Antibiotic-Free Nanoplasmids as Promising Alternatives for Conventional DNA Vectors
Abstract
:1. Background
2. Methods
2.1. Recombinant Plasmid Preparation
2.2. Cell Line Culture
2.3. Electroporation for Plasmid Transfer into Host Cells
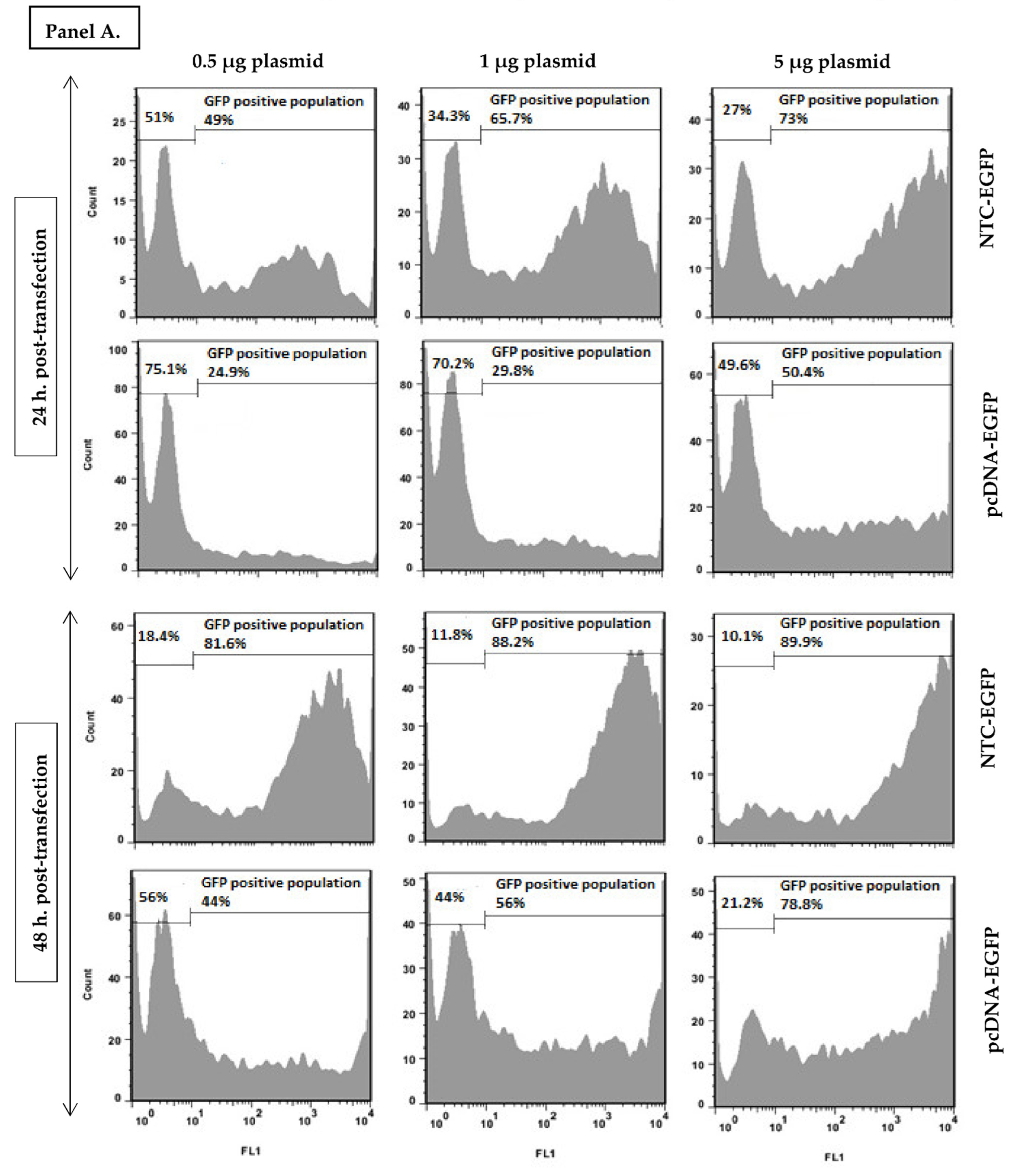
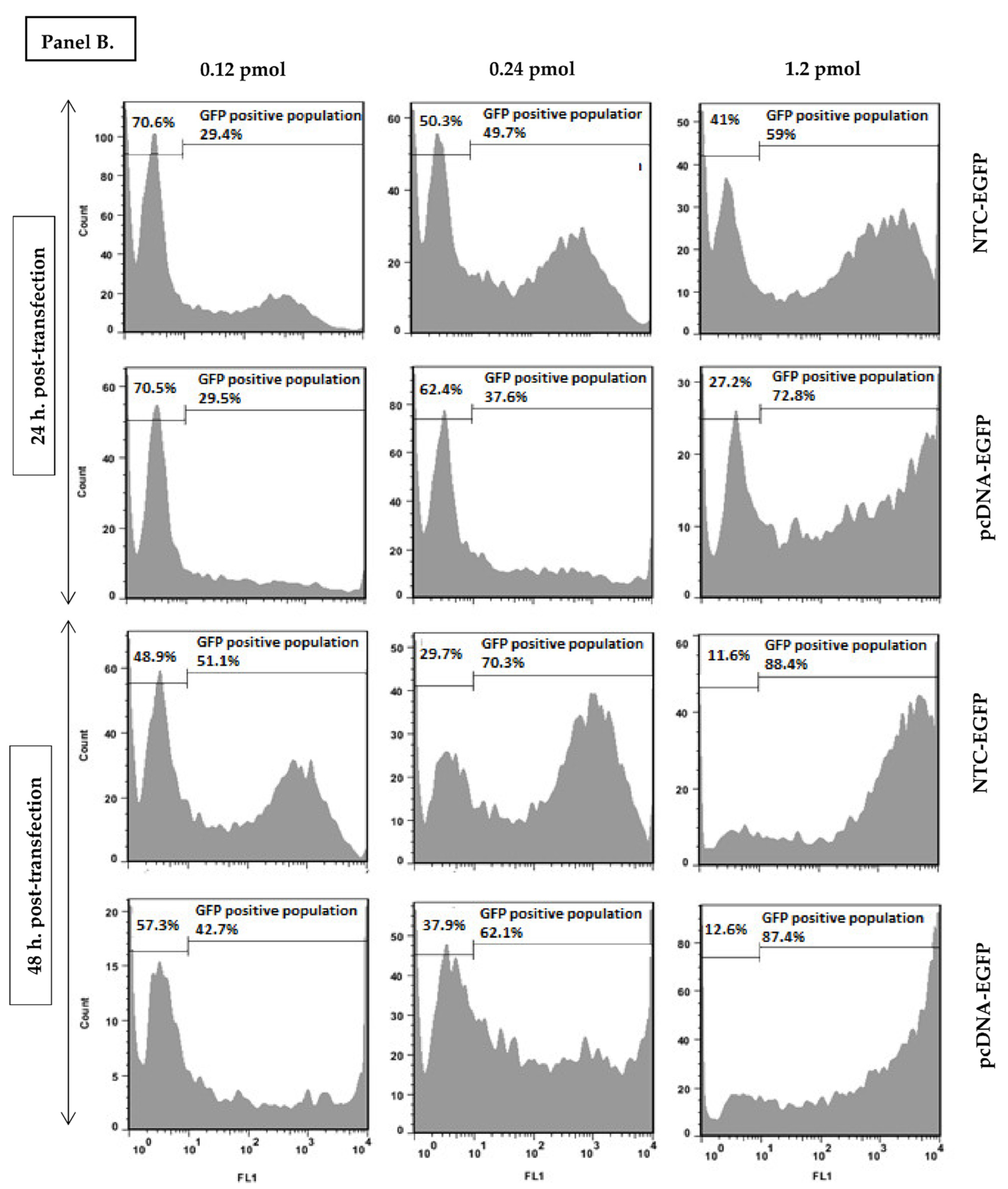
2.4. Quantification of EGFP Expressing Cells with Flow Cytometry
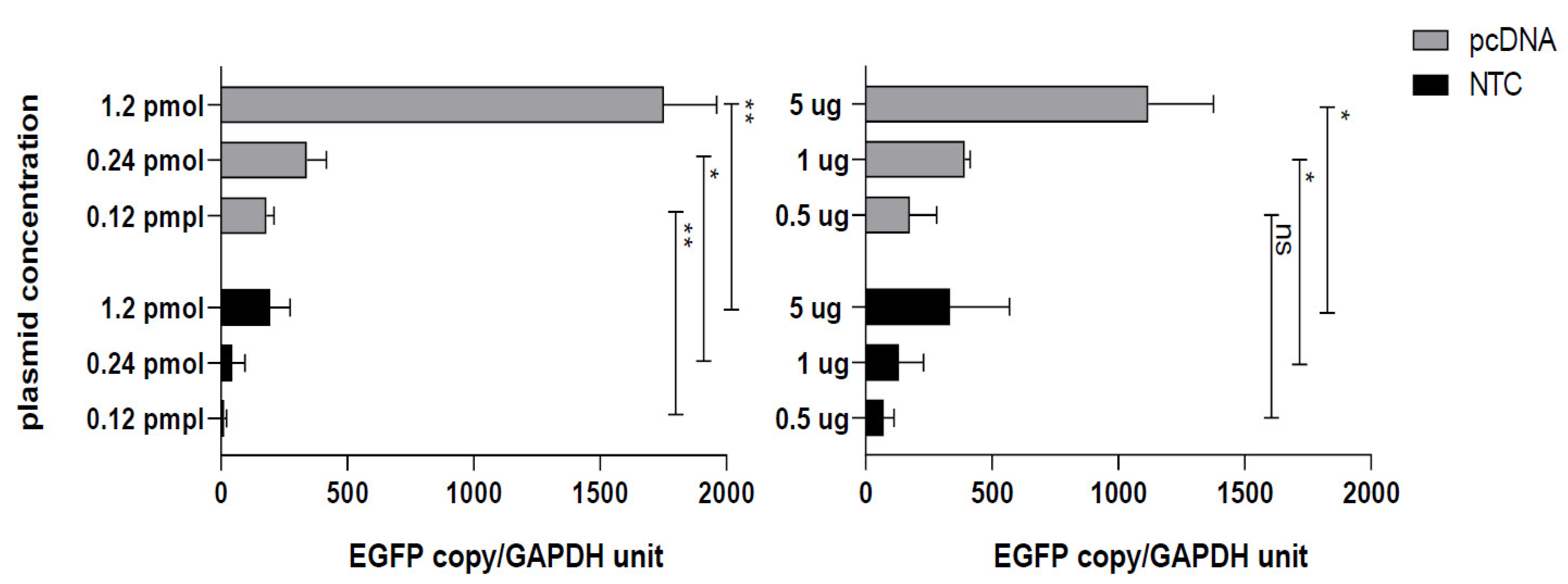
2.5. Quantitative Real Time PCR for Quantification of EGFP Copies in Transfected Cells
2.6. Statistical Analysis
3. Results a
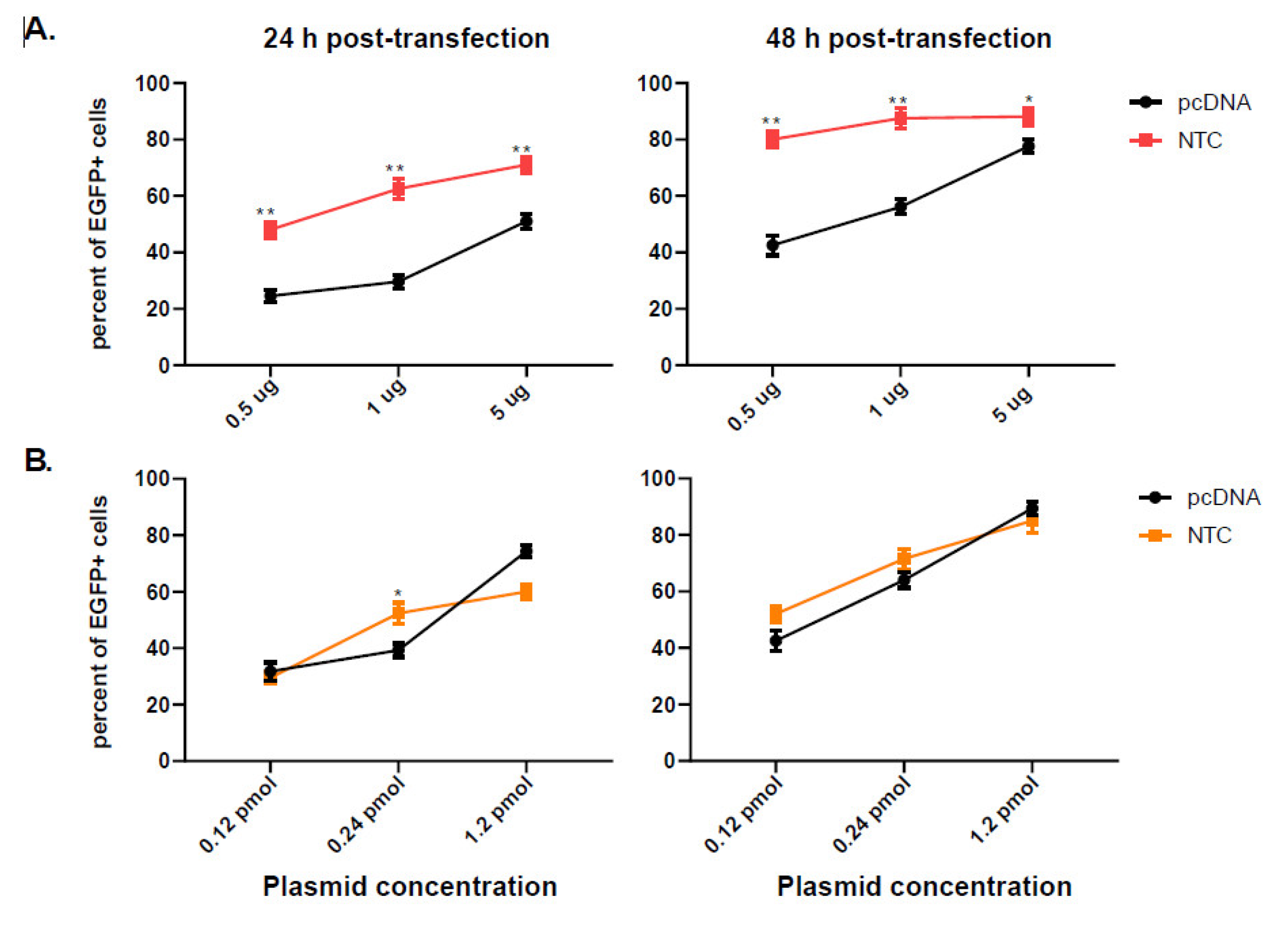
3.1. Nanoplasmid Efficiently Increases Protein Expression
3.2. Conventional CMV Based pcDNA Generates More EGFP mRNA Copies
4. Discussion
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hasson, S.S.A.A.; Al-Busaidi, J.K.Z.; Sallam, T.A. The past, current and future trends in DNA vaccine immunisations. Asian Pac. J. Trop. Biomed. 2015, 5, 344–353. [Google Scholar] [CrossRef] [Green Version]
- Gurunathan, S.; Wu, C.Y.; Freidag, B.L.; Seder, R.A. DNA vaccines: A key for inducing long-term cellular immunity. Curr. Opin. Immunol. 2000, 12, 442–447. [Google Scholar] [CrossRef]
- Jakob, T.; Walker, P.S.; Krieg, A.M.; Udey, M.C.; Vogel, J.C. Activation of cutaneous dendritic cells by CpG-containing oligodeoxynucleotides: A role for dendritic cells in the augmentation of Th1 responses by immunostimulatory DNA. J. Immunol. 1998, 161, 3042–3049. [Google Scholar] [PubMed]
- Sparwasser, T.; Koch, E.S.; Vabulas, R.M.; Heeg, K.; Lipford, G.B.; Ellwart, J.W.; Wagner, H. Bacterial DNA and immunostimulatory CpG oligonucleotides trigger maturation and activation of murine dendritic cells. Eur. J. Immunol. 1998, 28, 2045–2054. [Google Scholar] [CrossRef]
- Kayraklioglu, N.; Horuluoglu, B.; Klinman, D.M. CpG Oligonucleotides as Vaccine Adjuvants. Methods Mol. Biol. 2021, 2197, 51–85. [Google Scholar]
- Wolff, J.A.; Ludtke, J.J.; Acsadi, G.; Williams, P.; Jani, A. Long-term persistence of plasmid DNA and foreign gene expression in mouse muscle. Hum. Mol. Genet. 1992, 1, 363–369. [Google Scholar] [CrossRef]
- Suschak, J.J.; Williams, J.A.; Schmaljohn, C.S. Advancements in DNA vaccine vectors, non-mechanical delivery methods, and molecular adjuvants to increase immunogenicity. Hum. Vaccin. Immunother. 2017, 13, 2837–2848. [Google Scholar] [CrossRef] [Green Version]
- Mignon, C.; Sodoyer, R.; Werle, B. Antibiotic-free selection in biotherapeutics: Now and forever. Pathogens 2015, 4, 157–181. [Google Scholar] [CrossRef] [Green Version]
- Vandermeulen, G.; Marie, C.; Scherman, D.; Préat, V. New generation of plasmid backbones devoid of antibiotic resistance marker for gene therapy trials. Mol. Ther. J. Am. Soc. Gene Ther. 2011, 19, 1942–1949. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, P.H.; Mairhofer, J. Marker-free plasmids for biotechnological applications—Implications and perspectives. Trends Biotechnol. 2013, 31, 539–547. [Google Scholar] [CrossRef]
- Williams, J.A. Vector Design for Improved DNA Vaccine Efficacy, Safety and Production. Vaccines 2013, 1, 225–249. [Google Scholar] [CrossRef] [PubMed]
- Pastor, M.; Quiviger, M.; Pailloux, J.; Scherman, D.; Marie, C. Reduced Heterochromatin Formation on the pFAR4 Miniplasmid Allows Sustained Transgene Expression in the Mouse Liver. Mol. Ther. Nucleic Acids 2020, 21, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Schakowski, F.; Gorschlüter, M.; Junghans, C.; Schroff, M.; Buttgereit, P.; Ziske, C.; Schöttker, B.; König-Merediz, S.A.; Sauerbruch, T.; Wittig, B.; et al. A novel minimal-size vector (MIDGE) improves transgene expression in colon carcinoma cells and avoids transfection of undesired DNA. Mol. Ther. J. Am. Soc. Gene Therapy. 2001, 3, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Fuertes, L.; Perez-Jimenez, E.; Vila-Coro, A.J.; Sack, F.; Moreno, S.; Konig, S.A.; Junghans, C.; Wittig, B.; Timon, M.; Esteban, M. DNA vaccination with linear minimalistic (MIDGE) vectors confers protection against Leishmania major infection in mice. Vaccine 2002, 21, 247–257. [Google Scholar] [CrossRef]
- Das, S.; Freier, A.; Boussoffara, T.; Das, S.; Oswald, D.; Losch, F.O.; Selka, M.; Sacerdoti-Sierra, N.; Schönian, G.; Wiesmüller, K.H. Modular multiantigen T cell epitope-enriched DNA vaccine against human leishmaniasis. Sci. Transl. Med. 2014, 6, 234ra56. [Google Scholar] [CrossRef]
- Riede, O.; Seifert, K.; Oswald, D.; Endmann, A.; Hock, C.; Winkler, A.; Salguero, F.J.; Schroff, M.; Croft, S.L.; Juhls, C. Preclinical safety and tolerability of a repeatedly administered human leishmaniasis DNA vaccine. Gene Ther. 2015, 22, 628–635. [Google Scholar] [CrossRef] [Green Version]
- Hardee, C.L.; Arévalo-Soliz, L.M.; Hornstein, B.D.; Zechiedrich, L. Advances in non-viral DNA vectors for gene therapy. Genes 2017, 8, 65. [Google Scholar] [CrossRef] [Green Version]
- Schleef, M.; Schirmbeck, R.; Reiser, M.; Michel, M.L.; Schmeer, M. Minicircle: Next Generation DNA Vectors for Vaccination. Methods Mol. Biol. 2015, 1317, 327–339. [Google Scholar]
- Munye, M.M.; Tagalakis, A.D.; Barnes, J.L.; Brown, R.E.; McAnulty, R.J.; Howe, S.J.; Hart, S.L. Minicircle DNA Provides Enhanced and Prolonged Transgene Expression Following Airway Gene Transfer. Sci. Rep. 2016, 6, 23125. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Gao, X.; Xu, K.; Wang, J.; Huang, H.; Shi, C.; Yang, W.; Kang, Y.; Curtiss, R., III; Yang, G.; et al. A Novel Cre Recombinase-Mediated In Vivo Minicircle DNA (CRIM) Vaccine Provides Partial Protection against Newcastle Disease Virus. Appl. Environ. Microbiol. 2019, 85, e00407-19. [Google Scholar] [CrossRef] [Green Version]
- Suschak, J.J.; Dupuy, L.C.; Shoemaker, C.J.; Six, C.; Kwilas, S.A.; Spik, K.W.; Williams, J.A.; Schmaljohn, C.S. Nanoplasmid Vectors Co-expressing Innate Immune Agonists Enhance DNA Vaccines for Venezuelan Equine Encephalitis Virus and Ebola Virus. Mol. Ther. Methods Clin. Dev. 2020, 17, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Borggren, M.; Nielsen, J.; Bragstad, K.; Karlsson, I.; Krog, J.S.; Williams, J.A.; Fomsgaard, A. Vector optimization and needle-free intradermal application of a broadly protective polyvalent influenza A DNA vaccine for pigs and humans. Hum. Vaccines Immunother. 2015, 11, 1983–1990. [Google Scholar] [CrossRef] [PubMed]
- Luke, J.M.; Vincent, J.M.; Du, S.X.; Gerdemann, U.; Leen, A.M.; Whalen, R.G.; Hodgson, C.P.; Williams, J.A. Improved antibiotic-free plasmid vector design by incorporation of transient expression enhancers. Gene Ther. 2011, 18, 334–343. [Google Scholar] [CrossRef]
- Barouch, D.H.; Yang, Z.Y.; Kong, W.P.; Korioth-Schmitz, B.; Sumida, S.M.; Truitt, D.M.; Kishko, M.G.; Arthur, J.C.; Miura, A.; Mascola, J.R. A human T-cell leukemia virus type 1 regulatory element enhances the immunogenicity of human immunodeficiency virus type 1 DNA vaccines in mice and nonhuman primates. J. Virol. 2005, 79, 8828–8834. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seyed, N.; Zahedifard, F.; Habibzadeh, S.; Yousefi, R.; Lajevardi, M.S.; Gholami, E.; Rafati, S. Antibiotic-Free Nanoplasmids as Promising Alternatives for Conventional DNA Vectors. Vaccines 2022, 10, 1710. https://doi.org/10.3390/vaccines10101710
Seyed N, Zahedifard F, Habibzadeh S, Yousefi R, Lajevardi MS, Gholami E, Rafati S. Antibiotic-Free Nanoplasmids as Promising Alternatives for Conventional DNA Vectors. Vaccines. 2022; 10(10):1710. https://doi.org/10.3390/vaccines10101710
Chicago/Turabian StyleSeyed, Negar, Farnaz Zahedifard, Sima Habibzadeh, Roya Yousefi, Mahya Sadat Lajevardi, Elham Gholami, and Sima Rafati. 2022. "Antibiotic-Free Nanoplasmids as Promising Alternatives for Conventional DNA Vectors" Vaccines 10, no. 10: 1710. https://doi.org/10.3390/vaccines10101710



