Comparative Reverse Vaccinology of Piscirickettsia salmonis, Aeromonas salmonicida, Yersinia ruckeri, Vibrio anguillarum and Moritella viscosa, Frequent Pathogens of Atlantic Salmon and Lumpfish Aquaculture
Abstract
1. Introduction
2. Materials and Methods
2.1. Acquisition of Proteomes
2.2. Subcellular Localization
2.3. Adhesin and Antigenic Probability Scores
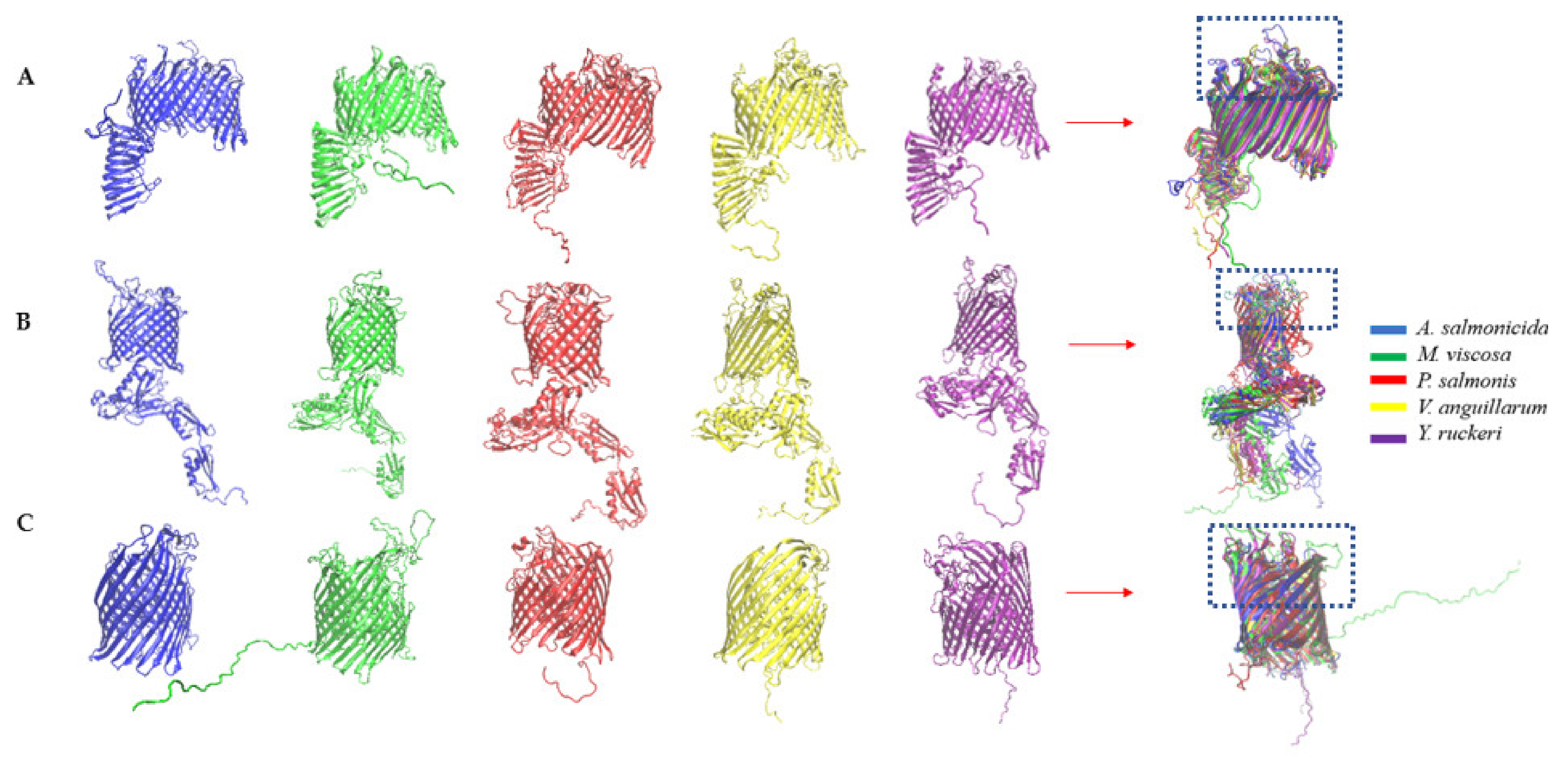
2.4. Sequence and Structural Alignment
2.5. Identification of Putative Domains
2.6. Detection of B and T Cell Epitopes
2.7. Atlantic Salmon and Lumpfish Major Histocompatibility Homology Modelling
2.8. Epitope Docking of Common Epitopes with Atlantic salmon and Lumpfish MHC Complex
3. Results
3.1. Pipeline Development
3.2. Subcellular Localization
3.3. Adhesin and Antigenicity
3.4. Comparative Topology
3.5. Epitope Prediction and Docking to Fish MHC Complexes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lafferty, K.D.; Harvell, C.D.; Conrad, J.M.; Friedman, C.S.; Kent, M.L.; Kuris, A.M.; Powell, E.N.; Rondeau, D.; Saksida, S.M. Infectious Diseases Affect Marine Fisheries and Aquaculture Economics. Annu. Rev. Mar. Sci. 2015, 7, 471–496. [Google Scholar] [CrossRef]
- Kumagai, A.; Sugimoto, K.; Itou, D.; Kamaishi, T.; Miwa, S.; Iida, T. Atypical Aeromonas salmonicida Infection in Cultured Marbled Sole Pleuronectes yokohamae. Fish. Pathol. 2006, 41, 7–12. [Google Scholar] [CrossRef][Green Version]
- Pedersen, K.; Larsen, J.L. First report on outbreak of furunculosis in turbot Scophthalmus maximus caused by Aeromonas salmonicida subsp. salmonicida in Denmark. Bull. Eur. Assoc. Fish. Pathol. 1996, 16, 129–133. [Google Scholar]
- Reddy, T.V.; Ravindranath, K.; Sreeraman, P.K.; Rao, M.V. Aeromonas salmonicida associated with mass mortality of Cyprinus carpio and Oreochromis mossambicus in a freshwater reservoir in Andhra Pradesh, India. J. Aquac. Trop. 1994, 9, 259–268. [Google Scholar]
- Jean, B. Practical experience with furunculosis in an Atlantic salmon hatchery in Washington State. Bull. Aquac. Assoc. Can. 1992, 1, 47–48. [Google Scholar]
- Rottereng, P.J. Control of furunculosis in fresh and seawater farms in Norway. In Parasites and Diseases in Natural Waters and Aquaculture in Nordic Countries; Stenmark, A.M.G., Ed.; Naturhistoriska Riksmuseet: Stockholm, Sweeden, 1987; pp. 129–130. [Google Scholar]
- Balcázar, J.L.; Vendrell, D.; De Blas, I.; Ruiz-Zarzuela, I.; Múzquiz, J.L. Effect of Lactococcus lactis CLFP 100 and Leuconostoc mesenteroides CLFP 196 on Aeromonas salmonicida Infection in Brown Trout (Salmo trutta). J. Mol. Microbiol. Biotechnol. 2009, 17, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Coyne, R.; Smith, P.; Dalsgaard, I.; Nilsen, H.; Kongshaug, H.; Bergh, Ø.; Samuelsen, O. Winter ulcer disease of post-smolt Atlantic salmon: An unsuitable case for treatment? Aquaculture 2006, 253, 171–178. [Google Scholar] [CrossRef]
- Coyne, R.; Bergh, O.; Samuelsen, O.; Andersen, K.; Lunestad, B.T.; Nilsen, H.; Dalsgaard, I.; Smith, P. Attempt to validate breakpoint MIC values estimated from pharmacokinetic data obtained during oxolinic acid therapy of winter ulcer disease in Atlantic salmon (Salmo salar). Aquaculture 2004, 238, 51–66. [Google Scholar] [CrossRef]
- Gaggero, A.; Castro, H.; Sandino, A.M. First isolation of Piscirickettsia salmonis from coho salmon, Oncorhynchus kisutch (Walbaum), and rainbow trout, Oncorhynchus mykiss (Walbaum), during the freshwater stage of their life cycle. J. Fish. Dis. 2006, 18, 277–280. [Google Scholar] [CrossRef]
- Mauel, M.J.; Miller, D.; Frazier, K.; Liggett, A.D.; Styer, L.; Montgomery-Brock, D.; Brock, J. Characterization of a piscirickettsiosis-like disease in Hawaiian tilapia. Dis. Aquat. Org. 2003, 53, 249–255. [Google Scholar] [CrossRef]
- Iregui, C.A.; Vasquez, G.M.; Rey, A.L.; Verjan, N. Piscirickettsia-Like Organisms as a Cause of Acute Necrotic Lesions in Colombian Tilapia larvae. J. Veter. Diagn. Investig. 2011, 23, 147–151. [Google Scholar] [CrossRef]
- Yiagnisis, M.V.I.; Kyriakou, C.; Alexis, M. First report of Vibrio anguillarum isolation from diseased big scale sand smelt, Atherina boyeri Risso 1810, in Limnos, Greece. Bull. Eur. Assoc. Fish. Pathol. 2007, 27, 61–69. [Google Scholar]
- Sakai, T.Y.H.; Shimizu, H.; Yuasa, K.; Kamaishi, T.; Oseko, N. Characteristics and pathogenicity of brown pigment-producing Vibrio anguillarum isolated from Japanese flounder [Paralichthys olivaceus]. Fish. Pathol. Jpn. 2006, 41, 77–79. [Google Scholar] [CrossRef]
- Guguianu, E.V.V.; Lazar, M.; Rimbu, C. Cercetari Agronomice in Moldova. Yersiniosis outbreak in rainbow trout (Oncorhynchus mykis) at a fish farm from northern Romania. Cercet. Agron. Mold. 2009, 42, 75–80. [Google Scholar]
- Oraić, D.; Zrnčić, S.; Šoštarić, B.; Al, E. Occurrence of enteric redmouth disease in rainbow trout (Oncorhynchus mykiss) on farms in Croatia. Acta Vet. Hung. 2002, 50, 283–291. [Google Scholar] [CrossRef]
- Rodriguez, L.A.; Castillo, A.; Gallardo, C.S.; Nieto, T.P. Outbreaks of Yersinia ruckeri in rainbow trout in north west of Spain. Bull. Eur. Assoc. Fish. Pathol. 1999, 19, 130–132. [Google Scholar]
- Eissa, A.E.; Moustafa, M.; Abdelaziz, M.; Ezzeldeen, N. Yersinia ruckeri infection in cultured Nile tilapia, Oreochromis niloticus, at a semi-intensive fish farm in lower Egypt. Afr. J. Aquat. Sci. 2008, 33, 283–286. [Google Scholar] [CrossRef]
- Denn, G. Yersinia ruckeri isolated from Atlantic salmon in Scotland. Bull. Eur. Assoc. Fish. Pathol. 1988, 8, 18–20. [Google Scholar]
- Sparboe, O.; Poppe, T.T.; Koren, C.; Stenwig, H. Yersinia ruckeri infection in salmon demonstrated for the first time in Norway. Nor. Vet. 1986, 98, 189–192. [Google Scholar]
- Einarsdottir, T.; Sigurdardottir, H.; Bjornsdottir, T.S.; Einarsdottir, E. Moritella viscosa in lumpfish (Cyclopterus lumpus) and Atlantic salmon (Salmo salar). J. Fish. Dis. 2018, 41, 1751–1758. [Google Scholar] [CrossRef]
- Marcos-López, M.; Ruane, N.; Scholz, F.; Bolton-Warberg, M.; Mitchell, O.S.; O’Sullivan, S.M.; Moore, A.I.; Rodger, H. Piscirickettsia salmonis infection in cultured lumpfish (Cyclopterus lumpus L.). J. Fish. Dis. 2017, 40, 1625–1634. [Google Scholar] [CrossRef] [PubMed]
- Vasquez, I.; Cao, T.; Chakraborty, S.; Gnanagobal, H.; O’Brien, N.; Monk, J.; Boyce, D.; Westcott, J.D.; Santander, J. Comparative Genomics Analysis of Vibrio anguillarum Isolated from Lumpfish (Cyclopterus lumpus) in Newfoundland Reveal Novel Chromosomal Organizations. Microorganism 2020, 8, 1666. [Google Scholar] [CrossRef]
- Kumar, G.; Menanteau-Ledouble, S.; Saleh, M.; El-Matbouli, M. Yersinia ruckeri, the causative agent of enteric redmouth disease in fish. Vet. Res. 2015, 46, 103. [Google Scholar] [CrossRef] [PubMed]
- Ormsby, M.J.; Caws, T.; Burchmore, R.; Wallis, T.; Verner-Jeffreys, D.W.; Davies, R.L. Yersinia ruckeri Isolates Recovered from Diseased Atlantic Salmon (Salmo salar) in Scotland Are More Diverse than Those from Rainbow Trout (Oncorhynchus mykiss) and Represent Distinct Subpopulations. Appl. Environ. Microbiol. 2016, 82, 5785–5794. [Google Scholar] [CrossRef] [PubMed]
- Valderrama, K.; Soto-Dávila, M.; Segovia, C.; Vásquez, I.; Dang, M.; Santander, J. Aeromonas salmonicidainfects Atlantic salmon (Salmo salar) erythrocytes. J. Fish. Dis. 2019, 42, 1601–1608. [Google Scholar] [CrossRef]
- Assefa, A.; Abunna, F. Maintenance of Fish Health in Aquaculture: Review of Epidemiological Approaches for Prevention and Control of Infectious Disease of Fish. Vet. Med. Int. 2018, 2018, 5432497. [Google Scholar] [CrossRef]
- Gudding, R.; Van Muiswinkel, W.B. A history of fish vaccination: Science-based disease prevention in aquaculture. Fish. Shellfish Immunol. 2013, 35, 1683–1688. [Google Scholar] [CrossRef]
- Bravo, S.; Midtlyng, P.J. The use of fish vaccines in the Chilean salmon industry 1999–2003. Aquaculture 2007, 270, 36–42. [Google Scholar] [CrossRef]
- Ma, J.; Bruce, T.J.; Jones, E.M.; Cain, K.D. A Review of Fish Vaccine Development Strategies: Conventional Methods and Modern Biotechnological Approaches. Microorganisms 2019, 7, 569. [Google Scholar] [CrossRef]
- Adams, A.; Subasinghe, R. Use of fish vaccines in aquaculture (including methods of administration). In Veterinary Vaccines for Livestock, 1st ed.; The Food and Agriculture Organization of the United Nations: Rome, Italy, 2019. [Google Scholar]
- Shefat, S.H.T. Vaccines for Use in Finfish Aquaculture. Acta Sci. Pharm. Sci. 2018, 2, 15–19. [Google Scholar]
- Duff, D.C.B. The oral immunization of trout against Bacterium salmonicida. J. Immunol. 1942, 44, 87–94. [Google Scholar]
- Snieszko, S.; Kocylowski, B.; Marek, K. Badania bakteriologiczne i serogiczne nad bakteriami posocznicy karpi. Mem. L’institut D’ichtyobiologie Piscic. Stn. Piscic. Exp. A Mydlniki L’universite Jagiellonienne Crac. 1938, 38. [Google Scholar]
- Brudeseth, B.E.; Wiulsrød, R.; Fredriksen, B.N.; Lindmo, K.; Løkling, K.-E.; Bordevik, M.; Steine, N.; Klevan, A.; Gravningen, K. Status and future perspectives of vaccines for industrialised fin-fish farming. Fish. Shellfish Immunol. 2013, 35, 1759–1768. [Google Scholar] [CrossRef]
- Adams, A. Progress, challenges and opportunities in fish vaccine development. Fish. Shellfish Immunol. 2019, 90, 210–214. [Google Scholar] [CrossRef]
- Figueroa, C.; Bustos, P.; Torrealba, D.; Dixon, B.; Soto, C.; Conejeros, P.; Gallardo, J.A. Coinfection takes its toll: Sea lice override the protective effects of vaccination against a bacterial pathogen in Atlantic salmon. Sci. Rep. 2017, 7, 17817. [Google Scholar] [CrossRef]
- Lee, N.-H.; Lee, J.-A.; Park, S.-Y.; Song, C.-S.; Choi, I.-S.; Lee, J.-B. A review of vaccine development and research for industry animals in Korea. Clin. Exp. Vaccine Res. 2012, 1, 18–34. [Google Scholar] [CrossRef]
- Zepp, F. Principles of vaccine design—Lessons from nature. Vaccine 2010, 28, C14–C24. [Google Scholar] [CrossRef]
- Kennedy, D.; Kurath, G.; Brito, I.; Purcell, M.; Read, A.F.; Winton, J.R.; Wargo, A.R. Potential drivers of virulence evolution in aquaculture. Evol. Appl. 2016, 9, 344–354. [Google Scholar] [CrossRef]
- Gandon, S.; Mackinnon, M.J.; Nee, S.; Read, A.F. Imperfect vaccines and the evolution of pathogen virulence. Nature 2001, 414, 751–756. [Google Scholar] [CrossRef]
- Mackinnon, M.; Gandon, S.; Read, A. Virulence evolution in response to vaccination: The case of malaria. Vaccine 2008, 26, C42–C52. [Google Scholar] [CrossRef]
- Lidder, P.; Sonnino, A. Chapter 1–Biotechnologies for the Management of Genetic Resources for Food and Agriculture. In Advances in Genetics; Goodwin, S.F., Friedmann, T., Dunlap, J.C., Eds.; Academic Press: Cambridge, MA, USA, 2012; pp. 1–167. [Google Scholar]
- Elanco Canada. Forte Micro. 2020. Available online: https://www.drugs.com/vet/forte-micro-can.html (accessed on 14 March 2022).
- Alpha Ject Micro 4. Available online: https://www.drugs.com/vet/alpha-ject-micro-4-can.html (accessed on 14 March 2022).
- Agricultural Analytics Division for the USDA National Organic Program. Vaccines for Aquaculture; USDA: Washington, DC, USA, 2014.
- Vasquez, I.; Retamales, J.; Parra, B.; Robeson, J.; Santander, J. Genomic Analysis of the Polyvalent Bacteriophage FP012019. Preprints 2019, 2019070169. [Google Scholar] [CrossRef]
- Nikoskelainen, S.; Verho, S.; Jarvinen, S.; Madetoja, J.; Wiklund, T.; Lilius, E. Multiple whole bacterial antigens in polyvalent vaccine may result in inhibition of specific responses in rainbow trout (Oncorhynchus mykiss). Fish. Shellfish Immunol. 2007, 22, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Baliga, P.; Shekar, M.; Venugopal, M.N. Potential Outer Membrane Protein Candidates for Vaccine Development Against the Pathogen Vibrio anguillarum: A Reverse Vaccinology Based Identification. Curr. Microbiol. 2018, 75, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Servín-Blanco, R.; Zamora-Alvarado, R.; Gevorkian, G.; Manoutcharian, K. Antigenic variability: Obstacles on the road to vaccines against traditionally difficult targets. Hum. Vaccines Immunother. 2016, 12, 2640–2648. [Google Scholar] [CrossRef]
- Sette, A.; Rappuoli, R. Reverse Vaccinology: Developing Vaccines in the Era of Genomics. Immunity 2010, 33, 530–541. [Google Scholar] [CrossRef]
- Rappuoli, R. Reverse vaccinology. Curr. Opin. Microbiol. 2000, 3, 445–450. [Google Scholar] [CrossRef]
- Pizza, M.; Scarlato, V.; Masignani, V.; Giuliani, M.M.; Aricò, B.; Comanducci, M.; Jennings, G.T.; Baldi, L.; Bartolini, E.; Capecchi, B. Identification of Vaccine Candidates Against Serogroup B Meningococcus by Whole-Genome Sequencing. Science 2000, 287, 1816–1820. [Google Scholar] [CrossRef]
- Vazquez-Juarez, R.C.; Gomez-Chiarri, M.; Barrera-Saldaña, H.; Hernandez-Saavedra, N.; Dumas, S.; Ascencio, F. Evaluation of DNA vaccination of spotted sand bass (Paralabrax maculatofasciatus) with two major outer-membrane protein-encoding genes from Aeromonas veronii. Fish. Shellfish Immunol. 2005, 19, 153–163. [Google Scholar] [CrossRef]
- Mao, Z.; Yu, L.; You, Z.; Wei, Y.; Liu, Y. Cloning, expression and immunogenicty analysis of five outer membrane proteins of Vibrio parahaemolyticus zj2003. Fish. Shellfish Immunol. 2007, 23, 567–575. [Google Scholar] [CrossRef]
- Qian, R.-H.; Xiao, Z.-H.; Zhang, C.-W.; Chu, W.-Y.; Wang, L.-S.; Zhou, H.-H.; Wei, Y.-W.; Yu, L. A conserved outer membrane protein as an effective vaccine candidate from Vibrio alginolyticus. Aquaculture 2008, 278, 5–9. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, E.; He, Y.; Wang, K.; Yang, Q.; Wang, J.; Geng, Y.; Chen, D.; Huang, X.; Ouyang, P.; et al. Identification and screening of effective protective antigens for channel catfish against Streptococcus iniae. Oncotarget 2017, 8, 30793–30804. [Google Scholar] [CrossRef][Green Version]
- Ong, E.; Wong, M.U.; He, Y. Identification of New Features from Known Bacterial Protective Vaccine Antigens Enhances Rational Vaccine Design. Front. Immunol. 2017, 8, 1382. [Google Scholar] [CrossRef]
- Andreoni, F.; Amagliani, G.; Magnani, M. Selection of Vaccine Candidates for Fish Pasteurellosis Using Reverse Vaccinology and an In Vitro Screening Approach. Adv. Struct. Saf. Stud. 2016, 1404, 181–192. [Google Scholar] [CrossRef]
- He, Y.; Xiang, Z.; Mobley, H.L.T. Vaxign: The First Web-Based Vaccine Design Program for Reverse Vaccinology and Applications for Vaccine Development. J. Biomed. Biotechnol. 2010, 2010, 297505. [Google Scholar] [CrossRef]
- Xiang, Z.; He, Y. Vaxign: A web-based vaccine target design program for reverse vaccinology. Procedia Vaccinol. 2009, 1, 23–29. [Google Scholar] [CrossRef]
- Xiang, Z.; He, Y. Genome-wide prediction of vaccine targets for human herpes simplex viruses using Vaxign reverse vaccinology. BMC Bioinform. 2013, 14, S2. [Google Scholar] [CrossRef]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. Identifying candidate subunit vaccines using an alignment-independent method based on principal amino acid properties. Vaccine 2007, 25, 856–866. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Eargle, J.; Wright, D.; Luthey-Schulten, Z. Multiple Alignment of protein structures and sequences for VMD. Bioinformatics 2005, 22, 504–506. [Google Scholar] [CrossRef]
- Roberts, E.; Eargle, J.; Wright, D.; Luthey-Schulten, Z. MultiSeq: Unifying sequence and structure data for evolutionary analysis. BMC Bioinform. 2006, 7, 382. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Larsen, J.E.P.; Lund, O.; Nielsen, M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006, 2, 2. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Thornton, J.M. PROCHECK: Validation of protein-structure coordinates. Int. Tables Crystallogr. 2012, F, 684–687. [Google Scholar] [CrossRef]
- Kurcinski, M.; Jamroz, M.; Blaszczyk, M.; Kolinski, A.; Kmiecik, S. CABS-dock web server for the flexible docking of peptides to proteins without prior knowledge of the binding site. Nucleic Acids Res. 2015, 43, W419–W424. [Google Scholar] [CrossRef]
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: A web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 2016, 32, 3676–3678. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Green, E.R.; Mecsas, J. Bacterial Secretion Systems: An Overview. Microbiol. Spectr. 2016, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Peng, B.; Li, H.; Peng, X.-X. TolC plays a crucial role in immune protection conferred by Edwardsiella tarda whole-cell vaccines. Sci. Rep. 2016, 6, 29488. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K.; Sahoo, P.K.; Dixit, A. Characterization of immune response elicited by the recombinant outer membrane protein OmpF of Aeromonas hydrophila, a potential vaccine candidate in murine model. Mol. Biol. Rep. 2014, 41, 1837–1848. [Google Scholar] [CrossRef] [PubMed]
- Maiti, B.; Dubey, S.; Munang’Andu, H.M.; Karunasagar, I.; Karunasagar, I.; Evensen, Ø. Application of Outer Membrane Protein-Based Vaccines Against Major Bacterial Fish Pathogens in India. Front. Immunol. 2020, 11, 1362. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Galperin, M.Y. Sequence—Evolution—Function: Computational Approaches in Comparative Genomics; Kluwer Academic: Dodrecht, The Netherlands, 2003; ISBN 1-40207-274-0. [Google Scholar]
- Gutlapalli, V.R.; Nireekshana-Acet, H.C.; Sykam, A.; Nayarisseri, A.; Suneetha, S.; Suneetha, L.M. Insights from the predicted epitope similarity between Mycobacterium tuberculosis virulent factors and its human homologs. Bioinformation 2015, 11, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Gentle, I.E.; Burri, L.; Lithgow, T. Molecular architecture and function of the Omp85 family of proteins. Mol. Microbiol. 2005, 58, 1216–1225. [Google Scholar] [CrossRef]
- Singh, R.; Capalash, N.; Sharma, P. Immunoprotective potential of BamA, the outer membrane protein assembly factor, against MDR Acinetobacter baumannii. Sci. Rep. 2017, 7, 12411. [Google Scholar] [CrossRef]
- Zha, Z.; Li, C.; Li, W.; Ye, Z.; Pan, J. LptD is a promising vaccine antigen and potential immunotherapeutic target for protection against Vibrio species infection. Sci. Rep. 2016, 6, 38577. [Google Scholar] [CrossRef]
- Soltan, M.; Elbassiouny, N.; Gamal, H.; Elkaeed, E.; Eid, R.; Eldeen, M.; Al-Karmalawy, A. In Silico Prediction of a Multitope Vaccine against Moraxella catarrhalis: Reverse Vaccinology and Immunoinformatics. Vaccines 2021, 9, 669. [Google Scholar] [CrossRef]
- Zielke, R.A.; Wierzbicki, I.H.; Baarda, B.I.; Gafken, P.R.; Soge, O.O.; Holmes, K.K.; Jerse, A.E.; Unemo, M.; Sikora, A.E. Proteomics-driven Antigen Discovery for Development of Vaccines Against Gonorrhea. Mol. Cell. Proteom. 2016, 15, 2338–2355. [Google Scholar] [CrossRef]
- Li, C.; Ye, Z.; Wen, L.; Chen, R.; Tian, L.; Zhao, F.; Pan, J. Identification of a novel vaccine candidate by immunogenic screening of Vibrio parahaemolyticus outer membrane proteins. Vaccine 2014, 32, 6115–6121. [Google Scholar] [CrossRef]
- Mukherjee, S.; Gangopadhay, K.; Mukherjee, S.B. Identification of potential new vaccine candidates in Salmonella typhi using reverse vaccinology and subtractive genomics-based approach. bioRxiv 2019, 521518. [Google Scholar] [CrossRef]
- Wang, J.; Xiong, K.; Pan, Q.; He, W.; Cong, Y. Application of TonB-Dependent Transporters in Vaccine Development of Gram-Negative Bacteria. Front. Cell. Infect. Microbiol. 2021, 10, 2235–2988. [Google Scholar] [CrossRef]
- Hu, Y.-H.; Dang, W.; Sun, L. A TonB-dependent outer membrane receptor of Pseudomonas fluorescens: Virulence and vaccine potential. Arch. Microbiol. 2012, 194, 795–802. [Google Scholar] [CrossRef]
- Reeves, S.A.; Torres, A.G.; Payne, S.M. TonB Is Required for Intracellular Growth and Virulence of Shigella dysenteriae. Infect. Immun. 2000, 68, 6329–6336. [Google Scholar] [CrossRef]
- Mott, T.M.; Vijayakumar, S.; Sbrana, E.; Endsley, J.J.; Torres, A.G. Characterization of the Burkholderia mallei tonB Mutant and Its Potential as a Backbone Strain for Vaccine Development. PLoS Negl. Trop. Dis. 2015, 9, e0003863. [Google Scholar] [CrossRef]
- Leduc, I.; Banks, K.E.; Fortney, K.R.; Patterson, K.B.; Billings, S.D.; Katz, B.P.; Spinola, S.M.; Elkins, C. Evaluation of the Repertoire of the TonB-Dependent Receptors of Haemophilus ducreyi for Their Role in Virulence in Humans. J. Infect. Dis. 2008, 197, 1103–1109. [Google Scholar] [CrossRef]
- Afonina, G.; Leduc, I.; Nepluev, I.; Jeter, C.; Routh, P.; Almond, G.; Orndorff, P.E.; Hobbs, M.; Elkins, C. Immunization with the Haemophilus ducreyi Hemoglobin Receptor HgbA Protects against Infection in the Swine Model of Chancroid. Infect. Immun. 2006, 74, 2224–2232. [Google Scholar] [CrossRef] [PubMed]
- Hynes, N. Bacterial Flagellin—A Novel Adjuvant for Vaccine Strategies. Ph.D. Thesis, University of Tromsø, Faculty of Biosciences, Fisheries and Economics, Department of Fishery Science, Tromsø, Norway, 2011. [Google Scholar]
- Wilhelm, V.; Miquel, A.; Burzio, L.O.; Rosemblatt, M.; Engel, E.; Valenzuela, S.; Parada, G.; Valenzuela, P.D. A vaccine against the salmonid pathogen Piscirickettsia salmonis based on recombinant proteins. Vaccine 2006, 24, 5083–5091. [Google Scholar] [CrossRef] [PubMed]
- Beck, B.R.; Lee, S.H.; Kim, D.; Park, J.H.; Lee, H.K.; Kwon, S.-S.; Lee, K.H.; Lee, J.I.; Song, S.K. A Lactococcus lactis BFE920 feed vaccine expressing a fusion protein composed of the OmpA and FlgD antigens from Edwardsiella tarda was significantly better at protecting olive flounder (Paralichthys olivaceus) from edwardsiellosis than single antigen vaccines. Fish. Shellfish Immunol. 2017, 68, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wu, H.; Li, X.; Yang, M.; Chen, T.; Wang, Q.; Liu, Q.; Zhang, Y. Edwardsiella tarda flagellar protein FlgD: A protective immunogen against edwardsiellosis. Vaccine 2012, 30, 3849–3856. [Google Scholar] [CrossRef]
- Clem, A.S. Fundamentals of vaccine immunology. J. Glob. Infect. Dis. 2011, 3, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Chen, Y.-H. High epitope density in a single protein molecule significantly enhances antigenicity as well as immunogenicity: A novel strategy for modern vaccine development and a preliminary investigation about B cell discrimination of monomeric proteins. Eur. J. Immunol. 2005, 35, 505–514. [Google Scholar] [CrossRef]
- Kastritis, P.; Moal, I.; Hwang, H.; Weng, Z.; Bates, P.; Bonvin, A.M.; Janin, J. A structure-based benchmark for protein-protein binding affinity. Protein Sci. 2011, 20, 482–491. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Xia, Y.-L.; Ai, S.-M.; Liang, J.; Sang, P.; Ji, X.-L.; Liu, S.-Q. Insights into Protein–Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef]
- Dubey, S.; Avadhani, K.; Mutalik, S.; Sivadasan, S.M.; Maiti, B.; Girisha, S.K.; Venugopal, M.N.; Mutoloki, S.; Evensen, Ø.; Karunasagar, I. Edwardsiella tarda OmpA Encapsulated in Chitosan Nanoparticles Shows Superior Protection over Inactivated Whole Cell Vaccine in Orally Vaccinated Fringed-Lipped Peninsula Carp (Labeo fimbriatus). Vaccines 2016, 4, 40. [Google Scholar] [CrossRef]
- Li, H.; Chu, X.; Li, D.; Zeng, Z.-H.; Peng, X.-X. Construction and immune protection evaluation of recombinant polyvalent OmpAs derived from genetically divergent ompA by DNA shuffling. Fish. Shellfish Immunol. 2016, 49, 230–236. [Google Scholar] [CrossRef]
- Maiti, B.; Shetty, M.; Shekar, M.; Karunasagar, I.; Karunasagar, I. Recombinant outer membrane protein A (OmpA) of Edwardsiella tarda, a potential vaccine candidate for fish, common carp. Microbiol. Res. 2011, 167, 1–7. [Google Scholar] [CrossRef]
- Mahendran, R.; Jeyabaskar, S.; Michael, R.D.; Paul, A.V.; Sitharaman, G. Computer-aided vaccine designing approach against fish pathogens Edwardsiella tarda and Flavobacterium columnare using bioinformatics softwares. Drug Des. Dev. Ther. 2016, 10, 1703–1714. [Google Scholar] [CrossRef]
- Zhu, Z.; Dong, C.; Weng, S.; He, J. Identification of outer membrane protein TolC as the major adhesin and potential vaccine candidate for Vibrio harveyi in hybrid grouper, Epinephelus fuscoguttatus (♀) × E. lanceolatus (♂). Fish. Shellfish Immunol. 2019, 86, 143–151. [Google Scholar] [CrossRef]
- Wang, X.; Yang, X.; Yang, C.; Wu, Z.; Xu, H.; Shen, Y. Crystal Structure of Outer Membrane Protein NMB0315 from Neisseria meningitidis. PLoS ONE 2011, 6, e26845. [Google Scholar] [CrossRef]
- Tidhar, A.; Flashner, Y.; Cohen, S.; Levi, Y.; Zauberman, A.; Gur, D.; Aftalion, M.; Elhanany, E.; Zvi, A.; Shafferman, A.; et al. The NlpD Lipoprotein Is a Novel Yersinia pestis Virulence Factor Essential for the Development of Plague. PLoS ONE 2009, 4, e7023. [Google Scholar] [CrossRef]
- Kaur, D.; Gandhi, S.; Mukhopadhaya, A. Salmonella Typhimurium Adhesin OmpV Activates Host Immunity to Confer Protection against Systemic and Gastrointestinal Infection in Mice. Infect. Immun. 2021, 89, e00121-21. [Google Scholar] [CrossRef]
- Zhang, D.; Xu, D.; Shoemaker, A.C. Immunization with recombinant aerolysin and haemolysin protected channel catfish against virulent Aeromonas hydrophila. Aquac. Res. 2017, 48, 875–882. [Google Scholar] [CrossRef]
- Wang, N.; Wu, Y.; Pang, M.; Liu, J.; Lu, C.; Liu, Y. Protective efficacy of recombinant hemolysin co-regulated protein (Hcp) of Aeromonas hydrophila in common carp (Cyprinus carpio). Fish. Shellfish Immunol. 2015, 46, 297–304. [Google Scholar] [CrossRef]
- Zhang, D.; Pridgeon, J.W.; Klesius, P.H. Vaccination of channel catfish with extracellular products of Aeromonas hydrophila provides protection against infection by the pathogen. Fish. Shellfish Immunol. 2014, 36, 270–275. [Google Scholar] [CrossRef]
- Duarte, A.S.; Correia, A.; Esteves, A.C. Bacterial collagenases—A review. Crit. Rev. Microbiol. 2016, 42, 106–126. [Google Scholar] [CrossRef]
- Askarian, F.; Uchiyama, S.; Masson, H.; Sørensen, H.V.; Golten, O.; Bunæs, A.C.; Mekasha, S.; Røhr, A.K.; Kommedal, E.; Ludviksen, J.A.; et al. The lytic polysaccharide monooxygenase CbpD promotes Pseudomonas aeruginosa virulence in systemic infection. Nat. Commun. 2021, 12, 1230. [Google Scholar] [CrossRef]
- Flores-Díaz, M.; Monturiol-Gross, L.; Naylor, C.; Alape-Girón, A.; Flieger, A. Bacterial Sphingomyelinases and Phospholipases as Virulence Factors. Microbiol. Mol. Biol. Rev. 2016, 80, 597–628. [Google Scholar] [CrossRef]
- Champion, M.D. Host-Pathogen O-Methyltransferase Similarity and Its Specific Presence in Highly Virulent Strains of Francisella tularensis Suggests Molecular Mimicry. PLoS ONE 2011, 6, e20295. [Google Scholar] [CrossRef]
- Stehr, F.; Kretschmar, M.; Kröger, C.; Hube, B.; Schäfer, W. Microbial lipases as virulence factors. J. Mol. Catal. B Enzym. 2003, 22, 347–355. [Google Scholar] [CrossRef]
- Sudhakara, P.; Sellamuthu, I.; Aruni, A.W. Bacterial sialoglycosidases in Virulence and Pathogenesis. Pathogens 2019, 8, 39. [Google Scholar] [CrossRef]
- Sumby, P.; Barbian, K.D.; Gardner, D.J.; Whitney, A.R.; Welty, D.M.; Long, R.D.; Bailey, J.R.; Parnell, M.J.; Hoe, N.P.; Adams, G.G.; et al. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc. Natl. Acad. Sci. USA 2005, 102, 1679–1684. [Google Scholar] [CrossRef]
- Rice, K.; Batul, K.; Whiteside, J.; Kelso, J.; Papinski, M.; Schmidt, E.; Pratasouskaya, A.; Wang, D.; Sullivan, R.; Bartlett, C.; et al. The predominance of nucleotidyl activation in bacterial phosphonate biosynthesis. Nat. Commun. 2019, 10, 369. [Google Scholar] [CrossRef]
- Teng, J.L.L.; Luo, R.; Tang, B.S.F.; Fong, J.Y.H.; Wang, L.; Jia, L.; Wong, C.K.S.; Chan, E.; Leung, A.W.S.; Siu, G.K.H.; et al. High Prevalence and Mechanism Associated with Extended Spectrum Beta-Lactamase-Positive Phenotype in Laribacter hongkongensis. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- Baker, P.; Ricer, T.; Moynihan, P.; Kitova, E.N.; Walvoort, M.; Little, D.J.; Whitney, J.C.; Dawson, K.; Weadge, J.T.; Robinson, H.; et al. P. aeruginosa SGNH Hydrolase-Like Proteins AlgJ and AlgX Have Similar Topology but Separate and Distinct Roles in Alginate Acetylation. PLoS Pathog. 2014, 10, e1004334. [Google Scholar] [CrossRef]








| Species | Genomes Used in This Study (NCBI Accession Numbers) |
|---|---|
| Piscirickettsia salmonis | LF 89, EM 90, AY3800B, AY3864B, AY6297B, PM25344B, AY6532B, PM31429B, PM32597B1, PSCGR01, PM49811B, PM58386B, PM22180B, AY6492A, PM15972A1, PM23019A, PM37984A, PM51819A, PM21567A. |
| Aeromonas salmonicida | J223, A449, O23A, S121, S68, S44, RFAS1, 34mel, A527, 01-B526. |
| Moritella viscosa | MVIS1 |
| Yersinia ruckeri | SC09, Big Creek 74, YRB, QMA0440, NHV_3758 |
| Vibrio anguillarum | JLL237, NB10, MVAV6203, ATCC-68554, M3, 90-11-286, S3 4/9, MHK3, VIB43 |
| Bacteria | Accession Number | Common Antigenic Proteins | Antigenic Probability Cutoff (0.4) | Adhesin Probability Cutoff (0.51) | Location |
|---|---|---|---|---|---|
| P. salmonis A. salmonicida M. viscosa Y. ruckeri V. anguillarum | WP_036771893 WP_011898993 WP_045111816 WP_038245171 WP_019282310 | Outer membrane protein assembly factor BamA | 0.484 0.656 0.533 0.577 0.559 | 0.811 0.613 0.608 0.716 0.612 | Outer membrane |
| P. salmonis A. salmonicida M. viscosa Y. ruckeri V. anguillarum | WP_005316218 WP_017377460 CED58662 WP_080748387 WP_019282793 | TonB dependent siderophore receptor | 0.567 0.606 0.599 0.576 0.631 | 0.832 0.497 0.525 0.424 0.714 | Outer membrane |
| P. salmonis A. salmonicida M. viscosa Y. ruckeri V. anguillarum | WP_048876074 WP_005311708 CED61712 WP_004721574 WP_029388238 | LPS assembly protein LptD | 0.514 0.727 0.600 0.546 0.580 | 0.739 0.517 0.472 0.671 0.740 | Outer membrane |
| P. salmonis A. salmonicida M. viscosa Y. ruckeri V. anguillarum | WP_027242990 WP_005320202 CED58793 WP_004720726 WP_010319379 | Flagellar basal-body rod protein FlgG | 0.593 0.628 0.477 0.579 0.578 | 0.764 0.725 0.751 0.791 0.723 | Secreted |
| P. salmonis A. salmonicida M. viscosa Y. ruckeri V. anguillarum | WP_027242992 WP_005320209 CED61365 WP_038242029 WP_013856252 | Flagellar hook assembly protein FlgD | 0.405 0.633 0.547 0.555 0.466 | 0.510 0.795 0.780 0.710 0.606 | Secreted |
| Outer Membrane Protein Assembly BamA | LPS-Assembly Protein LptD | TonB-Dependent Siderophore Receptor | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Bacteria Pathogens | QH | RMSD | Identity (%) | QH | RMSD | Identity (%) | QH | RMSD | Identity (%) |
| M.viscosa vs A. salmonicida | 0.52 | 4.42 | 41.80 | 0.71 | 2.35 | 30.69 | 0.64 | 1.96 | 20.20 |
| M. viscosa vs P. salmonis | 0.49 | 4.62 | 22.62 | 0.63 | 2.72 | 21.06 | 0.55 | 2.51 | 11.85 |
| M.viscosa vs V. anguillarum | 0.26 | 5.68 | 14.05 | 0.76 | 1.89 | 29.31 | 0.63 | 2.03 | 18.10 |
| M.viscosa vs Y.ruckeri | 0.25 | 6.27 | 14.75 | 0.74 | 1.80 | 29.84 | 0.84 | 0.82 | 19.50 |
| A.salmonicida vs P. salmonis | 0.67 | 2.06 | 27.41 | 0.72 | 1.63 | 21.74 | 0.58 | 2.69 | 14.05 |
| A.salmonicida vs V. anguillarum | 0.25 | 5.08 | 19.76 | 0.81 | 1.49 | 28.62 | 0.86 | 0.91 | 27.10 |
| A.salmonicida vs Y.ruckeri | 0.24 | 5.84 | 19.69 | 0.82 | 1.65 | 28.07 | 0.69 | 1.94 | 27.25 |
| P. salmonis vs V. anguillarum | 0.27 | 4.58 | 11.41 | 0.68 | 2.07 | 17.95 | 0.56 | 2.83 | 10.64 |
| P. salmonis vs Y.ruckeri | 0.25 | 5.66 | 11.89 | 0.68 | 2.21 | 20.70 | 0.58 | 2.53 | 11.14 |
| V. anguillarum vs Y.ruckeri | 0.75 | 2.41 | 53.08 | 0.88 | 0.90 | 31.81 | 0.67 | 2.07 | 26.34 |
| Flagellar Hook Assembly Protein FlgD | Flagellar Basal-Body Rod Protein FlgG | |||||
|---|---|---|---|---|---|---|
| Bacteria Pathogen | QH | RMSD | Identity (%) | QH | RMSD | Identity (%) |
| M.viscosa vs. A. salmonicida | 0.81 | 2.95 | 15.89 | 0.71 | 2.35 | 30.69 |
| M. viscosa vs. P. salmonis | 0.88 | 1.97 | 19.18 | 0.63 | 2.72 | 21.06 |
| M.viscosa vs. V. anguillarum | 0.86 | 2.18 | 25.17 | 0.76 | 1.89 | 29.31 |
| M.viscosa vs. Y.ruckeri | 0.75 | 3.19 | 13.90 | 0.74 | 1.80 | 29.84 |
| A.salmonicida vs. P. salmonis | 0.80 | 3.03 | 16.55 | 0.72 | 1.63 | 21.74 |
| A.salmonicida vs. V. anguillarum | 0.80 | 2.70 | 18.48 | 0.81 | 1.49 | 28.62 |
| A.salmonicida vs. Y.ruckeri | 0.82 | 2.16 | 14.85 | 0.82 | 1.65 | 28.07 |
| P. salmonis vs. V. anguillarum | 0.87 | 1.64 | 21.16 | 0.68 | 2.07 | 17.95 |
| P. salmonis vs. Y.ruckeri | 0.73 | 2.84 | 15.59 | 0.68 | 2.21 | 20.70 |
| V. anguillarum vs. Y.ruckeri | 0.66 | 2.59 | 17.83 | 0.88 | 0.90 | 31.81 |
| Protein | Common Epitopes | No of Residues | Atlantic Salmon | Lumpfish | ||
|---|---|---|---|---|---|---|
| Kd (M) at 10.0 °C | ΔG (kcal mol−1) | Kd (M) at 10.0 °C | ΔG (kcal mol−1) | |||
| LPS assembly protein LptD | PYYLNLAPNYD QTLEPRLYYLYVP | 11 13 | 2.3 × 10−7 2.3 × 10−7 | −8.6 −8.6 | 2.5 × 10−7 5.8 × 10−8 | −8.6 −9.4 |
| Outer membrane protein assembly factor BamA | IEGLQRL DEVLRR LNTLGYF LQWMSPLGP | 7 7 7 10 | 1.5 × 10−6 9.4 × 10−6 5.1 × 10−7 7.9 × 10−7 | −7.5 −6.5 −8.1 −7.9 | 5.5 × 10−6 9.4 × 10−6 2.2 × 10−7 1.9 × 10−7 | −6.8 −6.5 −8.6 −8.7 |
| TonB-dependent siderophore receptor | EKIDVRGGAAVQYG | 15 | 5.1 × 10−9 | −10.3 | 6.8 × 10−8 | −9.3 |
| Flagellar hook assembly protein FlgD | LKNQDPPTNP LQASALLG WDGNDQNGN | 10 8 9 | 1.8 × 10−7 1.9 × 10−7 2.5 × 10−8 | −8.7 −8.7 −9.8 | 5.4 × 10−9 6.4 × 10−7 1.5 × 10−7 | −10.7 −8.0 −8.8 |
| Flagellar basal-body rod protein FlgG | LLTQLAQQDP ALQASALVG WDGKL | 10 9 5 | 1.1 × 10−7 1.6 × 10−7 1.4 × 10−6 | −9.0 −8.8 −7.6 | 8.0 × 10−8 4.0 × 10−7 4.7 × 10−6 | −7.9 −8.3 −6.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chukwu-Osazuwa, J.; Cao, T.; Vasquez, I.; Gnanagobal, H.; Hossain, A.; Machimbirike, V.I.; Santander, J. Comparative Reverse Vaccinology of Piscirickettsia salmonis, Aeromonas salmonicida, Yersinia ruckeri, Vibrio anguillarum and Moritella viscosa, Frequent Pathogens of Atlantic Salmon and Lumpfish Aquaculture. Vaccines 2022, 10, 473. https://doi.org/10.3390/vaccines10030473
Chukwu-Osazuwa J, Cao T, Vasquez I, Gnanagobal H, Hossain A, Machimbirike VI, Santander J. Comparative Reverse Vaccinology of Piscirickettsia salmonis, Aeromonas salmonicida, Yersinia ruckeri, Vibrio anguillarum and Moritella viscosa, Frequent Pathogens of Atlantic Salmon and Lumpfish Aquaculture. Vaccines. 2022; 10(3):473. https://doi.org/10.3390/vaccines10030473
Chicago/Turabian StyleChukwu-Osazuwa, Joy, Trung Cao, Ignacio Vasquez, Hajarooba Gnanagobal, Ahmed Hossain, Vimbai Irene Machimbirike, and Javier Santander. 2022. "Comparative Reverse Vaccinology of Piscirickettsia salmonis, Aeromonas salmonicida, Yersinia ruckeri, Vibrio anguillarum and Moritella viscosa, Frequent Pathogens of Atlantic Salmon and Lumpfish Aquaculture" Vaccines 10, no. 3: 473. https://doi.org/10.3390/vaccines10030473
APA StyleChukwu-Osazuwa, J., Cao, T., Vasquez, I., Gnanagobal, H., Hossain, A., Machimbirike, V. I., & Santander, J. (2022). Comparative Reverse Vaccinology of Piscirickettsia salmonis, Aeromonas salmonicida, Yersinia ruckeri, Vibrio anguillarum and Moritella viscosa, Frequent Pathogens of Atlantic Salmon and Lumpfish Aquaculture. Vaccines, 10(3), 473. https://doi.org/10.3390/vaccines10030473








