Effectiveness and Efficacy of Vaccine on Mutated SARS-CoV-2 Virus and Post Vaccination Surveillance: A Narrative Review
Abstract
:1. Introduction
2. Methods
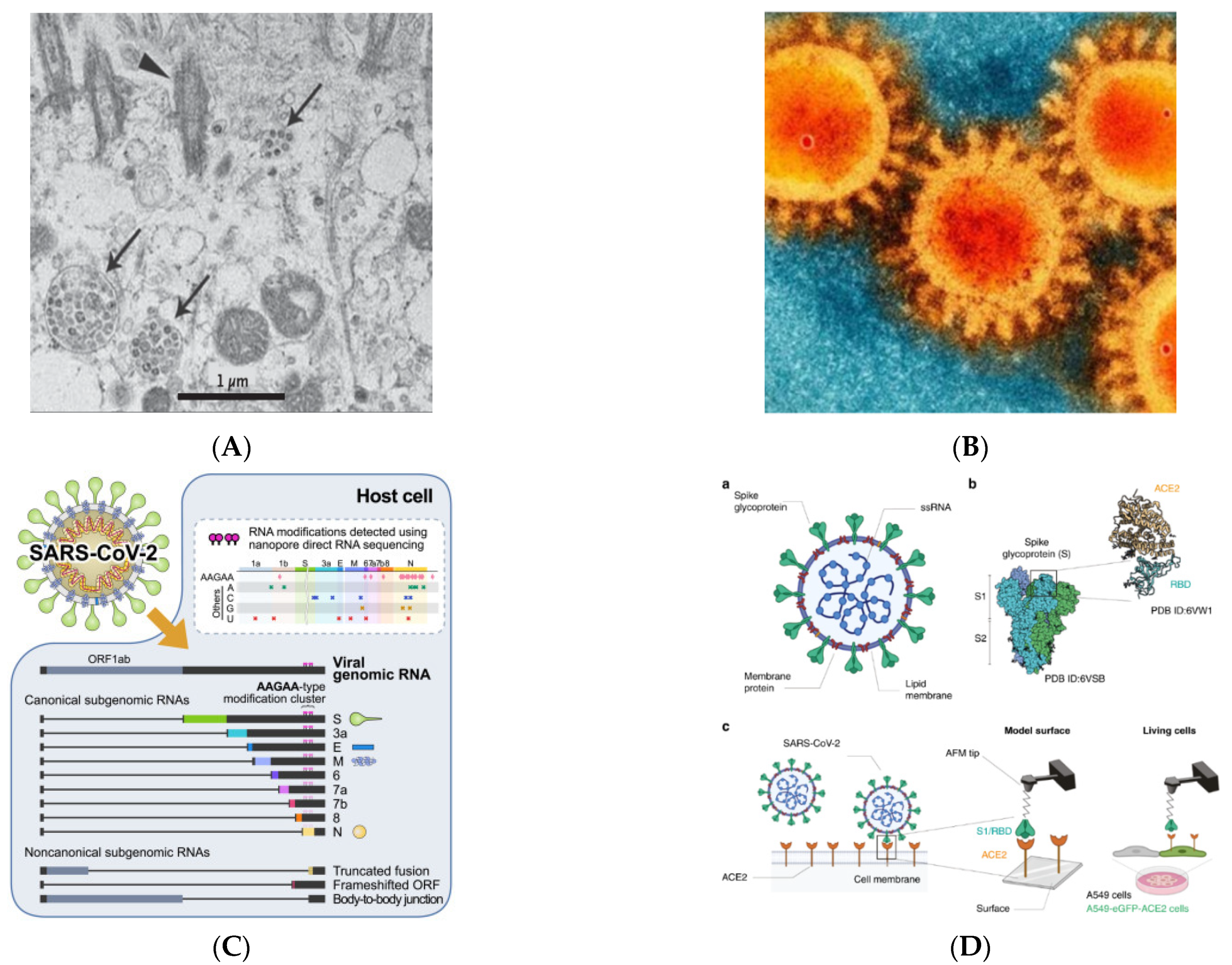
3. SARS-CoV-2 and Receptors
4. Mutation in the SARS-CoV-2
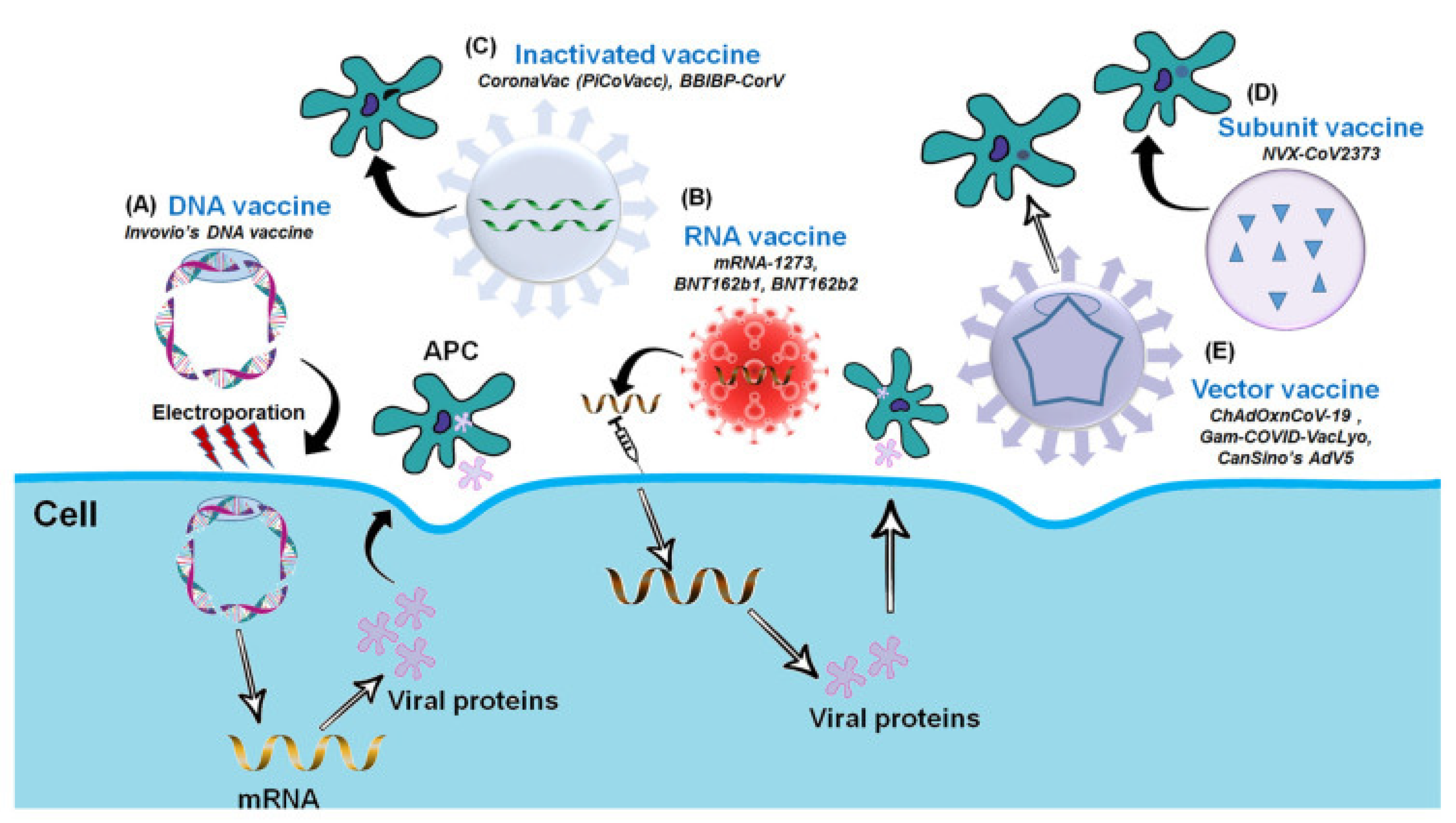
5. Types of COVID-19 Vaccines
5.1. Inactivated Virus-Based Vaccines
5.2. DNA-Based Vaccines
5.3. mRNA-Based Vaccines
5.4. Protein Subunit-Based Vaccine
5.5. Viral Vector-Based Vaccine
6. Efficacy and Effectiveness of COVID-19 Vaccines
- (1)
- Vaccinated individuals are estimated to have lower viral loads compared to the unvaccinated;
- (2)
- Vaccinated individuals exhibit milder symptoms compared to unvaccinated ones;
- (3)
- The UK implemented a fairly rapid vaccination program to protect the population (currently, it has reached 70%–80% of the total population, with vaccines used including Pfizer, Moderna, AstraZeneca, and Johnson & Johnson).
7. Post-Vaccination Surveillance Strategy
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef]
- Baloch, S.; Baloch, M.A.; Zheng, T.; Pei, X. The Coronavirus Disease 2019 (COVID-19) Pandemic. Tohoku J. Exp. Med. 2020, 250, 271–278. [Google Scholar] [CrossRef] [Green Version]
- WHO. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants (accessed on 5 September 2021).
- Davies, N.G.; Jarvis, C.I.; CMMID COVID-19 Working Group; Edmunds, W.J.; Jewell, N.P.; Diaz-Ordaz, K.; Keogh, R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature 2021, 593, 270–274. [Google Scholar] [CrossRef]
- Burki, T. Understanding variants of SARS-CoV-2. Lancet 2021, 397, 462. [Google Scholar] [CrossRef]
- Herlihy, R.; Bamberg, W.; Burakoff, A.; Alden, N.; Severson, R.; Bush, E.; Kawasaki, B.; Berger, B.; Austin, E.; Shea, M.; et al. Rapid Increase in Circulation of the SARS-CoV-2 B.1.617.2 (Delta) Variant—Mesa County, Colorado, April–June 2021. Morb. Mortal. Wkly. Rep. 2021, 70, 1084–1087. [Google Scholar] [CrossRef]
- Platto, S.; Wang, Y.; Zhou, J.; Carafoli, E. History of the COVID-19 pandemic: Origin, explosion, worldwide spreading. Biochem. Biophys. Res. Commun. 2020, 538, 14–23. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Kim, D.; Lee, J.-Y.; Yang, J.-S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e10. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, A.; Wahi, N.; Saxena, A.; Chaudhary, D. Proteome Organization of COVID-19: Illustrating Targets for Vaccine Development. J. Pure Appl. Microbiol. 2020, 14, 831–840. [Google Scholar] [CrossRef]
- Yang, J.; Petitjean, S.J.L.; Koehler, M.; Zhang, Q.; Dumitru, A.C.; Chen, W.; Derclaye, S.; Vincent, S.P.; Soumillion, P.; Alsteens, D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020, 11, 4541. [Google Scholar] [CrossRef]
- Hu, B.; Huang, S.; Yin, L. The cytokine storm and COVID-19. J. Med. Virol. 2021, 93, 250–256. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020, 80, 607–613. [Google Scholar] [CrossRef]
- Das, G.; Ghosh, S.; Garg, S.; Ghosh, S.; Jana, A.; Samat, R.; Mukherjee, N.; Roy, R.; Ghosh, S. An overview of key potential therapeutic strategies for combat in the COVID-19 battle. RSC Adv. 2020, 10, 28243–28266. [Google Scholar] [CrossRef]
- Carcaterra, M.; Caruso, C. Alveolar Epithelial Cell Type II as Main Target of SARS-CoV-2 Virus and COVID-19 Development via NF-Kb Pathway Deregulation: A Physio-Pathological Theory. Med. Hyphotheses 2021, 146, 110412. [Google Scholar] [CrossRef]
- Lukkasen, S.; Chua, R.L.; Trefzer, T.; Kahn, N.C.; Schneider, M.A.; Muley, T.; Winter, H.; Mesiter, M.; Veith, C.; Boots, A.W.; et al. SARS-CoV-2 Receptor ACE2 and TMPRSS2 Are Primarily Expressed in Bronchial Transient. EMBO J. 2020, 39, e105114. [Google Scholar] [CrossRef]
- Anonymous. SARS-CoV-2 Variant Classifications and Definitions. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html (accessed on 28 October 2021).
- Firestone, M.J.; Lorentz, A.J.; Wang, X.; Como-Sabetti, K.; Vetter, S.; Smith, K.; Holzbauer, S.; Meyer, S.; Ehresmann, K.; Danila, R.; et al. First Identified Cases of SARS-CoV-2 Variant B.1.1.7 in Minnesota—December 2020–January 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 278–279. [Google Scholar] [CrossRef]
- Planas, D.; Bruel, T.; Grzelak, L.; Guivel-Benhassine, F.; Staropoli, I.; Porrot, F.; Planchais, C.; Buchrieser, J.; Rajah, M.M.; Bishop, E.; et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021, 27, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, K.; Mannell, M.; Naqvi, O.; Matson, D.; Stone, J. SARS-CoV-2 B.1.617.2 (Delta) Variant COVID-19 Outbreak Associated with a Gymnastics Facility—Oklahoma, April–May 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1004–1007. [Google Scholar] [CrossRef]
- Torjesen, I. Covid-19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ 2021, 375, n2943. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. Lineage B.1.621. Available online: https://cov-lineages.org/lineage.html?lineage=B.1.621 (accessed on 5 September 2021).
- Anonymous. B.1.621 Lineage Report. Available online: https://outbreak.info/situation-reports?pango=B.1.621 (accessed on 5 September 2021).
- Messali, S.; Bertelli, A.; Campisi, G.; Zani, A.; Ciccozzi, M.; Caruso, A.; Caccuri, F. A cluster of the new SARS-CoV-2 B.1.621 lineage in Italy and sensitivity of the viral isolate to the BNT162b2 vaccine. J. Med. Virol. 2021, 93, 6468–6470. [Google Scholar] [CrossRef]
- Lurie, N.; Saville, M.; Hatchett, R.; Halton, J. Developing Covid-19 Vaccines at Pandemic Speed. N. Engl. J. Med. 2020, 382, 1969–1973. [Google Scholar] [CrossRef]
- Frederiksen, L.S.F.; Zhang, Y.; Foged, C.; Thakur, A. The Long Road Toward COVID-19 Herd Immunity: Vaccine Platform Technologies and Mass Immunization Strategies. Front. Immunol. 2020, 11, 1817. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.; Kim, Y.; Kumar, S.; Seo, D.; Ashraf, M.; Bae, Y.-S. COVID-19 Vaccines (Revisited) and Oral-Mucosal Vector System as a Potential Vaccine Platform. Vaccines 2021, 9, 171. [Google Scholar] [CrossRef]
- WHO. Draft Landscape and Tracker of COVID-19 Candidate Vaccines. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 16 December 2021).
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z.; et al. Development of an Inactivated Vaccine Candidate for SARS-CoV-2. Science 2020, 369, 77–81. [Google Scholar] [CrossRef]
- Fadlyana, E.; Rusmil, K.; Tarigan, R.; Rahmadi, A.R.; Prodjosoewojo, S.; Sofiatin, Y.; Khrisna, C.V.; Sari, R.M.; Setyaningsih, L.; Surachman, F.; et al. A phase III, observer-blind, randomized, placebo-controlled study of the efficacy, safety, and immunogenicity of SARS-CoV-2 inactivated vaccine in healthy adults aged 18–59 years: An interim analysis in Indonesia. Vaccine 2021, 39, 6520–6528. [Google Scholar] [CrossRef]
- Kaur, S.P.; Gupta, V. COVID-19 Vaccine: A comprehensive status report. Virus Res. 2020, 288, 198114. [Google Scholar] [CrossRef]
- FDA. Long Term Follow-Up After Administration of Human Gene Therapy Products, Guidance for Industry; FDA.gov: Silver Spring, MD, USA, 2020.
- Dortant, P.; Claassen, I.; Van Kreyl, C.; Van Steenis, G.; Wester, P. Risk Assessment on the Carcinogenic Potential of Hybridoma Cell DNA: Implications for Residual Contaminating Cellular DNA in Biological Products. Biologicals 1997, 25, 381–390. [Google Scholar] [CrossRef]
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020, 383, 2427–2438. [Google Scholar] [CrossRef]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Thomas, S.J.; Moreira, E.D., Jr.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Marc, G.P.; Polack, F.P.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine through 6 Months. N. Engl. J. Med. 2021, 385, 1761–1773. [Google Scholar] [CrossRef] [PubMed]
- Cowling, B.J.; The University of Hong Kong. Third Dose of mRNA Vaccination to Boost COVID-19 Immunity. Available online: https://clinicaltrials.gov/ct2/show/NCT05057182 (accessed on 16 December 2021).
- Guanaccia, B.J.; Browne, F.; Yale-Griffin Prevention Research Cente. COVID-19 Booster Vaccination in Persons with Multiple Sclerosis. Available online: https://clinicaltrials.gov/ct2/show/record/NCT05081271 (accessed on 16 December 2021).
- Pfizer. Study to Evaluate the Safety and Efficacy of a Booster Dose of BNT162b2 Against COVID-19 in Participants ≥16 Years of Age. Available online: https://clinicaltrials.gov/ct2/show/NCT04955626?term=vaccine&recrs=abdf&cond=COVID-19&phase=012345&sort=nwst&draw=2 (accessed on 16 December 2021).
- Heath, P.T.; Galiza, E.P.; Baxter, D.N.; Boffito, M.; Browne, D.; Burns, F.; Chadwick, D.R.; Clark, R.; Cosgrove, C.; Galloway, J.; et al. Safety and Efficacy of NVX-CoV2373 Covid-19 Vaccine. N. Engl. J. Med. 2021, 385, 1172–1183. [Google Scholar] [CrossRef]
- Thanh Le, T.; Andreadakis, Z.; Kumar, A.; Gómez Román, R.; Tollefsen, S.; Saville, M.; Mayhew, S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020, 19, 305–306. [Google Scholar] [CrossRef]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef]
- Groves, D.C.; Rowland-Jones, S.L.; Angyal, A. The D614G mutations in the SARS-CoV-2 spike protein: Implications for viral infectivity, disease severity and vaccine design. Biochem. Biophys. Res. Commun. 2020, 538, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef]
- Weissman, D.; Alameh, M.-G.; de Silva, T.; Collini, P.; Hornsby, H.; Brown, R.; LaBranche, C.C.; Edwards, R.J.; Sutherland, L.; Santra, S.; et al. D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. Cell Host Microbe 2021, 29, 23–31.e4. [Google Scholar] [CrossRef]
- Collier, D.A.; De Marco, A.; Ferreira, I.A.T.M.; Meng, B.; Datir, R.P.; Walls, A.C.; Kemp, S.A.; Bassi, J.; Pinto, D.; Silacci-Fregni, C.; et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 2021, 593, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Arora, P.; Grob, R.; Seidel, A.; Hörnich, B.F.; Hahn, A.S.; Krüger, N.; Graichen, L.; Hofmann-Winkler, H.; Kempf, A.; et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021, 184, 2384–2393.e12. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Casner, R.G.; Nair, M.S.; Wang, M.; Yu, J.; Cerutti, G.; Liu, L.; Kwong, P.D.; Huang, Y.; Shapiro, L.; et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe 2021, 29, 747–751.e4. [Google Scholar] [CrossRef]
- Liu, C.; Ginn, H.M.; Dejnirattisai, W.; Supasa, P.; Wang, B.; Tuekprakhon, A.; Nutalai, R.; Zhou, D.; Mentzer, A.J.; Zhao, Y.; et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell 2021, 184, 4220–4236.e13. [Google Scholar] [CrossRef]
- Wall, E.C.; Wu, M.; Harvey, R.; Kelly, G.; Warchal, S.; Sawyer, C.; Daniels, R.; Hobson, P.; Hatipoglu, E.; Ngai, Y.; et al. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet 2021, 397, 2331–2333. [Google Scholar] [CrossRef]
- Anonymous. Vaccinations in United Kingdom. Available online: https://coronavirus.data.gov.uk/details/vaccinations (accessed on 5 September 2021).
- Elliot, P.; Haw, D.; Wang, H.; Eales, O.; Walters, C.E. REACT-1 Round 13 Final Report: Exponential Growth, High Prevalence of SARS-CoV-2 and Vaccine Effectiveness Associated with Delta Variant in England during May to July 2021. Available online: https://spiral.imperial.ac.uk/handle/10044/1/90800 (accessed on 6 September 2021).
- Flanagan, K.L.; MacIntyre, C.R.; McIntyre, P.B.; Nelson, M.R. SARS-CoV-2 Vaccines: Where Are We Now? J. Allergy Clin. Immunol. Pract. 2021, 9, 3535–3543. [Google Scholar] [CrossRef]
- Cevik, M.; Grubaugh, N.D.; Iwasaki, A.; Openshaw, P. COVID-19 vaccines: Keeping pace with SARS-CoV-2 variants. Cell 2021, 184, 5077–5081. [Google Scholar] [CrossRef]
- Abu-Raddad, L.J.; Chemaitelly, H.; Butt, A.A. Effectiveness of the BNT162b2 Covid-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N. Engl. J. Med. 2021, 385, 187–189. [Google Scholar] [CrossRef]
- Lopez Bernal, J.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G.; et al. Effectiveness of COVID-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021, 385, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Bueno, S.M.; Abarca, K.; González, P.A.; Gálvez, N.M.S.; Soto, J.A.; Duarte, L.F.; Schultz, B.M.; Pacheco, G.A.; González, L.A.; Vázquez, Y.; et al. Safety and Immunogenicity of an Inactivated Severe Acute Respiratory Syndrome Coronavirus 2 Vaccine in a Subgroup of Healthy Adults in Chile. Clin. Infect. Dis. 2021, ciab823. [Google Scholar] [CrossRef]
- Tanriover, M.D.; Doğanay, H.L.; Akova, M.; Güner, H.R.; Azap, A.; Akhan, S.; Köse, Ş.; Erdinç, F.Ş.; Akalın, E.H.; Tabak, Ö.F.; et al. Efficacy and safety of an inactivated whole-virion SARS-CoV-2 vaccine (CoronaVac): Interim results of a double-blind, randomised, placebo-controlled, phase 3 trial in Turkey. Lancet 2021, 398, 213–222. [Google Scholar] [CrossRef]
- Al Kaabi, N.; Zhang, Y.; Xia, S.; Yang, Y.; Al Qahtani, M.M.; Abdulrazzaq, N.; Al Nusair, M.; Hassany, M.; Jawad, J.S.; Abdalla, J.; et al. Effect of 2 Inactivated SARS-CoV-2 Vaccines on Symptomatic COVID-19 Infection in Adults. JAMA 2021, 326, 35–45. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Marc, G.P.; Moreira, E.D.; Zerbini, C.; et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, M.N.; Minassian, A.M.; Ewer, K.J.; Flaxman, A.L.; Folegatti, P.M.; Owens, D.R.; Voysey, M.; Aley, P.K.; Angus, B.; Babbage, G.; et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): A single-blind, randomised, controlled, phase 2/3 trial. Lancet 2020, 396, 1979–1993. [Google Scholar] [CrossRef]
- Madhi, S.A.; Baillie, V.; Cutland, C.L.; Voysey, M.; Koen, A.L.; Fairlie, L.; Padayachee, S.D.; Dheda, K.; Barnabas, S.L.; Bhorat, Q.E.; et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021, 384, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cárdenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. N. Engl. J. Med. 2021, 384, 2187–2201. [Google Scholar] [CrossRef]
- Logunov, D.Y.; Dolzhikova, I.V.; Shcheblyakov, D.V.; Tukhvatulin, A.I.; Zubkova, O.V.; Dzharullaeva, A.S.; Kovyrshina, A.V.; Lubenets, N.L.; Grousova, D.M.; Erokhova, A.S.; et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: An interim analysis of a randomised controlled phase 3 trial in Russia. Lancet 2021, 397, 671–681. [Google Scholar] [CrossRef]
- Faillie, J.-L.; Montastruc, F.; Montastruc, J.-L.; Pariente, A. Pharmacoepidemiology and its input to pharmacovigilance. Therapies 2016, 71, 211–216. [Google Scholar] [CrossRef]
- AlOmar, M.; Tawfiq, A.; Hassan, N.; Palaian, S. Post marketing surveillance of suspected adverse drug reactions through spontaneous reporting: Current status, challenges and the future. Ther. Adv. Drug Saf. 2020, 11. [Google Scholar] [CrossRef]
- Ibrahim, H.; Saad, A.; Abdo, A.; Eldin, A.S. Mining association patterns of drug-interactions using post marketing FDA’s spontaneous reporting data. J. Biomed. Inform. 2016, 60, 294–308. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Liu, M.; Hu, Y.; Melton, B.L.; Matheny, M.; Xu, H.; Duan, L.; Waitman, L.R. Identification of adverse drug-drug interactions through causal association rule discovery from spontaneous adverse event reports. Artif. Intell. Med. 2017, 76, 7–15. [Google Scholar] [CrossRef] [Green Version]
- Farcaş, A.; Măhălean, A.; Bulik, N.B.; Leucuta, D.; Mogoșan, C. New safety signals assessed by the Pharmacovigilance Risk Assessment Committee at EU level in 2014–2017. Expert Rev. Clin. Pharmacol. 2018, 11, 1045–1051. [Google Scholar] [CrossRef]
- BPOM. Pengawalan Keamanan, Khasiat, dan Mutu Vaksin COVID-19. Available online: https://www.pom.go.id/new/view/more/pers/572/Pengawalan-Keamanan--Khasiat--dan-Mutu-Vaksin-COVID-19.html (accessed on 6 September 2021).
- Bahri, P.; Tsintis, P. Pharmacovigilance-related topics at the level of the International Conference on Harmonisation (ICH). Pharmacoepidemiol. Drug Saf. 2004, 14, 377–387. [Google Scholar] [CrossRef] [PubMed]
- WHO. What Is VigiBase? Available online: https://www.who-umc.org/vigibase/vigibase/ (accessed on 6 September 2021).
- Ontario. COVID-19 Vaccine Surveillance Plan. Available online: https://www.health.gov.on.ca/en/pro/programs/publichealth/coronavirus/docs/vaccine/COVID-19_vaccine_surveillance_plan.pdf (accessed on 20 November 2021).
- UK Health Security Agency. COVID-19 Vaccine Surveillance Report Week 42; UK Health Security Agency: London, UK, 2021. [Google Scholar]
- England, P.H. COVID-19: Vaccine Surveillance Strategy. Available online: https://www.gov.uk/government/publications/covid-19-vaccine-surveillance-strategy (accessed on 6 September 2021).
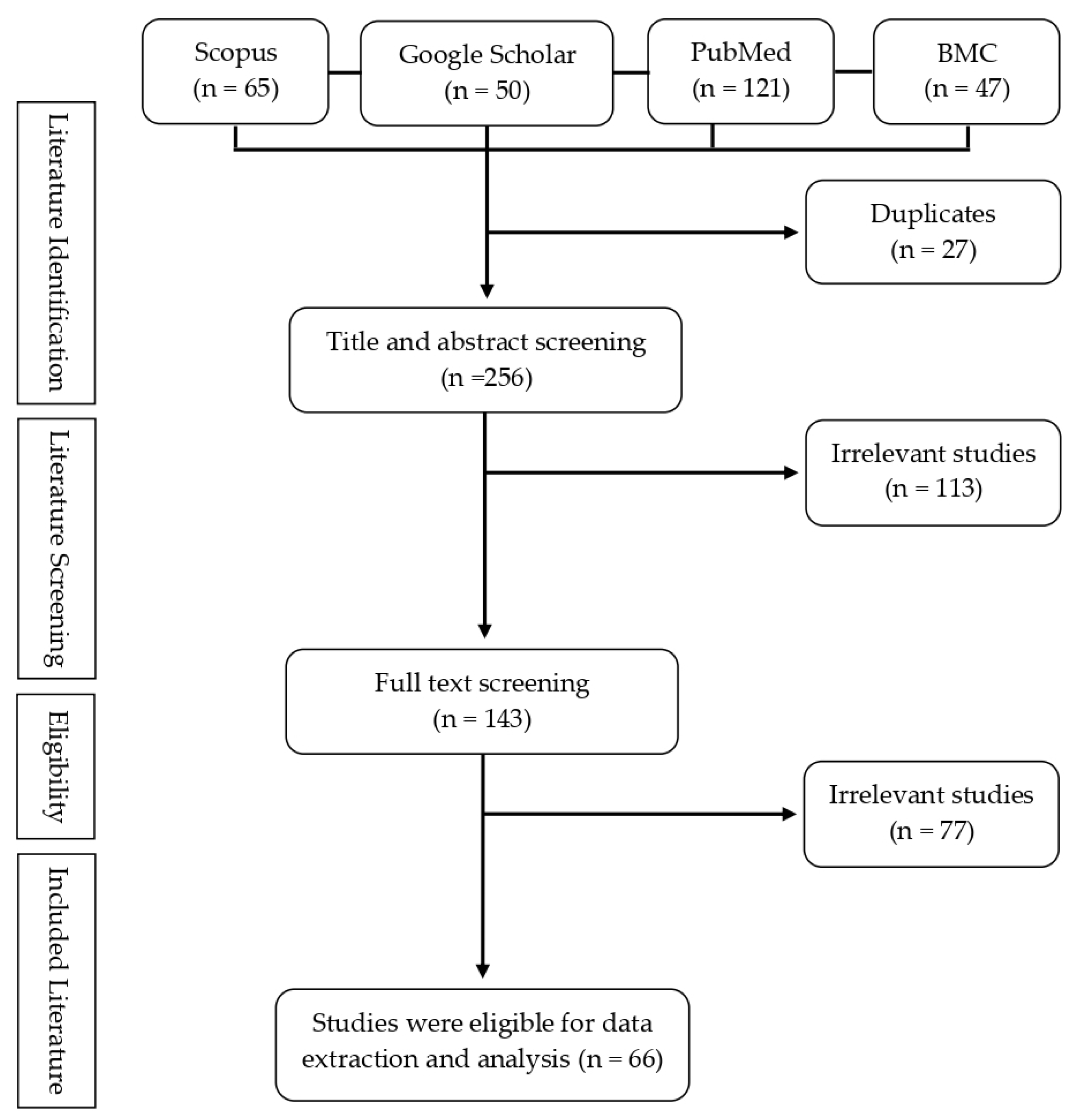
| Screening Stages | Questions | Screening Outcome |
|---|---|---|
| Title and abstract screening |
| Studies are included if they satisfy all questions |
| Full-text screening |
| Studies are included if they satisfy at least two screening questions |
| Vaccine Platform | Vaccine Developers | Approval Status (Number of Countries) | Efficacy (%) (Number of Participants) | Effectiveness to Reduce Infection (%) | |||
|---|---|---|---|---|---|---|---|
| Alpha | Beta | Delta | Gamma | ||||
| Inactivated Virus-Based Vaccine | Sinovac (CoronaVac) | 49 | 82.22–84.44 (18–59 years old) and 62.69–70.37 (>60 years old) from 434 participants [57]; 83.5 from 10,214 participants (6646 vaccinated, 3470 placebo) [58]; 65.30 (1620 participants) [30] | n/a | 65.9 [54] | n/a | 65.9 [54] |
| Sinopharm (BBIBP-CorV) | 77 | 78.1 (40,382 participats) [59] | – | – | – | – | |
| mRNA-Based Vaccine | Pfizer-BioNTech (BNT162b2) | 113 | 95 (18,198 vaccinated, 18,325 Placebo) [60] | 89.5 (50/19,354 vaccinated/unvaccinated PCR-positive) and (465/15,939 vaccinated/unvaccinated PCR-negative) [55] | 75 (179/19,396 vaccinated/unvaccinated PCR-positive) and (698/18,877 vaccinated/unvaccinated PCR-negative) [55] | 88 (122 case/15,749 control) after second dose [56] | 85 [54] |
| Moderna (mRNA-1273) | 77 | 94.1 (15,181 vaccinated, 15,170 placebo) [61] | 91 [54] | 78 [54] | 70 [54] | 78 [54] | |
| Viral Vector-Based Vaccine | AstraZeneca-Oxford (ChAdOx1-S) | 172 | 70.4 (5807 vaccinated, 5829 placebo) [62] | 74.5 (94 case/8244 control) after second dose [56] | 75.4 (3/944 vaccinated, 12/938 placebo) [63] | 67.0 (218 case/8244 control) after second dose [56] | <70 [54] |
| Janssen (Ad26.COV2.S) | 79 | 66.9 (19,630 vaccinated, 19,691 placebo) [64] | – | 52 [54] | – | 52 [54] | |
| Gamaleya (Sputnik V) | 74 | 91.6 (14,964 vaccinated, 4902 placebo) [65] | – | – | – | – | |
| CanSino (Convidecia) | 9 | 65.7 [53] | – | – | – | – | |
| Protein Sub-unit-Based Vaccine | Novavax (NVX-CoV-2373) | Not Approved | 89.7 (15,187 participant) [40] | 86.3 (15,187 participant) [40] | 60 [54] | 60 [54] | 60 [54] |
| Vaccine Developers | Effectiveness to Reduce Severe Disease/Death (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Alpha | Participants | Beta | Participants | Delta | Participants | Gamma | Participants | |
| Sinovac (CoronaVac) | n/a | n/a | 87.5 | n/a | n/a | n/a | 87.5 | n/a |
| Pfizer-BioNTech (BNT162b2) | 54.1 (after one dose) [55] 100.0 (after second dose) [55] | PCR-positive: 30 vaccinated/468 unvaccinated PCR-negative: 61/437 PCR-positive: 0/401 PCR negative: 20/381 | 100.0 (after second dose) [55] | PCR-positive: 0/300 PCR-negative: 14/246 | 95 | n/a | 98 | n/a |
| Moderna (mRNA-1273) | 94 | n/a | 94 | n/a | 96 | n/a | 94 | n/a |
| AstraZeneca-Oxford (ChAdOx1-S) | 95 | n/a | n/a | n/a | 95 | n/a | n/a | n/a |
| Janssen (Ad26.COV2.S) | n/a | n/a | 65–66 (hospitalization) 91–95 (mortality) | n/a | 71 | n/a | 65–66 (hospitalization) 91–95 (mortality) | n/a |
| Outcome | Vaccine Effectiveness * | ||
|---|---|---|---|
| Pfizer-BioNTech Cominarty | AstraZeneca Vaxzevria | Moderna Spikevax | |
| Infection | 75–85% | 60–70% | – |
| Symptomatic disease | 80–90% | 65–75% | 90–99% |
| Hospitalization | 95–99% | 90–99% | 95–99% |
| Mortality | 90–99% | 90–95% | – |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hafiz, I.; Illian, D.N.; Meila, O.; Utomo, A.R.H.; Susilowati, A.; Susetya, I.E.; Desrita, D.; Siregar, G.A.; Basyuni, M. Effectiveness and Efficacy of Vaccine on Mutated SARS-CoV-2 Virus and Post Vaccination Surveillance: A Narrative Review. Vaccines 2022, 10, 82. https://doi.org/10.3390/vaccines10010082
Hafiz I, Illian DN, Meila O, Utomo ARH, Susilowati A, Susetya IE, Desrita D, Siregar GA, Basyuni M. Effectiveness and Efficacy of Vaccine on Mutated SARS-CoV-2 Virus and Post Vaccination Surveillance: A Narrative Review. Vaccines. 2022; 10(1):82. https://doi.org/10.3390/vaccines10010082
Chicago/Turabian StyleHafiz, Ihsanul, Didi Nurhadi Illian, Okpri Meila, Ahmad Rusdan Handoyo Utomo, Arida Susilowati, Ipanna Enggar Susetya, Desrita Desrita, Gontar Alamsyah Siregar, and Mohammad Basyuni. 2022. "Effectiveness and Efficacy of Vaccine on Mutated SARS-CoV-2 Virus and Post Vaccination Surveillance: A Narrative Review" Vaccines 10, no. 1: 82. https://doi.org/10.3390/vaccines10010082








