SARS-CoV-2 Doggybone DNA Vaccine Produces Cross-Variant Neutralizing Antibodies and Is Protective in a COVID-19 Animal Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. dbDNA Construction
2.3. Animal Vaccinations
2.4. Other Animal Procedures
2.5. SARS-CoV-2 Challenge Stock
2.6. SARS-CoV-2 D614G Variant Stock
2.7. Viral RNA Assay
2.8. Pseudovirion Production and Pseudovirion Assay (PsVNA)
2.9. Plaque Assay
2.10. Plaque Reduction Neutralization Test (PRNT)
2.11. Hematology
2.12. Preparation of Tissues for Histology
2.13. In Situ Hybridization
2.14. Statistical Analyses
3. Results
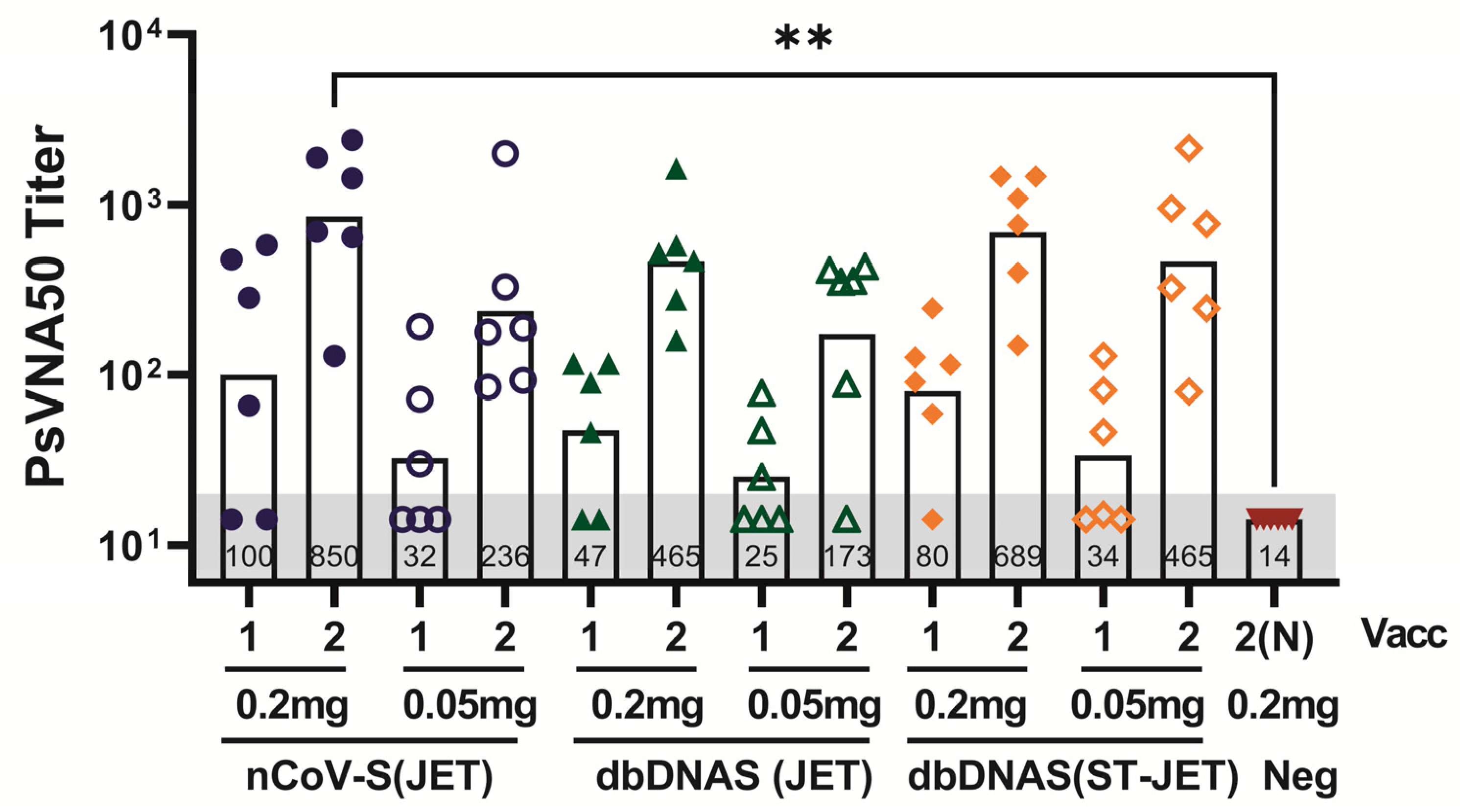
3.1. Immunogenicity of SARS-CoV-2 DNA Vaccines Administered by Jet Injection in Syrian Hamsters
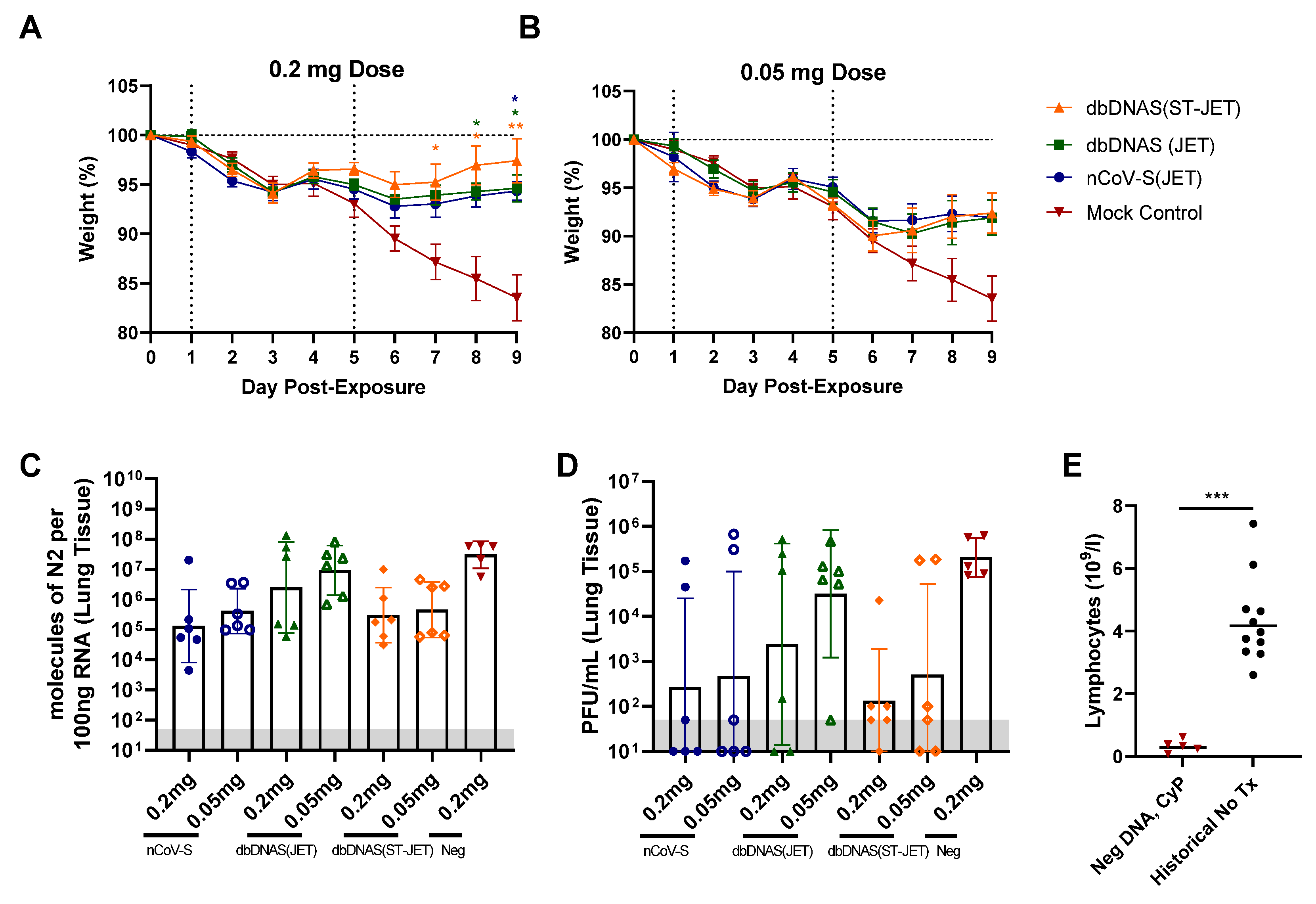
3.2. Protective Efficacy of DNA Vaccines in Immunosuppressed SARS-CoV-2 Exposed Hamsters
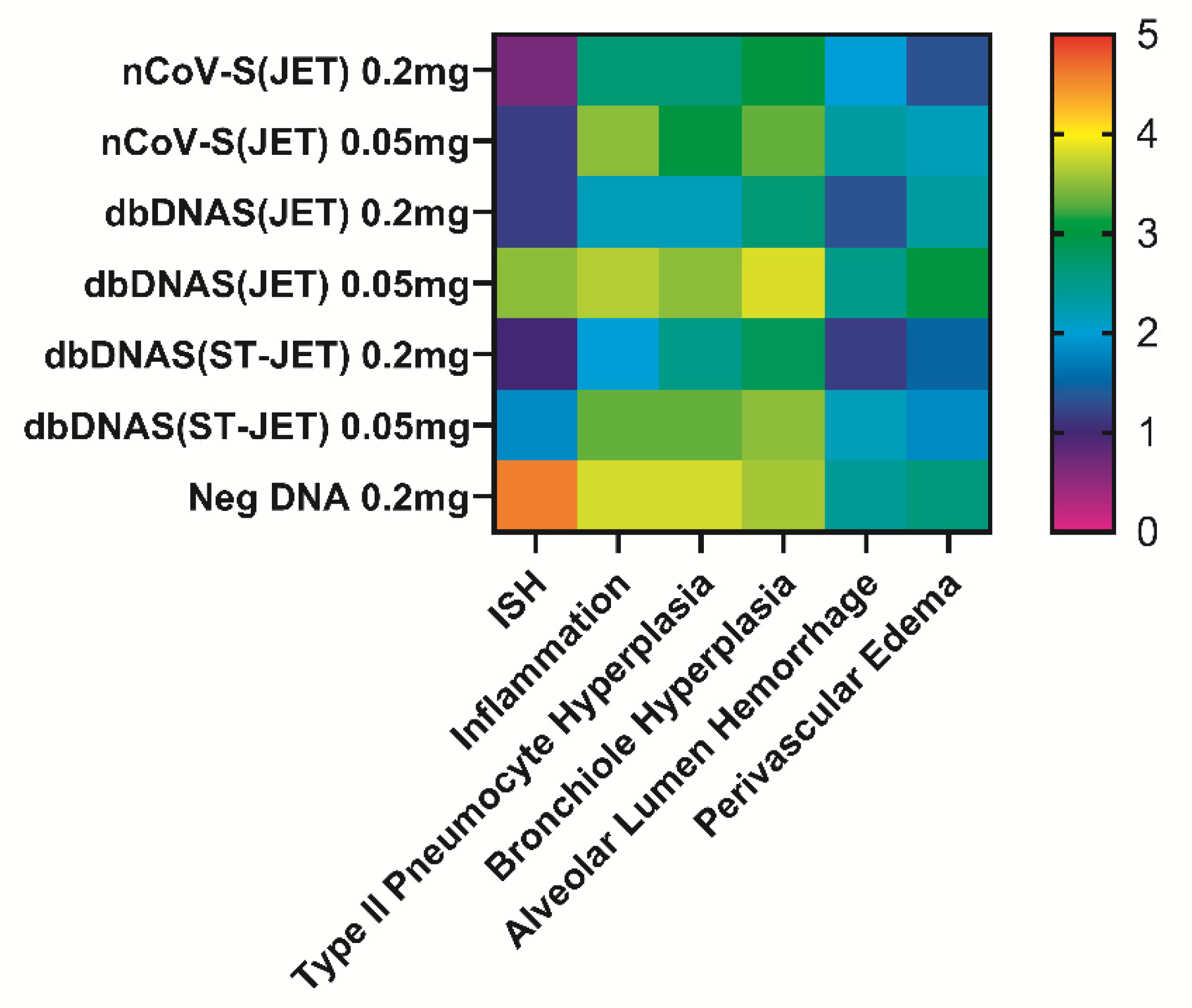
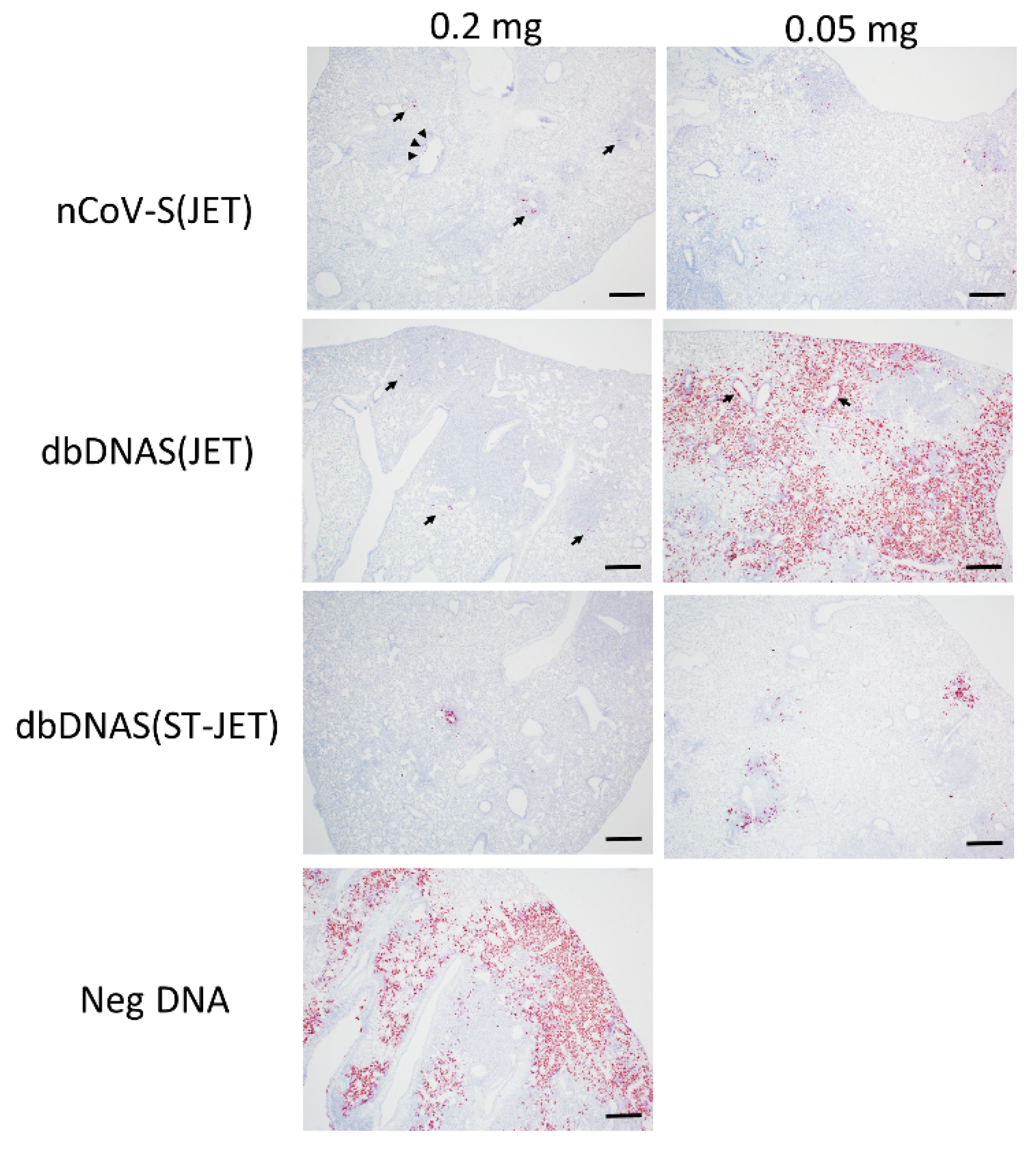
3.3. Lung Pathology of SARS-CoV-2 DNA Vaccinated Hamsters Following Virus Exposure
3.4. Serum Neutralization Efficacy against Novel SARS-CoV-2 Variants
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Mucker, E.M.; Karmali, P.P.; Vega, J.; Kwilas, S.A.; Wu, H.; Joselyn, M.; Ballantyne, J.; Sampey, D.; Mukthavaram, R.; Sullivan, E.; et al. Lipid Nanoparticle Formulation Increases Efficiency of DNA-Vectored Vaccines/Immunoprophylaxis in Animals Including Transchromosomic Bovines. Sci. Rep. 2020, 10, 8764. [Google Scholar] [CrossRef] [PubMed]
- Morrow, M.P.; Weiner, D.B. DNA drugs come of age. Sci. Am. 2010, 303, 48–53. [Google Scholar] [CrossRef]
- Mallapaty, S. India’s DNA COVID vaccine is a world first—More are coming. Nature 2021, 597, 161–162. [Google Scholar] [CrossRef]
- Brocato, R.L.; Kwilas, S.A.; Josleyn, M.D.; Long, S.; Zeng, X.; Perley, C.C.; Principe, L.M.; Somerville, B.; Cohen, M.V.; Hooper, J.W. Small animal jet injection technique results in enhanced immunogenicity of hantavirus DNA vaccines. Vaccine 2021, 39, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Brocato, R.L.; Kwilas, S.A.; Kim, R.K.; Zeng, X.; Principe, L.M.; Smith, J.M.; Hooper, J.W. Protective efficacy of a SARS-CoV-2 DNA vaccine in wild-type and immunosuppressed Syrian hamsters. NPJ Vaccines 2021, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Muthumani, K.; Block, P.; Flingai, S.; Muruganantham, N.; Chaaithanya, I.K.; Tingey, C.; Wise, M.; Reuschel, E.L.; Chung, C.; Muthumani, A.; et al. Rapid and Long-Term Immunity Elicited by DNA-Encoded Antibody Prophylaxis and DNA Vaccination against Chikungunya Virus. J. Infect. Dis. 2016, 214, 369–378. [Google Scholar] [CrossRef] [Green Version]
- WHO. DRAFT Landscape of COVID-19 Vaccine Candidates—10 August 2020. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 27 May 2022).
- Gaudinski, M.R.; Houser, K.V.; Morabito, K.M.; Hu, Z.; Yamshchikov, G.; Rothwell, R.S.; Berkowitz, N.; Mendoza, F.; Saunders, J.G.; Novik, L.; et al. Safety, tolerability, and immunogenicity of two Zika virus DNA vaccine candidates in healthy adults: Randomised, open-label, phase 1 clinical trials. Lancet 2018, 391, 552–562. [Google Scholar] [CrossRef] [Green Version]
- Graham, B.S.; Koup, R.A.; Roederer, M.; Bailer, R.T.; Enama, M.E.; Moodie, Z.; Martin, J.E.; McCluskey, M.M.; Chakrabarti, B.K.; Lamoreaux, L.; et al. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 DNA candidate vaccine. J. Infect. Dis. 2006, 194, 1650–1660. [Google Scholar] [CrossRef]
- Hannaman, D.; Dupuy, L.C.; Ellefsen, B.; Schmaljohn, C.S. A Phase 1 clinical trial of a DNA vaccine for Venezuelan equine encephalitis delivered by intramuscular or intradermal electroporation. Vaccine 2016, 34, 3607–3612. [Google Scholar] [CrossRef]
- Martin, J.E.; Sullivan, N.J.; Enama, M.E.; Gordon, I.J.; Roederer, M.; Koup, R.A.; Bailer, R.T.; Chakrabarti, B.K.; Bailey, M.A.; Gomez, P.L.; et al. A DNA vaccine for Ebola virus is safe and immunogenic in a phase I clinical trial. Clin. Vaccine Immunol. 2006, 13, 1267–1277. [Google Scholar] [CrossRef] [Green Version]
- Tebas, P.; Yang, S.; Boyer, J.D.; Reuschel, E.L.; Patel, A.; Christensen-Quick, A.; Andrade, V.M.; Morrow, M.P.; Kraynyak, K.; Agnes, J.; et al. Safety and immunogenicity of INO-4800 DNA vaccine against SARS-CoV-2: A preliminary report of an open-label, Phase 1 clinical trial. EClinicalMedicine 2020, 31, 100689. [Google Scholar] [CrossRef] [PubMed]
- Belakova, J.; Horynova, M.; Krupka, M.; Weigl, E.; Raska, M. DNA vaccines: Are they still just a powerful tool for the future? Arch. Immunol. Ther. Exp. 2007, 55, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Allen, A.; Wang, C.; Caproni, L.J.; Sugiyarto, G.; Harden, E.; Douglas, L.R.; Duriez, P.J.; Karbowniczek, K.; Extance, J.; Rothwell, P.J.; et al. Linear doggybone DNA vaccine induces similar immunological responses to conventional plasmid DNA independently of immune recognition by TLR9 in a pre-clinical model. Cancer Immunol. Immunother. 2018, 67, 627–638. [Google Scholar] [CrossRef] [Green Version]
- Scott, V.L.; Patel, A.; Villarreal, D.O.; Hensley, S.E.; Ragwan, E.; Yan, J.; Sardesai, N.Y.; Rothwell, P.J.; Extance, J.P.; Caproni, L.J.; et al. Novel synthetic plasmid and Doggybone DNA vaccines induce neutralizing antibodies and provide protection from lethal influenza challenge in mice. Hum. Vaccine Immunother. 2015, 11, 1972–1982. [Google Scholar] [CrossRef] [Green Version]
- Walters, A.A.; Kinnear, E.; Shattock, R.J.; McDonald, J.U.; Caproni, L.J.; Porter, N.; Tregoning, J.S. Comparative analysis of enzymatically produced novel linear DNA constructs with plasmids for use as DNA vaccines. Gene Ther. 2014, 21, 645–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brocato, R.L.; Principe, L.M.; Kim, R.K.; Zeng, X.; Williams, J.A.; Liu, Y.; Li, R.; Smith, J.M.; Golden, J.W.; Gangemi, D.; et al. Disruption of Adaptive Immunity Enhances Disease in SARS-CoV-2 Infected Syrian Hamsters. J. Virol. 2020, 94, e01683-20. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Surman, S.; Amaro-Carambot, E.; Kabatova, B.; Mackow, N.; Lingemann, M.; Yang, L.; McLellan, J.S.; Graham, B.S.; Kwong, P.D.; et al. Enhanced Neutralizing Antibody Response Induced by Respiratory Syncytial Virus Prefusion F Protein Expressed by a Vaccine Candidate. J. Virol. 2015, 89, 9499–9510. [Google Scholar] [CrossRef] [Green Version]
- McLellan, J.S.; Chen, M.; Joyce, M.G.; Sastry, M.; Stewart-Jones, G.B.; Yang, Y.; Zhang, B.; Chen, L.; Srivatsan, S.; Zheng, A.; et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 2013, 342, 592–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Niu, L.; Wittrock, K.N.; Clabaugh, G.C.; Srivastava, V.; Cho, M.W. A Structural Landscape of Neutralizing Antibodies against SARS-CoV-2 Receptor Binding Domain. Front. Immunol. 2021, 12, 647934. [Google Scholar] [CrossRef]
- Krarup, A.; Truan, D.; Furmanova-Hollenstein, P.; Bogaert, L.; Bouchier, P.; Bisschop, I.J.M.; Widjojoatmodjo, M.N.; Zahn, R.; Schuitemaker, H.; McLellan, J.S.; et al. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat. Commun. 2015, 6, 8143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karda, R.; Counsell, J.R.; Karbowniczek, K.; Caproni, L.J.; Tite, J.P.; Waddington, S.N. Production of lentiviral vectors using novel, enzymatically produced, linear DNA. Gene Ther. 2019, 26, 86–92. [Google Scholar] [CrossRef] [Green Version]
- Thornton, C.D.; Fielding, S.; Karbowniczek, K.; Roig-Merino, A.; Burrows, A.E.; FitzPatrick, L.M.; Sharaireh, A.; Tite, J.P.; Mole, S.E.; Harbottle, R.P.; et al. Safe and stable generation of induced pluripotent stem cells using doggybone DNA vectors. Mol. Ther. Methods Clin. Dev. 2021, 23, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Hornung, R.W.; Reed, L.D. Estimation of average concentration in the presence of non-detectable values. Appl. Occup. Environ. Hyg. 1990, 5, 46–51. [Google Scholar] [CrossRef]
- Liu, J.; Babka, A.M.; Kearney, B.J.; Radoshitzky, S.R.; Kuhn, J.H.; Zeng, X. Molecular detection of SARS-CoV-2 in formalin fixed paraffin embedded specimens. JCI Insight 2020, 5, e139042. [Google Scholar] [CrossRef]
- Sia, S.F.; Yan, L.M.; Chin, A.W.H.; Fung, K.; Choy, K.T.; Wong, A.Y.L.; Kaewpreedee, P.; Perera, R.; Poon, L.L.M.; Nicholls, J.M.; et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020, 583, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Tostanoski, L.H.; Wegmann, F.; Martinot, A.J.; Loos, C.; McMahan, K.; Mercado, N.B.; Yu, J.; Chan, C.N.; Bondoc, S.; Starke, C.E.; et al. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat. Med. 2020, 26, 1694–1700. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e819. [Google Scholar] [CrossRef] [PubMed]
- Wise, J. COVID-19: New coronavirus variant is identified in UK. BMJ 2020, 371, m4857. [Google Scholar] [CrossRef] [PubMed]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183, 739–751.e738. [Google Scholar] [CrossRef] [PubMed]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirchdoerfer, R.N.; Wang, N.; Pallesen, J.; Wrapp, D.; Turner, H.L.; Cottrell, C.A.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018, 8, 15701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jassat, W.; Mudara, C.; Ozougwu, L.; Tempia, S.; Blumberg, L.; Davies, M.A.; Pillay, Y.; Carter, T.; Morewane, R.; Wolmarans, M.; et al. Difference in mortality among individuals admitted to hospital with COVID-19 during the first and second waves in South Africa: A cohort study. Lancet Glob. Health 2021, 9, e1216–e1225. [Google Scholar] [CrossRef]
- Abu-Raddad, L.J.; Chemaitelly, H.; Butt, A.A.; National Study Group for COVID-19 Vaccination. Effectiveness of the BNT162b2 COVID-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N. Engl. J. Med. 2021, 385, 187–189. [Google Scholar] [CrossRef] [PubMed]
- Kandeel, M.; Mohamed, M.E.M.; Abd El-Lateef, H.M.; Venugopala, K.N.; El-Beltagi, H.S. Omicron variant genome evolution and phylogenetics. J. Med. Virol. 2022, 94, 1627–1632. [Google Scholar] [CrossRef] [PubMed]
- Momin, T.; Kansagra, K.; Patel, H.; Sharma, S.; Sharma, B.; Patel, J.; Mittal, R.; Sanmukhani, J.; Maithal, K.; Dey, A.; et al. Safety and Immunogenicity of a DNA SARS-CoV-2 vaccine (ZyCoV-D): Results of an open-label, non-randomized phase I part of phase I/II clinical study by intradermal route in healthy subjects in India. EClinicalMedicine 2021, 38, 101020. [Google Scholar] [CrossRef]
- Kwilas, S.; Kishimori, J.M.; Josleyn, M.; Jerke, K.; Ballantyne, J.; Royals, M.; Hooper, J.W. A hantavirus pulmonary syndrome (HPS) DNA vaccine delivered using a spring-powered jet injector elicits a potent neutralizing antibody response in rabbits and nonhuman primates. Curr. Gene Ther. 2014, 14, 200–210. [Google Scholar] [CrossRef] [Green Version]
- Lassauniere, R.; Polacek, C.; Gram, G.J.; Frische, A.; Tingstedt, J.L.; Kruger, M.; Dorner, B.G.; Cook, A.; Brown, R.; Orekov, T.; et al. Preclinical evaluation of a candidate naked plasmid DNA vaccine against SARS-CoV-2. NPJ Vaccines 2021, 6, 156. [Google Scholar] [CrossRef]
- Alamri, S.S.; Alluhaybi, K.A.; Alhabbab, R.Y.; Basabrain, M.; Algaissi, A.; Almahboub, S.; Alfaleh, M.A.; Abujamel, T.S.; Abdulaal, W.H.; ElAssouli, M.Z.; et al. Synthetic SARS-CoV-2 Spike-Based DNA Vaccine Elicits Robust and Long-Lasting Th1 Humoral and Cellular Immunity in Mice. Front. Microbiol. 2021, 12, 727455. [Google Scholar] [CrossRef]
- Ohlson, J. Plasmid manufacture is the bottleneck of the genetic medicine revolution. Drug Discov. Today 2020, 25, 1891–1893. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mucker, E.M.; Brocato, R.L.; Principe, L.M.; Kim, R.K.; Zeng, X.; Smith, J.M.; Kwilas, S.A.; Kim, S.; Horton, H.; Caproni, L.; et al. SARS-CoV-2 Doggybone DNA Vaccine Produces Cross-Variant Neutralizing Antibodies and Is Protective in a COVID-19 Animal Model. Vaccines 2022, 10, 1104. https://doi.org/10.3390/vaccines10071104
Mucker EM, Brocato RL, Principe LM, Kim RK, Zeng X, Smith JM, Kwilas SA, Kim S, Horton H, Caproni L, et al. SARS-CoV-2 Doggybone DNA Vaccine Produces Cross-Variant Neutralizing Antibodies and Is Protective in a COVID-19 Animal Model. Vaccines. 2022; 10(7):1104. https://doi.org/10.3390/vaccines10071104
Chicago/Turabian StyleMucker, Eric M., Rebecca L. Brocato, Lucia M. Principe, Robert K. Kim, Xiankun Zeng, Jeffrey M. Smith, Steven A. Kwilas, Sungwon Kim, Helen Horton, Lisa Caproni, and et al. 2022. "SARS-CoV-2 Doggybone DNA Vaccine Produces Cross-Variant Neutralizing Antibodies and Is Protective in a COVID-19 Animal Model" Vaccines 10, no. 7: 1104. https://doi.org/10.3390/vaccines10071104
APA StyleMucker, E. M., Brocato, R. L., Principe, L. M., Kim, R. K., Zeng, X., Smith, J. M., Kwilas, S. A., Kim, S., Horton, H., Caproni, L., & Hooper, J. W. (2022). SARS-CoV-2 Doggybone DNA Vaccine Produces Cross-Variant Neutralizing Antibodies and Is Protective in a COVID-19 Animal Model. Vaccines, 10(7), 1104. https://doi.org/10.3390/vaccines10071104





