1. Introduction
COVID-19 is a disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It emerged in December 2019 in Wuhan and since then has spread around the globe causing a pandemic that had devastating health and economic consequences worldwide [
1,
2]. An enormous effort made by the scientific community resulted in more than 300 new vaccine candidates in less than a year since the outbreak, some of them being approved for emergency use [
3], as well as the development of diagnosis methods for its detection [
4] and treatment [
5]. As of November 2021, more than 7 billion doses had been administrated [
6], with an associated significant reduction of transmission and mortality among vaccinated populations [
7]. Vaccination rollout offers a promising avenue for the pandemic and sanitary restrictions to come to an end. However, there are still some questions left to answer, like how long the immune memory lasts, the protective effect that current approved vaccines generate against emerging SARS-CoV-2 variants, or if it is possible to generate fully prophylactic vaccines against this new coronavirus [
8].
FDA- and EMA-approved COVID-19 vaccines can be classified into mRNA, adenovirus-based or recombinant. The first group delivers mRNA into dendritic cells (DCs) using a lipid nanoparticle (LNP) as a carrier. The second one delivers DNA into DCs using a non-replicating recombinant adenovirus vector. Both strategies result in DCs producing the genetically encoded SARS-CoV-2 spike (S) surface glycoprotein and presenting it on their own membrane, where it is then recognized by the immune system cells [
9]. The third strategy uses a saponin-based nanoparticle to present a recombinantly produced and purified spike glycoprotein, as well [
10].
Virus-like particles (VLPs) are a promising, robust, safe, versatile and highly immunogenic approach that can be used to produce novel vaccines for emerging pandemics or diseases, like COVID-19. VLPs are supramolecular structures that closely mimic the native conformation of viruses without carrying genetic material (DNA or RNA), so they are unable to infect, replicate or integrate. They are generated by taking advantage of the intrinsic ability of some viral proteins to self-assemble when expressed in heterologous production platforms. Two VLP-based vaccines have been proven successful and licensed for HPB and HPV [
11,
12,
13]. VLPs can be functionalized to present pathogenic epitopes to generate immunity against diseases like dengue, influenza, etc. HIV-1 Gag VLPs have shown great potential for this purpose, since they are composed by a core of Gag molecules surrounded by a lipid bilayer, a membrane that can be further functionalized [
14]. When administrated, due to their tridimensional configuration, VLPs drain and traffic within the immune system, interacting with cells such as DCs, B cells, T cells and macrophages [
15,
16]. Several proteins present at the VLP membrane interact with DC pattern-recognition receptors leading to a strong adaptative immune response, while multimeric epitopes promote the cross-linking of B cell receptors to induce antibody production [
15,
16]. Consequently, VLPs had been shown to induce potent humoral and cellular immune responses, and, although the use of adjuvants improves VLP-based vaccines’ immunogenicity, their nature make adjuvant co-administration optional [
15]. Therefore, VLP vaccination constitutes a promising approach compared to currently available vaccine technologies: VLPs are safer to manufacture and administrate than inactivated or attenuated vaccines due to their lack of viral genetical material, while they also provide a more potent and effective immune response compared to proteic or subunit vaccines since VLPs present conformationally authentic viral epitopes [
17].
Gag VLPs share a similar particle diameter (~145 nm) with wild-type SARS-CoV-2 virus (~80 nm), making them a suitable platform for the expression of its epitopes in order to generate a new vaccine candidate for COVID-19 [
14,
18]. For this purpose, the SARS-CoV-2 spike (S) protein is a promising lead, since is the major structural protein anchored at the exterior of the membrane of native viruses, carries B cell and T cell epitopes and is the main target for neutralizing antibodies generated from natural infection that protect against viral infection and currently approved COVID-19 vaccines [
19,
20]. S protein monomers are 180 kDa and assemble to form trimeric units at the surface of native virions, giving them their characteristic crown-shape [
21]. They contain a variable receptor-binding domain (RBD), responsible for binding to angiotensin-converting enzyme 2 receptor (ACE-2), facilitating viral entry into target cells [
21]. As the disruption of the RBD-ACE2 interaction can block SARS-CoV-2 cell entry [
22], most of the reported neutralizing antibodies against SARS-CoV-2 bind to the RBD [
23].
VLPs can be produced in prokaryotic and eukaryotic heterologous expression systems depending on their nature and use. Mammalian cells constitute an attractive production platform for enveloped or multimeric VLPs due to their capacity to perform complex post-translational modifications. There are several mammalian cell platforms in which VLPs can be produced, such as HeLa, Vero, CAP, CHO or HEK293 [
13]. HEK293 can be cultured in suspension in bioreactors using chemically defined media free of animal components and can also be easily transfected. This makes them a good choice to produce Gag-based COVID-19 VLPs in large-scale bioreactors, in order to satisfy the needs for pre-clinical trials, clinical trials, and eventually large-scale production for its manufacture. For this purpose, a robust and scalable downstream process (DSP) needs to be implemented in order to obtain a high-purity vaccine product in its final buffer formulation. Finally, for the initial steps of proof of concept, HEK 293 cells transient transfection methodologies have been well established to provide a robust and fast approach to the generation of VLPs for testing, of special relevance when developing and comparing several candidates.
In the last decades and especially after the COVID-19 outbreak, different published works have focused on the generation of SARS-CoV and SARS-CoV-2 VLPs by the co-expression of the coronavirus S, M and E proteins [
24]. This work focuses on the production, purification and characterization of a potential COVID-19 vaccine candidate, based on HIV-1 Gag-based SARS-CoV-2 spike VLPs (from now on S-VLPs), a never-before reported approach to our knowledge. S-VLP production scale-up and its DSP have been achieved by HEK293 transient transfection in a 1 L bioreactor and a purification process consisting of two clarification steps, an ion-exchange affinity step and a size-exclusion polishing and buffer exchange step. The production process and the obtained S-VLPs have been studied and characterized in this work.
2. Materials and Methods
2.1. Cell Line, Media and Culture Conditions
The serum-free suspension-adapted HEK293 cell line (HEK293SF-3F6) was used, kindly provided by Dr. Amine Kamen from the Biotechnology Research Institute at the National Research Council of Canada (Montreal, Canada) and McGill University. This cell line was derived from a current good manufacturing practice (cGMP) master cell bank available for manufacturing of clinical material.
The medium used for HEK293 cellular growth was the chemically defined and free from animal components HyCell TransFx-H from HyClone (GE Healthcare, Chicago, IL, USA) supplemented with 4 mM GlutaMAX (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) and 0.1% Pluronic F-68 Non-ionic Surfactant (Gibco, Thermo Fisher Scientific, Waltham, MA, USA).
Suspension cell cultures were maintained routinely in exponential growth phase in 125mL or 1L disposable polycarbonate Erlenmeyer flasks with a vent cap (Corning, Tewksbury, MA, USA) in a LT-X Kuhner shaker (LT-X Kuhner, Birsfelden, Switzerland) shaking at 130 rpm, at 37 °C, 5% CO2 and 85% RH. Cell counts and viability determinations were performed using the NucleoCounter NC-300 automatic cell counter (Chemometec, Lillerød, Denmark) following the manufacturer’s instructions.
2.2. Plasmids and Transfection
2.2.1. Plasmid Expression Vectors
The pGag::eGFP plasmid codes for a codon-optimized Rev-independent HIV-1 Gag protein fused in frame to the enhanced GFP driven by the CMV enhancer and promoter. The plasmid from the NIH AIDS Reagent Program (Cat 11468) (Hermida-Matsumoto and Resh, 2000) was constructed by cloning the Gag sequence from pCMV55M1-10 (Schwartz et al., 1992) into the pEGFP-N1 plasmid (Clontech, Palo Alto, CA, USA).
The pSpike plasmid codes for a mammalian cell codon optimized nucleotide sequence coding for the spike protein of SARS-CoV-2 driven by the CAG enhancer and β-actin promoter. It was produced under HHSN272201400008C and obtained through BEI Resources, NIAID, NIH: Vector pCAGGS Containing the SARS-Related Coronavirus 2, Wuhan-Hu-1 spike Glycoprotein Gene, NR-52310.
pMock plasmid does not have any mammalian promoter or coding DNA sequence (CDS). It was constructed by the ligation of the pGag::eGFP backbone.
2.2.2. Plasmid Amplification and Purification
Plasmids were amplified in Escherichia coli DH5α strain grown in LB medium (Conda, Madrid, Spain) supplemented with kanamycin (10 µg/mL, Sigma, St. Louis, MO, USA) or ampicillin (100 µg/mL, Sigma, St. Louis, MO, USA) depending on the E. coli antibiotic resistance present on each plasmid. Plasmid purification was carried out using the Endofree Plasmid Mega kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
2.2.3. PEI-Mediated Transient Transfection
Exponentially growing HEK293 cells were passaged in order to have a cell density of 2·106 cells/mL at transfection time. PEIpro (Polyplus-transfection SA, Illkirch-Graffenstaden, France) was used as a transfection reagent. PEI-DNA complexes were formed under sterile conditions, by adding PEI to a plasmid DNA mixture diluted for a total DNA concentration of 1 µg/mL in fresh culture media (10% of the total volume of cell culture to be transfected). The mixture was incubated for 15 min at RT and then added to cell culture. The ratio between plasmids and transfection reagent was optimized using a Box–Behnken design of experiments and described in the next section.
2.3. Box–Behnken Design
A Box–Behnken design was used in order to define the optimal concentration for three independent variables in the cell transfection step: pGag::eGFP, pSpike and PEI. These variables were screened at three levels: a low level, coded as −1; a medium level, coded as 0; and a high level coded as +1, as indicated in
Table 1. Box–Behnken experimental results were fitted to a second-order polynomial equation described below by non-linear regression analysis:
where Y is the response (in this work, the percentage of cells expressing simultaneously Gag::eGFP and spike at 72 hpt); β
0 is the offset term; β
i is the linear coefficient; β
ii is the quadratic coefficient; β
ij the interaction coefficient, and X
i and X
j are the independent variables (pGag::eGFP, pSpike and PEI). The equation was used to predict the concentration of the independent variables in order to maximize the desired response. Three-dimensional response surface plots were generated using Design Expert version V8.0.6 software (Stat-Ease Inc., Minneapolis, MN, USA). Statistical analyses of the model were performed using Design Expert. The coefficient values corresponding to the generated response model are shown at
Table 1.
Control groups to be transfected with just one plasmid coding for a protein, were co-transfected with pMock plasmid in order to deliver the same gene copies of the protein being expressed in the other conditions. Transfections associated with Box–Behnken optimization studies, validation and bioreactor production were carried out following the later described plasmid and PEI concentrations. Expression was analyzed at 0, 24, 48 and 72 hpt.
2.4. Stirred Tank Reactor (STR) Bioprocess
A BioStat B Plus bioreactor (Sartorius Stedim Biotech, Göttingen, Germany) equipped with a 3-blade segment dual impeller with UP-DP configuration [
25] was used for HEK293 cell cultivation and production. The agitation was set at 200 rpm; the temperature was set at 37 ºC, and the pH was set at 7.1, controlled with CO
2 and NaHCO
3 (7.5%
w/v). Dissolved oxygen was controlled at 40% of air saturation by supplementing air by sparger at a constant flow of 0.1 L/min and additional pure oxygen when needed. HEK293 growing exponentially in disposable polycarbonate 1 L shake flasks (Corning, Tewksbury, MA, USA) were used to seed the bioreactor at 0.5·10
6 cells/mL in 1 L of working volume.
2.5. Sucrose Cushion Small-Scale Purification
Culture harvests were performed at 72 hpt and centrifuged at 10,000× g for 10 min, and the supernatant was stored at −80 °C for further analysis or stored at 4 °C for its purification in less than 24 h. The supernatants containing VLPs were placed on a 30% (w/v) sucrose cushion for ultracentrifugation at 31,000 rpm for 2 h at 4 °C. The supernatant was carefully discarded, and pellets were resuspended and placed on a new sucrose cushion for a second ultracentrifugation following the same protocol. The pellets were collected and resuspended in PBS.
2.6. Downstream Processing
2.6.1. Cell Harvest and Culture Supernatant Clarification
After 2 h of sedimentation, the culture medium from transfected cells was subjected to primary clarification using Supracap 50 V100 depth filter capsules (Pall Corporation, Port Washington, NY, USA) to remove cellular debris and other contaminants. For secondary clarification, the Supor EAV—Mini Kleenpak 20 filter capsules (Pall Corporation, Port Washington, NY, USA) was used. For both clarifications, a K2Ri pump (Repligen, Waltham, MA, USA) with MasterFlex 96410-13 silicon tubes (Cole-Parmer, Vernon Hills, IL, USA) connected to the filter inlet and outlet; and a pressure sensor (Cole-Parmer, Vernon Hills, IL, USA) connected to the filter inlet. The turbidity of the clarification samples was measured using a portable Eutech TN-100 turbidimeter (Thermo Fisher Scientific, Waltham, MA, USA).
2.6.2. Ion-exchange chromatography (IEX)
A prepacked 0.86 mL Mustang Q XT Acrodisc column (Pall Corporation, Port Washington, NY, USA) was used to capture the S-VLPs from the secondary clarification. Before loading, the column was pre-equilibrated with 5 column volumes (CV) of 5% buffer B (50 mM HEPES, 2M NaCl, pH = 7.2: Buffer B). The sample was directly loaded into the column via the sample pump. After sample application, the column was washed with 5 CV of buffer B at 5%. Elution was achieved by a salt step gradient consisting of 20 CV of 15%, 35%, 45% and 65% of buffer B (300 mM NaCl, 700 mM NaCl, 900 mM NaCl and 1300 mM NaCl). Solutions were filtered using 0.22 μm filters. Chromatographic runs were performed with a flow rate of 1 mL·min−1, except for the sample application (10 mL·min−1). Fractions of 1 mL were collected and pooled according to the chromatograms.
2.6.3. Size-Exclusion Chromatography
The collected peak containing the desired product from the IEX was loaded into a sepharose 4 Fast Flow (GE Healthcare, Chicago, IL, USA) in-house packed XK 16/40 desalting column of 48 mL. A column performance test with 1% acetone confirmed the correct values of asymmetry 10% and height equivalent to a theoretical plate (HETP). The column was pre-equilibrated with 5 CV of the formulation buffer (20 mM NaH2PO4, 50 mM NaCl, 2 mM MgCl2, 2% sucrose, pH 7.5). Subsequently, the sample was injected onto the column via its sample pump. Elution was achieved with an isocratic elution (0–100%) of 2 CV of the formulation buffer. The column was sanitized with 5 CV of 0.5 M NaOH. The chromatographic run was performed at a 2 mL·min−¹ flow rate. Fractions of 1 mL were collected and pooled according to the chromatograms.
2.7. Immunocytochemistry Staining for Flow Citometry and Confocal Microscopy
For IF-ICC staining, cells were centrifuged 5 min at 300× g and rinsed with staining solution (1.5% (v/v) fetal bovine serum (FBS) 1X phosphate-buffered saline (PBS)) before primary antibody incubation for 20 min at 4 °C in the dark. After rinsing twice, cells were incubated with the corresponding secondary antibody for 20 min at 4 °C. After IF-ICC staining, fixation was performed using 2% (v/v) formaldehyde 1X PBS for 10 min at RT. Cells were resuspended in staining solution and stored at 4 ºC prior to analysis.
Primary human anti-SARS-CoV-2 spike glycoprotein RBD domain antibody (ab272854, AbCam, Cambridge, UK) was diluted 1:1000. The secondary antibody used for flow citometry analysis was an anti-human IgG (H+L) coupled with Cy™5, produced in donkey (709-175-149, Jackson ImmunoResearch, West Grove, PA, USA), diluted 1:400. The secondary antibody used for confocal microscopy imaging was an anti-human IgG (H+L) coupled with Alexa Fluor 568, produced in goat (#A-21090, Thermo Fisher Scientific, Waltham, MA, USA), diluted 1:400. All IF-ICC antibodies were diluted using staining solution.
2.7.1. Flow Cytometry
The transfected cellular populations of previously IF-ICC stained cells were assessed by flow cytometry using a BD FACS Canto flow cytometer (BD BioSciences, San Jose, CA, USA), at Servei de Cultius Cel·lulars, Producció d’Anticossos i Citometria (Universitat Autònoma de Barcelona, Bellaterra, Catalonia, Spain).
2.7.2. Confocal Microscopy
The imaging of previously IF-ICC-stained cells was performed using Leica TCS SP5 confocal fluorescence microscope (Leica Microsystems, Wetzlar, Germany) at Servei de Microscòpia (Universitat Autònoma de Barcelona, Bellaterra, Catalonia, Spain). Prior to visualization, cells were treated with 0.1% (v/v) of Hoechst 33342 (Thermo Fisher Scientific, Waltham, MA, USA) and 0.1% (v/v) of CellMask Deep Red (Thermo Fisher Scientific, Waltham, MA, USA) in order to stain cell nuclei and lipid membranes, respectively. Samples were placed in 35 mm glass bottom Petri dishes with 14 mm microwells (MatTek Corporation, Ashland, MA, USA) prior to their visualization under the microscope. 3D images were generated and analyzed using Imaris software (Bitplane, Oxford Instruments, Zurich, Switzerland).
2.8. Transmission Electron Microscopy
TEM analyses were performed at Servei de Microscòpia (Universitat Autònoma de Barcelona, Bellaterra, Catalonia, Spain). Samples were visualized in a JEOL 2011 transmission electron microscope (Jeol, Tokio, Japan) operating at an accelerating voltage of 200 kV. Electron micrographs were recorded with the Digital Micrograph software package (Gatan, Pleasanton, CA, USA). Images were recorded by a Gatan US4000 (Gatan, Pleasanton, CA, USA) cooled charge-coupled device (CCD) camera.
2.8.1. Transmission Electron Microscopy: Negative Staining
For negative staining, samples were prepared by means of the air-dried method. Briefly, an aliquot of purified VLPs was absorbed by flotation onto freshly glow discharged 400 mesh carbon film copper grids (22-1MC040-100, MicrotoNano, Haarlem, The Netherlands). After standing for 1 min at RT, excess sample was drained carefully off the grid using Whatman filter paper, Grade 1 (WHA1001325, Merck, Kenilworth, NJ, USA). Samples were then stained with 5µL of uranyl acetate (2%) by incubation for 1 min at RT. The excess uranyl acetate was drained off as previously described.
2.8.2. Transmission Electron Microscopy: Immunogold Labeling
For immunogold labeling, 8 µL of purified VLPs were loaded onto copper grids as previously described. After absorption, two wash cycles were performed. Each wash cycle consisted of adding by flotation 2% (w/v) BSA in PBS and removing the excess sample, followed by the addition of 1X PBS at RT. Then, primary human anti-SARS-CoV-2 spike glycoprotein antibody (ab272854, AbCam, Cambridge, UK) diluted 1:50 was added, and the grids were incubated for 1 h at RT. Following three wash cycles, grids were incubated with 6 nm gold-conjugated anti-human IgG (109-195-088, Jackson ImmunoResearch, West Grove, PA, USA) diluted 1:20 for 1 h at RT. After three wash cycles, grids were stained with uranyl acetate as mentioned before.
2.9. Nanoparticle Tracking Analysis
NTA-based Gag::eGFP VLP quantification and characterization was performed using a NanoSight NS300 (Nanosight Ltd., Amesbury, UK) at the soft material services of the Institut de Ciència de Materials de Barcelona (ICMAB-CSIC, Bellaterra, Catalonia, Spain).
2.10. Total Protein and dsDNA Quantification
A BCA Protein Assay (#23225, Thermo Fisher Scientific, Waltham, MA, USA) was performed following manufacturer’s instructions using the provided BSA as standard. Colorimetric absorbance at 562 nm was read on a Multilabel Plate Reader Victor3 (Perkin Elmer, Waltham, MA, USA).
A Quant-iT PicoGreen dsDNA Assay Kit (#P11496, Thermo Fisher Scientific, Waltham, MA, USA) was performed following the manufacturer’s instructions using the provided λDNA as standard. Fluorescence (λex= 488 nm, λem= 520 nm) was read on a Multilabel Plate Reader Victor3 (Perkin Elmer, Waltham, MA, USA). The fluorescence value of the reagent blank was subtracted for each sample before calculating the dsDNA concentration using the generated standard curve.
2.11. Western Blot and SDS-PAGE
Samples were mixed with 4x Laemili Buffer (#1610747, Bio-Rad, Hercules, CA, USA) and 1.4 M DTT (10708984001, Merck, Kenilworth, NJ, USA) to a final concentration of 1% v/v. Each sample was incubated at 96 °C for 20 min and stored at 4 ºC until gelled. Precision Plus Protein WesternC (#1610376, Bio-Rad, Hercules, CA, USA) was used as molecular weight standard. A total of 20 μL of sample per lane was loaded and ran on precast SDS-polyacrylamide (4–20%) gel electrophoresis (#4561093, Bio-Rad, Hercules, CA, USA) at 200V, 400mA for 45 min. Running buffer used was Tris/Glicine/SDS (25 mM Tris, 192 mM glycine, 0.1% SDS, pH 8.3) (#1610772, Bio-Rad, Hercules, CA, USA).
For SDS-PAGE, proteins were stained with Coomassie Brilliant Blue EZBlueTM Gel Staining Reagent (G1041, Sigma Aldrich, St. Louis, MO, USA).
For Western blot, electrophoresis gel was transferred onto a polyvinylidene difluoride membrane for 7 min using the Trans-Blot Turbo Transfer System (#17001918, Bio-Rad, Hercules, CA, USA) following the manufacturer’s instructions. Transferred membranes were then blocked with 5% (w/v) nonfat dry milk in wash buffer (1× PBS 0.1% Tween-20). All the incubations and wash steps between incubations were performed at 40 rpm in a Polymax 1040 rocker shaker (Polymax 1040, Heidolph Instruments, Schwabach, Germany). For anti-HIV-1 Gag WB, blocking was performed overnight at 4 °C and incubated 2 h at RT with primary antibody. For SARS-CoV-2 spike WB, blocking was performed 40 min at RT, and it was incubated overnight at 4 °C with primary antibody. Primary antibodies used were rabbit polyclonal Anti-SARS-CoV-2 spike glycoprotein antibody (ab272504, AbCam, Cambridge, UK) and mouse monoclonal antibody to HIV-1 p24 (A2-851-500, Icosagen, Tartu, Estonia), both diluted 1:1000 in wash buffer. After primary incubation, membranes were incubated using anti-mouse IgG coupled with alkaline phosphatase antibody produced in goat (A3562, Merck, Kenilworth, NJ, USA) or anti-rabbit IgG coupled with alkaline phosphatase antibody produced in goat (A9919, Merck, Kenilworth, NJ, USA), as required, in wash buffer for 1 h at RT. Protein bands were visualized using NBT-BCIP solution (#1706432, Bio-Rad, Hercules, CA, USA) after 2−3 min incubation. Membranes were let to dry and then scanned and analyzed using the software ImageJ2 Fiji (National Institutes of Health, Bethesda, MD, USA).
2.12. Dot Blot
Samples were charged into Bio-Dot Apparatus (#1706545, Bio-Rad, Hercules, CA, USA) while a low vacuum was applied. Nitrocellulose membrane (#88018, Thermo Fisher Scientific, Waltham, MA, USA) was placed at the top of humidified filter paper. Once samples were transferred, membrane was incubated with anti-SARS-CoV-2 spike glycoprotein S2 monoclonal antibody (Ab281312, AbCam, Cambridge, UK) and an anti-rabbit secondary antibody (A9919, Merck, Kenilworth, NJ, USA) following the same procedure previously mentioned for Western blot. Once dried, membranes were scanned, and the pixel density for each loaded sample was analyzed using software ImageJ2 Fiji (National Institutes of Health, Bethesda, MD, USA). The standard used for quantification was a recombinant human coronavirus SARS-CoV-2 spike glycoprotein S2 subunit (Ab272106, AbCam, Cambridge, UK).
4. Discussion
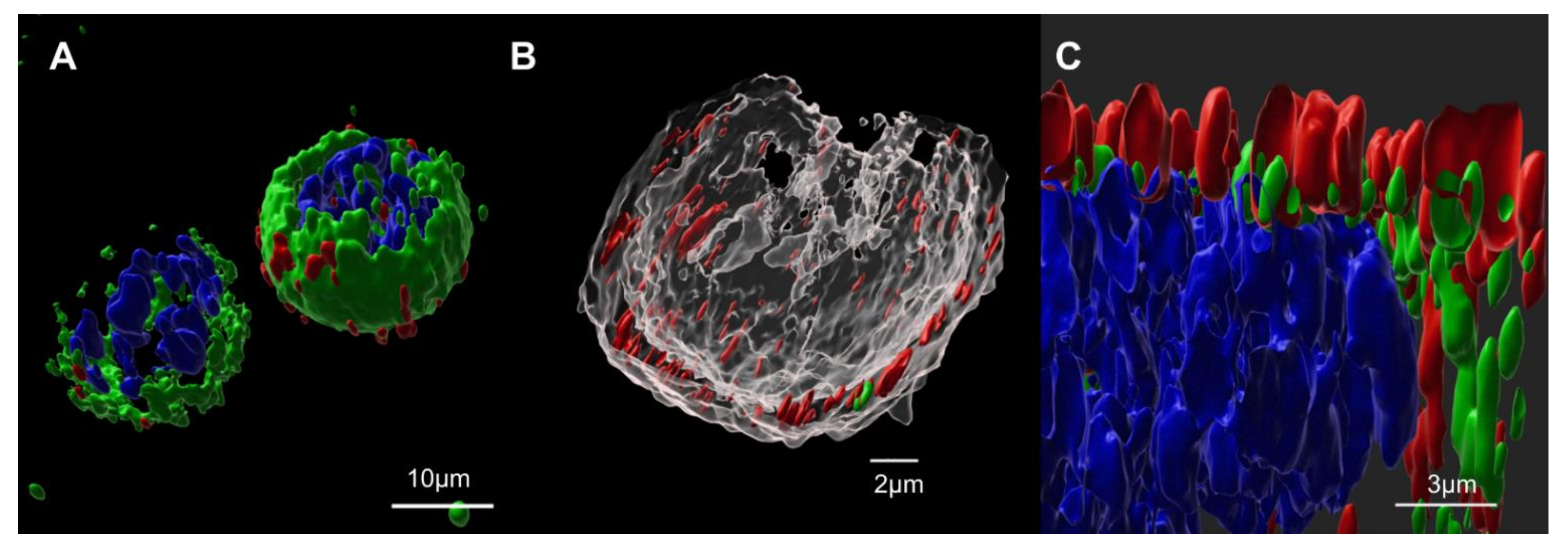
In this article, we first evaluated the cellular co-expression of SARS-CoV-2 spike glycoprotein with HIV-1 Gag, concluding that it has no significant negative effect in cell growth and viability. This suggests that it has no cytotoxic effect caused by protein secretory pathway failures. Confocal microscopy analysis showed that, after its expression, native envelope spike glycoprotein travels to the plasmatic membrane, wherein it co-localizes with Gag::eGFP. As Gag-based VLP generation occurs at the plasmatic membrane via budding, those results lay the groundwork to hypothesize that the S protein is incorporated to the VLPs. After that, we analyzed the produced and sucrose-cushion-purified VLPs by Western blot to find that S protein is present on the produced Gag-VLPs, confirming the incorporation of this SARS-CoV-2 antigen in our vaccine candidate. The produced VLPs had a mode diameter of 134.9±1.2 nm, as measured by NTA. EM observations led us to conclude that they had no significant structural differences from Gag-based non-functionalized G-VLPs. Further, S protein presence was confirmed by immunogold labeling at the surface of S-VLPs, a key feature in order to present immunogenic SARS-CoV-2 epitopes to a patient’s immune system when used as a vaccine. This was also relevant as, to the best of our knowledge, this study is the first report of Gag-based VLP functionalization with SARS-CoV-2 epitopes in order to generate a vaccine candidate against COVID-19.
Further, we optimized the production bioprocess using design of experiments in order to increase S-VLP productivity. We identified the transfection conditions maximizing the cellular population co-expressing simultaneously Gag and S proteins. This is important in order to maximize the percentage of cells responsible for the production of the S-VLPs and to minimize the single-expressing population that generates non-functionalized VLPs. The model predicted a double-positive population of 57.5±2.3% for the optimal transfection condition, which was validated and then implemented to transfect a 1L stirred tank bioreactor.
The bioprocess was carried out satisfactorily, achieving good cellular growth and viabilities comparable to the parallel Erlenmeyer shake flasks. The double-transfected population accounted for 55.1% of the total cells, which is concordant with what was predicted by the previously generated model. The bioreactor showed similar behavior with the parallel Erlenmeyer flasks at 72 hpt in terms of spike concentration, VLP concentration and the purity of the produced S-VLPs. This confirmed that production in a 1 L reactor was achieved successfully.
Finally, the 1 L bioreactor harvested product was purified using a downstream process consisting of two clarification steps, an anion-exchange capture step and a size-exclusion final polishing step. Clarification steps succeeded at reducing the turbidity of the sample by removing undesired contaminants, aggregates, intact cells and debris. Capture and polishing steps reduced the presence of undesired proteins, dsDNA and VLP aggregates while increasing VLP purity. The final purified product presents a significant reduction of dsDNA (1.03%) and host cell protein presence (0.22%) relative from the initial sample. Western blot analysis helped to track spike and Gag presence along the purification process, while dot blot analyses were also performed in order to quantify spike concentrations, obtaining a concentration of 2.198 ng/µL at the final purified product. Overall, the DSP process had a low yield in terms of VLP recovery but highly succeeded at concentrating and purifying the desired S-VLPs while generating a final product with little undesired contaminant presence.