Acetylsalicylic Acid and Salicylic Acid Inhibit SARS-CoV-2 Replication in Precision-Cut Lung Slices
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells Lines, Compounds and Viruses
2.2. Cellular Toxicity
2.3. RNA Purification and Determination of Viral Genome Copies
2.4. Human Precision-Cut Lung Slices
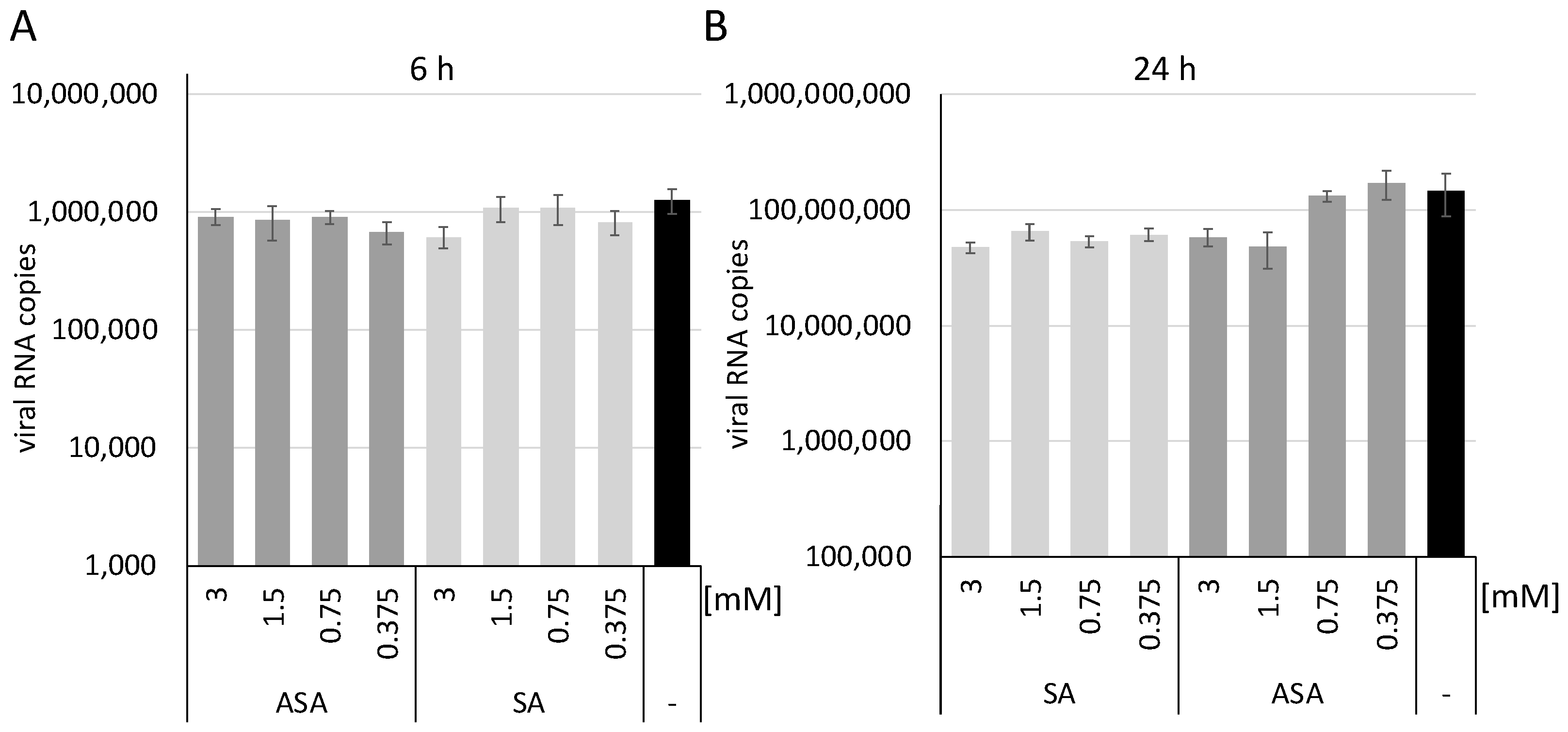
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- Khoo, S.H.; Fitzgerald, R.; Fletcher, T.; Ewings, S.; Jaki, T.; Lyon, R.; Downs, N.; Walker, L.; Tansley-Hancock, O.; Greenhalf, W.; et al. Optimal dose and safety of molnupiravir in patients with early SARS-CoV-2: A Phase I, open-label, dose-escalating, randomized controlled study. J. Antimicrob. Chemother. 2021, 76, 3286–3295. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Goldman, J.D.; Lye, D.C.B.; Hui, D.S.; Marks, K.M.; Bruno, R.; Montejano, R.; Spinner, C.D.; Galli, M.; Ahn, M.-Y.; Nahass, R.G.; et al. Remdesivir for 5 or 10 Days in Patients with Severe COVID-19. N. Engl. J. Med. 2020, 383, 1827–1837. [Google Scholar] [CrossRef] [PubMed]
- Zimniak, M.; Kirschner, L.; Hilpert, H.; Geiger, N.; Danov, O.; Oberwinkler, H.; Steinke, M.; Sewald, K.; Seibel, J.; Bodem, J. The serotonin reuptake inhibitor Fluoxetine inhibits SARS-CoV-2 in human lung tissue. Sci. Rep. 2021, 11, 5890. [Google Scholar] [CrossRef] [PubMed]
- Oskotsky, T.; Maric, I.; Tang, A.; Oskotsky, B.; Wong, R.J.; Aghaeepour, N.; Sirota, M.; Stevenson, D.K. Mortality Risk Among Patients With COVID-19 Prescribed Selective Serotonin Reuptake Inhibitor Antidepressants. JAMA Netw. Open 2021, 4, e2133090. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Mosbauer, K.; Hofmann-Winkler, H.; Kaul, A.; Kleine-Weber, H.; Kruger, N.; Gassen, N.C.; Muller, M.A.; Drosten, C.; Pohlmann, S. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature 2020, 585, 588–590. [Google Scholar] [CrossRef]
- Geiger, N.; Kersting, L.; Schlegel, J.; Stelz, L.; Fahr, S.; Diesendorf, V.; Roll, V.; Sostmann, M.; Konig, E.M.; Reinhard, S.; et al. The Acid Ceramidase Is a SARS-CoV-2 Host Factor. Cells 2022, 11, 2532. [Google Scholar] [CrossRef]
- Glatthaar-Saalmuller, B.; Mair, K.H.; Saalmuller, A. Antiviral activity of aspirin against RNA viruses of the respiratory tract-an in vitro study. Influenza Respir. Viruses 2017, 11, 85–92. [Google Scholar] [CrossRef]
- Di Bella, S.; Luzzati, R.; Principe, L.; Zerbato, V.; Meroni, E.; Giuffre, M.; Croce, L.S.; Merlo, M.; Perotto, M.; Dolso, E.; et al. Aspirin and Infection: A Narrative Review. Biomedicines 2022, 10, 263. [Google Scholar] [CrossRef]
- Liu, Q.; Huang, N.; Li, A.; Zhou, Y.; Liang, L.; Song, X.; Yang, Z.; Zhou, X. Effect of low-dose aspirin on mortality and viral duration of the hospitalized adults with COVID-19. Medicine 2021, 100, e24544. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.H.; Khanna, A.K.; Kethireddy, S.; Yamane, D.; Levine, A.; Jackson, A.M.; McCurdy, M.T.; Tabatabai, A.; Kumar, G.; Park, P.; et al. Aspirin Use Is Associated With Decreased Mechanical Ventilation, Intensive Care Unit Admission, and In-Hospital Mortality in Hospitalized Patients with Coronavirus Disease 2019. Anesth. Analg. 2021, 132, 930–941. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Han, K.; Blair, R.; Kenst, K.; Qin, Z.; Upcin, B.; Worsdorfer, P.; Midkiff, C.C.; Mudd, J.; Belyaeva, E.; et al. SARS-CoV-2 Infects Endothelial Cells In Vivo and In Vitro. Front. Cell. Infect. Microbiol. 2021, 11, 701278. [Google Scholar] [CrossRef] [PubMed]
- Avota, E.; Bodem, J.; Chithelen, J.; Mandasari, P.; Beyersdorf, N.; Schneider-Schaulies, J. The Manifold Roles of Sphingolipids in Viral Infections. Front. Physiol. 2021, 12, 715527. [Google Scholar] [CrossRef] [PubMed]
- Perlman, S.; Masters, P.S. Coronaviridae: The Viruses and Their Replication. In Fields Virology; Howley, P.M., Knipe, D.M., Eds.; Wolters Kluwer: Alphen aan den Rijn, The Netherlands, 2021; Volume 1, pp. 410–448. [Google Scholar]
- Schmidt, N.; Lareau, C.A.; Keshishian, H.; Ganskih, S.; Schneider, C.; Hennig, T.; Melanson, R.; Werner, S.; Wei, Y.; Zimmer, M.; et al. The SARS-CoV-2 RNA-protein interactome in infected human cells. Nat. Microbiol. 2021, 6, 339–353. [Google Scholar] [CrossRef]
- Neuhaus, V.; Danov, O.; Konzok, S.; Obernolte, H.; Dehmel, S.; Braubach, P.; Jonigk, D.; Fieguth, H.G.; Zardo, P.; Warnecke, G.; et al. Assessment of the Cytotoxic and Immunomodulatory Effects of Substances in Human Precision-cut Lung Slices. J. Vis. Exp. 2018, 135, e57042. [Google Scholar] [CrossRef]
- Maisonnasse, P.; Guedj, J.; Contreras, V.; Behillil, S.; Solas, C.; Marlin, R.; Naninck, T.; Pizzorno, A.; Lemaitre, J.; Goncalves, A.; et al. Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates. Nature 2020, 585, 584–587. [Google Scholar] [CrossRef]
- Kumar, N.; Xin, Z.T.; Liang, Y.; Ly, H.; Liang, Y. NF-kappaB signaling differentially regulates influenza virus RNA synthesis. J. Virol. 2008, 82, 9880–9889. [Google Scholar] [CrossRef]
- Jancso, G.; Cserepes, B.; Gasz, B.; Benko, L.; Ferencz, A.; Borsiczky, B.; Lantos, J.; Dureja, A.; Kiss, K.; Szeberenyi, J.; et al. Effect of acetylsalicylic acid on nuclear factor-kappaB activation and on late preconditioning against infarction in the myocardium. J. Cardiovasc. Pharm. 2005, 46, 295–301. [Google Scholar] [CrossRef]
- Mazur, I.; Wurzer, W.J.; Ehrhardt, C.; Pleschka, S.; Puthavathana, P.; Silberzahn, T.; Wolff, T.; Planz, O.; Ludwig, S. Acetylsalicylic acid (ASA) blocks influenza virus propagation via its NF-kappaB-inhibiting activity. Cell Microbiol. 2007, 9, 1683–1694. [Google Scholar] [CrossRef]
- Nilsson-Payant, B.E.; Uhl, S.; Grimont, A.; Doane, A.S.; Cohen, P.; Patel, R.S.; Higgins, C.A.; Acklin, J.A.; Bram, Y.; Chandar, V.; et al. The NF-kappaB Transcriptional Footprint Is Essential for SARS-CoV-2 Replication. J. Virol. 2021, 95, e0125721. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geiger, N.; König, E.-M.; Oberwinkler, H.; Roll, V.; Diesendorf, V.; Fähr, S.; Obernolte, H.; Sewald, K.; Wronski, S.; Steinke, M.; et al. Acetylsalicylic Acid and Salicylic Acid Inhibit SARS-CoV-2 Replication in Precision-Cut Lung Slices. Vaccines 2022, 10, 1619. https://doi.org/10.3390/vaccines10101619
Geiger N, König E-M, Oberwinkler H, Roll V, Diesendorf V, Fähr S, Obernolte H, Sewald K, Wronski S, Steinke M, et al. Acetylsalicylic Acid and Salicylic Acid Inhibit SARS-CoV-2 Replication in Precision-Cut Lung Slices. Vaccines. 2022; 10(10):1619. https://doi.org/10.3390/vaccines10101619
Chicago/Turabian StyleGeiger, Nina, Eva-Maria König, Heike Oberwinkler, Valeria Roll, Viktoria Diesendorf, Sofie Fähr, Helena Obernolte, Katherina Sewald, Sabine Wronski, Maria Steinke, and et al. 2022. "Acetylsalicylic Acid and Salicylic Acid Inhibit SARS-CoV-2 Replication in Precision-Cut Lung Slices" Vaccines 10, no. 10: 1619. https://doi.org/10.3390/vaccines10101619
APA StyleGeiger, N., König, E.-M., Oberwinkler, H., Roll, V., Diesendorf, V., Fähr, S., Obernolte, H., Sewald, K., Wronski, S., Steinke, M., & Bodem, J. (2022). Acetylsalicylic Acid and Salicylic Acid Inhibit SARS-CoV-2 Replication in Precision-Cut Lung Slices. Vaccines, 10(10), 1619. https://doi.org/10.3390/vaccines10101619




