Dolganov, G.; Peterson, M.; Riley, R.; Zucker, J.; Abeel, T.; et al. Comparative analysis of mycobacterium and related actinomycetes yields insight into the evolution of Mycobacterium tuberculosis pathogenesis. BMC Genom. 2012, 13, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Li, Q.; Yu, Z.; Zhou, M.; Xie, J. Mycobacterium tuberculosis PE13 (Rv1195) manipulates the host cell fate via p38-ERK-NF-κB axis and apoptosis. Apoptosis 2016, 21, 795–808. [Google Scholar] [CrossRef]
- Peng, X.; Luo, T.; Zhai, X.; Zhang, C.; Suo, J.; Ma, P.; Wang, C.; Bao, L. PPE11 of Mycobacterium tuberculosis can alter host inflammatory response and trigger cell death. Microb. Pathog. 2019, 126, 45–55. [Google Scholar] [CrossRef]
- Mitra, A.; Speer, A.; Lin, K.; Ehrt, S.; Niederweis, M. PPE Surface Proteins Are Required for Heme Utilization by Mycobacterium tuberculosis. mBio 2017, 8, e01720-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, J.; Chen, R.; Wang, H.; Zhang, X. Role of the PE/PPE Family in Host–Pathogen Interactions and Prospects for Anti-Tuberculosis Vaccine and Diagnostic Tool Design. Front. Cell. Infect. Microbiol. 2020, 10, 594288. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, H.; Johnston, J.B.; Ortiz de Montellano, P.R. Cholesterol catabolism as a therapeutic target in Mycobacterium tuberculosis. Trends Microbiol. 2011, 19, 530–539. [Google Scholar] [CrossRef] [Green Version]
- Fang, H.; Yu, D.; Hong, Y.; Zhou, X.; Li, C.; Sun, B. The LuxR family regulator Rv0195 modulates Mycobacterium tuberculosis dormancy and virulence. Tuberculosis 2013, 93, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Aguilo, N.; Marinova, D.; Gonzalo-Asensio, J. Update on TB Vaccine Pipeline. Appl. Sci. 2020, 10, 2632. [Google Scholar] [CrossRef] [Green Version]
- Hansen, S.G.; Zak, D.E.; Xu, G.; Ford, J.C.; Marshall, E.E.; Malouli, D.; Gilbride, R.M.; Hughes, C.M.; Ventura, A.B.; Ainslie, E.; et al. Prevention of tuberculosis in rhesus macaques by a cytomegalovirus-based vaccine. Nat. Med. 2018, 24, 130–143. [Google Scholar] [CrossRef]
- Roupie, V.; Romano, M.; Zhang, L.; Korf, H.; Lin, M.Y.; Franken, K.L.M.C.; Ottenhoff, T.H.M.; Klein, M.R.; Huygen, K. Immunogenicity of Eight Dormancy Regulon-Encoded Proteins of Mycobacterium tuberculosis in DNA-Vaccinated and Tuberculosis-Infected Mice. Infect. Immun. 2007, 75, 941–949. [Google Scholar] [CrossRef] [Green Version]
- Kassa, D.; Ran, L.; Geberemeskel, W.; Tebeje, M.; Alemu, A.; Selase, A.; Tegbaru, B.; Franken, K.L.M.C.; Friggen, A.H.; van Meijgaarden, K.E.; et al. Analysis of Immune Responses against a Wide Range of Mycobacterium tuberculosis Antigens in Patients with Active Pulmonary Tuberculosis. Clin. Vaccine Immunol. 2012, 19, 1907–1915. [Google Scholar] [CrossRef] [Green Version]
- Zvi, A.; Ariel, N.; Fulkerson, J.; Sadoff, J.C.; Shafferman, A. Whole genome identification of Mycobacterium tuberculosis vaccine candidates by comprehensive data mining and bioinformatic analyses. BMC Med. Genom. 2008, 1, 18. [Google Scholar] [CrossRef] [Green Version]
- Brosch, R.; Gordon, S.V.; Marmiesse, M.; Brodin, P.; Buchrieser, C.; Eiglmeier, K.; Garnier, T.; Gutierrez, C.; Hewinson, G.; Kremer, K.; et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 2002, 99, 3684–3689. [Google Scholar] [CrossRef] [Green Version]
- Guo, S.; Xue, R.; Li, Y.; Wang, S.M.; Ren, L.; Xu, J.J. The CFP10/ESAT6 complex of Mycobacterium tuberculosis may function as a regulator of macrophage cell death at different stages of tuberculosis infection. Med. Hypotheses 2012, 78, 389–392. [Google Scholar] [CrossRef]
- Fontán, P.; Aris, V.; Ghanny, S.; Soteropoulos, P.; Smith, I. Global Transcriptional Profile of Mycobacterium tuberculosis during THP-1 Human Macrophage Infection. Infect. Immun. 2008, 76, 717–725. [Google Scholar] [CrossRef] [Green Version]
- Pethe, K.; Sequeira, P.C.; Agarwalla, S.; Rhee, K.; Kuhen, K.; Phong, W.Y.; Patel, V.; Beer, D.; Walker, J.R.; Duraiswamy, J.; et al. A chemical genetic screen in Mycobacterium tuberculosis identifies carbon-source-dependent growth inhibitors devoid of in vivo efficacy. Nat. Commun. 2010, 1, 57–58. [Google Scholar] [CrossRef] [PubMed]
- Ugalde, S.O.; Boot, M.; Commandeur, J.N.M.; Jennings, P.; Bitter, W.; Vos, J.C. Function, essentiality, and expression of cytochrome P450 enzymes and their cognate redox partners in Mycobacterium tuberculosis: Are they drug targets? Appl. Microbiol. Biotechnol. 2019, 103, 3597–3614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLean, K.J.; Munro, A.W. Drug targeting of heme proteins in Mycobacterium tuberculosis. Drug Discov. Today 2017, 22, 566–575. [Google Scholar] [CrossRef] [PubMed]




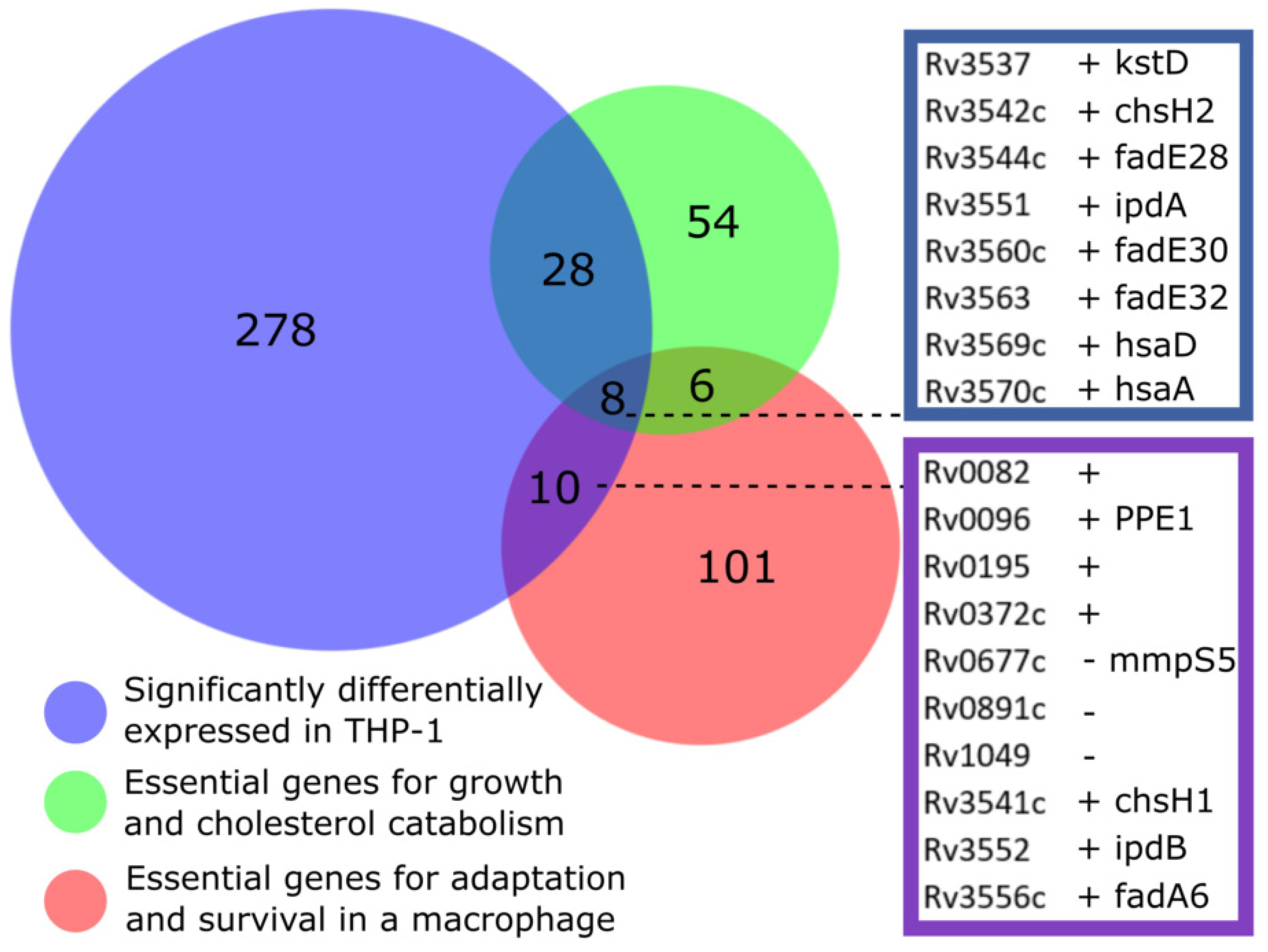
| M. bovis Gene ID | H37Rv Gene ID | Gene Name | L2FC | Functional Category | Prediction of Function |
|---|---|---|---|---|---|
| Mb0085 | Rv0082 | - | 2.4 | Intermediary metabolism and respiration | Probable oxidoreductase, member of DosR regulon |
| Mb0099 | Rv0096 | ppe1 | 1.39 | PE/PPE | PPE family protein |
| Mb0201 | Rv0195 | - | 1.95 | Regulatory | Possible transcriptional regulation |
| Mb0379c | Rv0372c | - | 1.55 | Conserved hypotheticals | Unknown |
| Mb3571c | Rv3541c | chsH1 | 2.28 | Conserved hypotheticals | Cholesterol side chain degradation |
| Mb3582 | Rv3552 | ipdB | 2.91 | Intermediary metabolism and respiration | Possible CoA-transferase |
| Mb3586c | Rv3556 | fadA6 | 2.04 | Lipid Metabolism | Catalyses the formation of 4-methyl-5-oxo-octanedioyl-CoA in steroid catabolic pathway |
| M. bovis Gene ID | H37Rv Gene ID | Gene Name | L2FC | Functional Category | Prediction of Function |
|---|---|---|---|---|---|
| Mb0082 | Rv0079 ** | - | 2.71 | Conserved hypothetical | Dormancy associated translation inhibitor |
| Mb0084 | Rv0081 | - | 2.09 | Regulatory protein | Transcriptional regulatory protein, member of DosR regulon |
| Mb1762c | Rv1733c ** | - | 1.46 | Cell wall and processes | Probable conserved transmembrane protein, member of DosR regulon |
| Mb1763c | Rv1734c | - | 2.21 | Conserved hypothetical | Unknown, member of DosR regulon |
| Mb2030c | Rv2007c | fdxA | 2.15 | Intermediary metabolism and respiration | Involved in electron transfer |
| Mb2053c | Rv2028c | - | 1.21 | Virulence, detoxification and adaptation | Universal stress protein, member of DosR regulon |
| Mb2660c | Rv2627c ** | - | 1.56 | Conserved hypothetical | Unknown, member of DosR regulon |
| Mb2410c | Rv2389c ** | rpfD | 2.17 | Cell wall and processes | Resuscitation-promoting factor |
| Mb1767 | Rv1738 * | - | 2.56 | Conserved hypothetical | Implicated in control of non-replicating persistence |
| Mb2057c | Rv2031c * | hspX | 4.28 | Virulence, detoxification, adaptation | Heat shock protein, induced under stress |
| Mb2058 | Rv2032 * | acg | 1.01 | Conserved hypothetical | Putative nitroreductase, induced under stress |
| Mb3154c | Rv3130c * | tgs1 | 2.47 | Lipid metabolism | Triacylglycerol synthase |
| Mb3155 | Rv3131 * | - | 2.38 | Conserved hypothetical | Putative nitroreductase |
| Mb1384 | Rv1349 * | irtb | 1.00 | Cell wall and processes | Involved in iron homeostasis |
| Mb2054c | Rv2029c * | pfkB | 1.30 | Intermediary metabolism and respiration | Involved in glycolysis |
| Mb2055c | Rv2030c * | - | 3.64 | Conserved hypothetical | Unknown, induced under stress |
| Mb0476 | Rv0467 * | icl | 2.67 | Intermediary metabolism and respiration | Involved in glyoxylate shunt |
| Mb1161 | Rv1130 * | prpD | 10.42 | Intermediary metabolism and respiration | Involved in methylcitrate cycle |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Medley, J.; Goff, A.; Bettencourt, P.J.G.; Dare, M.; Cole, L.; Cantillon, D.; Waddell, S.J. Dissecting the Mycobacterium bovis BCG Response to Macrophage Infection to Help Prioritize Targets for Anti-Tuberculosis Drug and Vaccine Discovery. Vaccines 2022, 10, 113. https://doi.org/10.3390/vaccines10010113
Medley J, Goff A, Bettencourt PJG, Dare M, Cole L, Cantillon D, Waddell SJ. Dissecting the Mycobacterium bovis BCG Response to Macrophage Infection to Help Prioritize Targets for Anti-Tuberculosis Drug and Vaccine Discovery. Vaccines. 2022; 10(1):113. https://doi.org/10.3390/vaccines10010113
Chicago/Turabian StyleMedley, Jamie, Aaron Goff, Paulo J. G. Bettencourt, Madelaine Dare, Liam Cole, Daire Cantillon, and Simon J. Waddell. 2022. "Dissecting the Mycobacterium bovis BCG Response to Macrophage Infection to Help Prioritize Targets for Anti-Tuberculosis Drug and Vaccine Discovery" Vaccines 10, no. 1: 113. https://doi.org/10.3390/vaccines10010113
APA StyleMedley, J., Goff, A., Bettencourt, P. J. G., Dare, M., Cole, L., Cantillon, D., & Waddell, S. J. (2022). Dissecting the Mycobacterium bovis BCG Response to Macrophage Infection to Help Prioritize Targets for Anti-Tuberculosis Drug and Vaccine Discovery. Vaccines, 10(1), 113. https://doi.org/10.3390/vaccines10010113







