Diagnostic Tools for Rapid Screening and Detection of SARS-CoV-2 Infection
Abstract
:1. Introduction
1.1. Onset of Disease and Transmission
1.2. Clinical Manifestation
2. Diagnosis
2.1. Nucleic Acid-Based Diagnosis
2.1.1. Reverse-Transcription–Polymerase Chain Reaction (RT-PCR)
2.1.2. Isothermal Nucleic Acid Amplification
Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP)
Transcription-Mediated Amplification (TMA)
Rolling Circle Amplification
CRISPR-Based Assays
2.1.3. Nucleic Acid Hybridization Microarray Assays
2.1.4. Nucleic Acid Sequencing
2.2. Serological and Immunological Assays
2.2.1. Enzyme-Linked Immunosorbent Assay (ELISA)
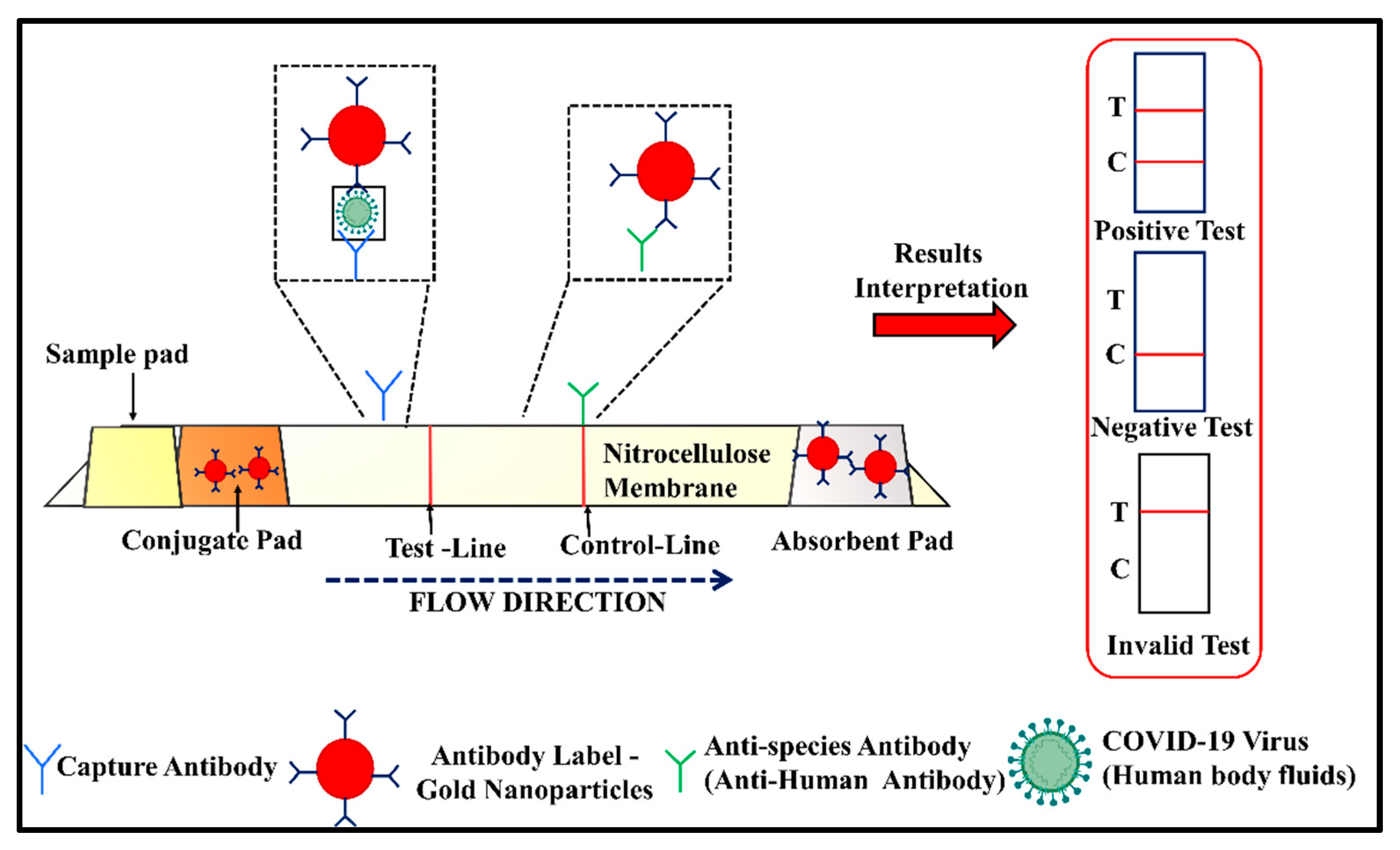
2.2.2. Lateral Flow Assay (LFA)
2.2.3. Luminescent Immunoassay
2.2.4. Protein Microarray
2.2.5. Agglutination Assay
2.2.6. Neutralization Assay
2.3. Imaging-Based Diagnosis
2.3.1. Chest Computed Tomography (CT) Scan
2.3.2. Lung Ultrasound (LUS)
3. Other Diagnostic Techniques
3.1. Biosensors
3.1.1. Colorimetric Biosensors
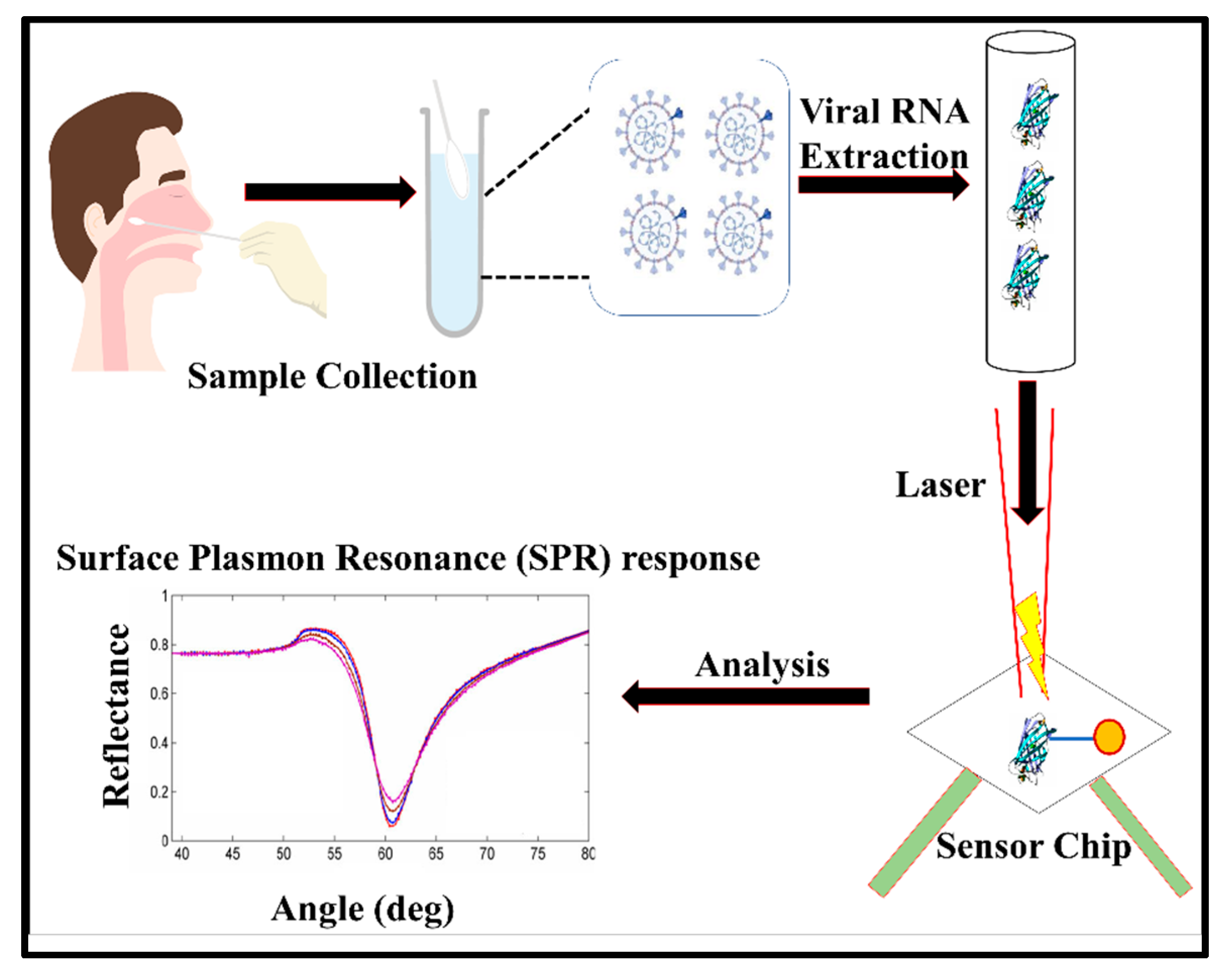
3.1.2. Plasmonic Biosensors
3.1.3. Electrochemical Biosensors
3.2. Spectroscopic Biosensors
| Name of Detection Technique | Type of Test | Infrastructure | Time of Detection | Accuracy | Qualitative OR Quantitative | Time Required after Infection for Detection | Medical Use | Reference |
|---|---|---|---|---|---|---|---|---|
| Reverse-transcription-polymerase chain reaction (RT-PCR) | Nucleic acid-based | Laboratory setup needed | 4 h | High | Both types | 1–8 days | Mostly used test | [49,50,53] |
| Isothermal nucleic acid amplification | Nucleic acid-based | Laboratory setup needed | >3 h | High | Both types | 1–8 days | Moderately used test | [67] |
| Reverse-transcription loop-mediated isothermal amplification (RT-LAMP) | Nucleic acid-based | Laboratory setup needed | 30 min−1 h | High | Both types | 1–8 days | Small use | [69,70,71] |
| Transcription-mediated amplification (TMA) | Nucleic acid-based | Laboratory setup needed | 30 min | High | Both types | 1–8 days | Small use | [76,78] |
| Rolling circle amplification | Nucleic acid-based | Laboratory setup needed | 90 min | High | Both types | 1–8 days | Small use | [79,80] |
| CRISPR-based assays | Nucleic acid-based | Laboratory setup needed | 1 h | High | Both types | 5–10 days | Moderately used test | [82,83,84] |
| Nucleic acid hybridization microarray assays | Nucleic acid-based | Laboratory setup needed | 2 h | High | Only quantitative | 5–10 days | Not in use: research stage | [90,91,92] |
| Nucleic acid sequencing | Nucleic acid-based | Laboratory setup needed | 2 h | High | Only qualitative | 5–10 days | Moderately used test | [95,97,98] |
| Enzyme-linked immunosorbent assay (ELISA) | Antibody detection | Laboratory setup needed | 2 h | High | Both | 5–10 days | Moderate | [110,111,112] |
| Lateral flow assay (LFA) | Rapid antigen detection | Point-of-Care Device | 10–30 min | High | Moderate to high | 10 days | Moderately used test | [115,117] |
| Luminescent immunoassay | IgM and IgG antibody detection in patients’ serum | Laboratory setup needed | 2–3 h | High | Both | 5–10 days | Moderately used test | [118,119,120] |
| Protein microarray | IgM and IgG antibody detection | Laboratory setup needed | 20–30 min | Moderate | Both | 5–10 days | Moderate to high | [121] |
| Agglutination assay | Antibody detection | Point-of-Care Device | 10–30 min | Moderate | Only qualitative | 5–10 days | Moderately used test | [122,123] |
| Chest computed tomography (CT) scan | Imaging-based diagnosis—radiography | Complex and costly test | 4–5 h | High (if lung infected) | Both | 4–15 days | Second after RT-PCR | [128,129] |
| Lung ultrasound (LUS) | Imaging-based diagnosis | Point-of-Care Device (portable device) | 1 h | High | Both | 4–15 days | Moderately used test | [131,133] |
| Plasmonic biosensors | Detection of viral pathogens | Point-of-Care Device | 10–30 min | Moderate | Both | 10 days | Not in use: research stage | [133,134] |
| Electrochemical biosensors | Antigen detection (SARS-CoV-2 spike proteins) | Point-of-Care Device | 30–60 min | Moderate | Both | 10 days | Not in use: research stage | [141,142,146,147] |
| Spectroscopic biosensors | Detection of viral pathogens/RNA | Point-of-Care Device | 10 min | Moderate | Both | 10 days | Not in use: research stage | [150,151] |
3.3. Piezoelectric Sensors
3.4. Microfluidic Sensor
4. Conclusions and Future Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jonsdottir, H.R.; Dijkman, R. Coronaviruses and the human airway: A universal system for virus-host interaction studies. Virol. J. 2016, 13, 24. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Lian, X.; Su, X.; Wu, W.; Marraro, G.A.; Zeng, Y. From SARS and MERS to COVID-19: A brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir. Res. 2020, 21, 224. [Google Scholar] [CrossRef]
- Cherry, J.D.; Krogstad, P. SARS: The First Pandemic of the 21st Century. Pediatr. Res. 2004, 56, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilgenfeld, R.; Peiris, M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antivir. Res. 2013, 100, 286–295. [Google Scholar] [CrossRef]
- Xiao, K.; Zhai, J.; Feng, Y.; Zhou, N.; Zhang, X.; Zou, J.-J.; Li, N.; Guo, Y.; Li, X.; Shen, X.; et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 2020, 583, 286–289. [Google Scholar] [CrossRef]
- Jalava, K. First respiratory transmitted food borne outbreak? Int. J. Hyg. Environ. Health 2020, 226, 113490. [Google Scholar] [CrossRef]
- Meyerowitz, E.A.; Richterman, A.; Gandhi, R.T.; Sax, P.E. Transmission of SARS-CoV-2: A Review of Viral, Host, and Environmental Factors. Ann. Intern. Med. 2020, 174, 69–79. [Google Scholar] [CrossRef]
- Greenhalgh, T.; Jimenez, J.L.; Prather, K.A.; Tufekci, Z.; Fisman, D.; Schooley, R. Ten scientific reasons in support of airborne transmission of SARS-CoV-2. Lancet 2021, 397, 1603–1605. [Google Scholar] [CrossRef]
- Coroneo, M.T.; Collignon, P.J. SARS-CoV-2: Eye protection might be the missing key. Lancet Microbe 2021, 2, e173–e174. [Google Scholar] [CrossRef]
- Liu, P.-Y.; He, S.; Rong, L.-B.; Tang, S.-Y. The effect of control measures on COVID-19 transmission in Italy: Comparison with Guangdong province in China. Infect. Dis. Poverty 2020, 9, 130. [Google Scholar] [CrossRef]
- Murano, Y.; Ueno, R.; Shi, S.; Kawashima, T.; Tanoue, Y.; Tanaka, S.; Nomura, S.; Shoji, H.; Shimizu, T.; Nguyen, H.; et al. Impact of domestic travel restrictions on transmission of COVID-19 infection using public transportation network approach. Sci. Rep. 2021, 11, 3109. [Google Scholar] [CrossRef]
- Tan, J.; Liu, S.; Zhuang, L.; Chen, L.; Dong, M.; Zhang, J.; Xin, Y. Transmission and clinical characteristics of asymptomatic patients with SARS-CoV-2 infection. Future Virol. 2020, 15, 373–380. [Google Scholar] [CrossRef]
- Mizumoto, K.; Kagaya, K.; Zarebski, A.; Chowell, G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance 2020, 25, 2000180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mbunge, E. Effects of COVID-19 in South African health system and society: An explanatory study. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 1809–1814. [Google Scholar] [CrossRef]
- Yuan, J.; Li, M.; Lv, G.; Lu, Z.K. Monitoring transmissibility and mortality of COVID-19 in Europe. Int. J. Infect. Dis. 2020, 95, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gayle, A.A.; Wilder-Smith, A.; Rocklöv, J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020, 27, taaa021. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Pei, S.; Chen, B.; Song, Y.; Zhang, T.; Yang, W.; Shaman, J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 2020, 368, 489–493. [Google Scholar] [CrossRef] [Green Version]
- Singhal, T. A Review of Coronavirus Disease-2019 (COVID-19). Indian J. Pediatrics 2020, 87, 281–286. [Google Scholar] [CrossRef] [Green Version]
- Billah, M.A.; Miah, M.M.; Khan, M.N. Reproductive number of coronavirus: A systematic review and meta-analysis based on global level evidence. PLoS ONE 2020, 15, e0242128. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Martinez, I.L.; Llinás, D.T.; Romero, M.B.; Salazar, L.M. High Mutation Rate in SARS-CoV-2: Will It Hit Us the Same Way Forever? J. Infect. Dis. Epidemiol. 2020, 6, 176. [Google Scholar] [CrossRef]
- Aleem, A.; Akbar Samad, A.B.; Slenker, A.K. Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19). In StatPearls; StatPearls Publishing LLC: Treasure Island, FL, USA, 2022. [Google Scholar]
- Thiruvengadam, R.; Binayke, A.; Awasthi, A. SARS-CoV-2 delta variant: A persistent threat to the effectiveness of vaccines. Lancet Infect. Dis. 2022, 22, 301–302. [Google Scholar] [CrossRef]
- Sandmann, F.G.; Jit, M. Rapid COVID-19 vaccine rollout: Immense success but challenges ahead. Lancet Infect. Dis. 2022, 22, 302–304. [Google Scholar] [CrossRef]
- Bálint, G.; Vörös-Horváth, B.; Széchenyi, A. Omicron: Increased transmissibility and decreased pathogenicity. Signal Transduct. Target. Ther. 2022, 7, 151. [Google Scholar] [CrossRef]
- Kumar, S.; Thambiraja, T.S.; Karuppanan, K.; Subramaniam, G. Omicron and Delta variant of SARS-CoV-2: A comparative computational study of spike protein. J. Med. Virol. 2022, 94, 1641–1649. [Google Scholar] [CrossRef]
- Harrison, A.G.; Lin, T.; Wang, P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020, 41, 1100–1115. [Google Scholar] [CrossRef] [PubMed]
- da Rosa Mesquita, R.; Francelino Silva Junior, L.C.; Santos Santana, F.M.; Farias de Oliveira, T.; Campos Alcântara, R.; Monteiro Arnozo, G.; Rodrigues da Silva Filho, E.; Galdino dos Santos, A.G.; Oliveira da Cunha, E.J.; Salgueiro de Aquino, S.H.; et al. Clinical manifestations of COVID-19 in the general population: Systematic review. Wien. Klin. Wochenschr. 2021, 133, 377–382. [Google Scholar] [CrossRef]
- Brodin, P. Immune determinants of COVID-19 disease presentation and severity. Nat. Med. 2021, 27, 28–33. [Google Scholar] [CrossRef] [PubMed]
- van Westen-Lagerweij, N.A.; Meijer, E.; Meeuwsen, E.G.; Chavannes, N.H.; Willemsen, M.C.; Croes, E.A. Are smokers protected against SARS-CoV-2 infection (COVID-19)? The origins of the myth. NPJ Prim. Care Respir. Med. 2021, 31, 10. [Google Scholar] [CrossRef]
- Hopkinson, N.S.; Rossi, N.; El-Sayed_Moustafa, J.; Laverty, A.A.; Quint, J.K.; Freidin, M.; Visconti, A.; Murray, B.; Modat, M.; Ourselin, S.; et al. Current smoking and COVID-19 risk: Results from a population symptom app in over 2.4 million people. Thorax 2021, 76, 714. [Google Scholar] [CrossRef]
- Zou, H.; Lu, J.; Liu, J.; Wong, J.H.-Y.; Cheng, S.; Li, Q.; Shen, Y.; Li, C.; Jia, X. Characteristics of pediatric multi-system inflammatory syndrome (PMIS) associated with COVID-19: A meta-analysis and insights into pathogenesis. Int. J. Infect. Dis. 2021, 102, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.-M.; Bai, P.; He, W.; Wu, F.; Liu, X.-F.; Han, D.-M.; Liu, S.; Yang, J.-K. Gender Differences in Patients with COVID-19: Focus on Severity and Mortality. Front. Public Health 2020, 8, 152. [Google Scholar] [CrossRef]
- Ortolan, A.; Lorenzin, M.; Felicetti, M.; Doria, A.; Ramonda, R. Does gender influence clinical expression and disease outcomes in COVID-19? A systematic review and meta-analysis. Int. J. Infect. Dis. 2020, 99, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Peckham, H.; de Gruijter, N.M.; Raine, C.; Radziszewska, A.; Ciurtin, C.; Wedderburn, L.R.; Rosser, E.C.; Webb, K.; Deakin, C.T. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun. 2020, 11, 6317. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.N.; Hussain, F.; Hashmi, S.K. Role of testosterone in COVID-19 patients—A double-edged sword? Med. Hypotheses 2020, 144, 110287. [Google Scholar] [CrossRef]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.-Y.; Chen, L.; Wang, M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA 2020, 323, 1406–1407. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, R.; He, Q.; Pascual, M. Quantifying asymptomatic infection and transmission of COVID-19 in New York City using observed cases, serology, and testing capacity. Proc. Natl. Acad. Sci. USA 2021, 118, e2019716118. [Google Scholar] [CrossRef]
- Pascarella, G.; Strumia, A.; Piliego, C.; Bruno, F.; Del Buono, R.; Costa, F.; Scarlata, S.; Agrò, F.E. COVID-19 diagnosis and management: A comprehensive review. J. Intern. Med. 2020, 288, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Vandenberg, O.; Martiny, D.; Rochas, O.; van Belkum, A.; Kozlakidis, Z. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 2021, 19, 171–183. [Google Scholar] [CrossRef]
- Fathi Karkan, S.; Maleki Baladi, R.; Shahgolzari, M.; Gholizadeh, M.; Shayegh, F.; Arashkia, A. The evolving direct and indirect platforms for the detection of SARS-CoV-2. J. Virol. Methods 2022, 300, 114381. [Google Scholar] [CrossRef]
- Naikoo, G.A.; Arshad, F.; Hassan, I.U.; Awan, T.; Salim, H.; Pedram, M.Z.; Ahmed, W.; Patel, V.; Karakoti, A.S.; Vinu, A. Nanomaterials-based sensors for the detection of COVID-19: A review. Bioeng. Transl. Med. 2022, e10305, Online ahead of print. [Google Scholar] [CrossRef]
- Kurkela, S.; Brown, D.W.G. Molecular diagnostic techniques. Medicine 2009, 37, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, M. Next-Generation Sequencing in Diagnostic Pathology. Pathobiology 2017, 84, 292–305. [Google Scholar] [CrossRef] [Green Version]
- Müller, K.; Wirth, M. Real-time RT-PCR detection of retroviral contaminations of cells and cell lines. Cytotechnology 2002, 38, 147–153. [Google Scholar] [CrossRef]
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef] [Green Version]
- Czumbel, L.M.; Kiss, S.; Farkas, N.; Mandel, I.; Hegyi, A.; Nagy, Á.; Lohinai, Z.; Szakács, Z.; Hegyi, P.; Steward, M.C.; et al. Saliva as a Candidate for COVID-19 Diagnostic Testing: A Meta-Analysis. Front. Med. 2020, 7, 465. [Google Scholar] [CrossRef]
- Hung, K.-F.; Sun, Y.-C.; Chen, B.-H.; Lo, J.-F.; Cheng, C.-M.; Chen, C.-Y.; Wu, C.-H.; Kao, S.-Y. New COVID-19 saliva-based test: How good is it compared with the current nasopharyngeal or throat swab test? J. Chin. Med. Assoc. 2020, 83, 891–894. [Google Scholar] [CrossRef]
- Malik, M.; Kunze, A.-C.; Bahmer, T.; Herget-Rosenthal, S.; Kunze, T. SARS-CoV-2: Viral Loads of Exhaled Breath and Oronasopharyngeal Specimens in Hospitalized Patients with COVID-19. Int. J. Infect. Dis. 2021, 110, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Sawano, M.; Takeshita, K.; Ohno, H.; Oka, H. RT-PCR diagnosis of COVID-19 from exhaled breath condensate: A clinical study. J. Breath Res. 2021, 15, 037103. [Google Scholar] [CrossRef]
- Giovannini, G.; Haick, H.; Garoli, D. Detecting COVID-19 from Breath: A Game Changer for a Big Challenge. ACS Sens. 2021, 6, 1408–1417. [Google Scholar] [CrossRef]
- Teymouri, M.; Mollazadeh, S.; Mortazavi, H.; Naderi Ghale-noie, Z.; Keyvani, V.; Aghababaei, F.; Hamblin, M.R.; Abbaszadeh-Goudarzi, G.; Pourghadamyari, H.; Hashemian, S.M.R.; et al. Recent advances and challenges of RT-PCR tests for the diagnosis of COVID-19. Pathol.-Res. Pract. 2021, 221, 153443. [Google Scholar] [CrossRef]
- Yilmaz Gulec, E.; Cesur, N.P.; Yesilyurt Fazlioğlu, G.; Kazezoğlu, C. Effect of different storage conditions on COVID-19 RT-PCR results. J. Med. Virol. 2021, 93, 6575–6581. [Google Scholar] [CrossRef] [PubMed]
- Tsang, N.N.Y.; So, H.C.; Ng, K.Y.; Cowling, B.J.; Leung, G.M.; Ip, D.K.M. Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: A systematic review and meta-analysis. Lancet Infect. Dis. 2021, 21, 1233–1245. [Google Scholar] [CrossRef]
- Pecoraro, V.; Negro, A.; Pirotti, T.; Trenti, T. Estimate false-negative RT-PCR rates for SARS-CoV-2. A systematic review and meta-analysis. Eur. J. Clin. Investig. 2022, 52, e13706. [Google Scholar] [CrossRef] [PubMed]
- Smet, K.D.; Smet, D.D.; Ryckaert, T.; Laridon, E.; Heremans, B.; Vandenbulcke, R.; Demedts, I.; Bouckaert, B.; Gryspeerdt, S.; Martens, G.A. Diagnostic Performance of Chest CT for SARS-CoV-2 Infection in Individuals with or without COVID-19 Symptoms. Radiology 2021, 298, E30–E37. [Google Scholar] [CrossRef] [PubMed]
- Ai, T.; Yang, Z.; Hou, H.; Zhan, C.; Chen, C.; Lv, W.; Tao, Q.; Sun, Z.; Xia, L. Correlation of Chest CT and RT-PCR Testing for Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology 2020, 296, E32–E40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasudevan, H.N.; Xu, P.; Servellita, V.; Miller, S.; Liu, L.; Gopez, A.; Chiu, C.Y.; Abate, A.R. Digital droplet PCR accurately quantifies SARS-CoV-2 viral load from crude lysate without nucleic acid purification. Sci. Rep. 2021, 11, 780. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Fan, D.; Wang, N.; Wang, F.; Wang, B.; Zhu, L.; Guo, Y. Applications of digital PCR in COVID-19 pandemic. VIEW 2021, 2, 20200082. [Google Scholar] [CrossRef]
- Tedim, A.P.; Almansa, R.; Domínguez-Gil, M.; González-Rivera, M.; Micheloud, D.; Ryan, P.; Méndez, R.; Blanca-López, N.; Pérez-García, F.; Bustamante, E.; et al. Comparison of real-time and droplet digital PCR to detect and quantify SARS-CoV-2 RNA in plasma. Eur. J. Clin. Investig. 2021, 51, e13501. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef]
- Jang, Y.O.; Lee, H.J.; Koo, B.; Cha, H.-H.; Kwon, J.-S.; Kim, J.Y.; Kim, M.G.; Kim, H.S.; Kim, S.-H.; Shin, Y. Rapid COVID-19 Molecular Diagnostic System Using Virus Enrichment Platform. Biosensors 2021, 11, 373. [Google Scholar] [CrossRef]
- Njiru, Z.K. Loop-Mediated Isothermal Amplification Technology: Towards Point of Care Diagnostics. PLoS Negl. Trop. Dis. 2012, 6, e1572. [Google Scholar] [CrossRef] [Green Version]
- Rhoads Daniel, D.; Cherian Sree, S.; Roman, K.; Stempak Lisa, M.; Schmotzer Christine, L.; Sadri, N.; McAdam Alexander, J. Comparison of Abbott ID Now, DiaSorin Simplexa, and CDC FDA Emergency Use Authorization Methods for the Detection of SARS-CoV-2 from Nasopharyngeal and Nasal Swabs from Individuals Diagnosed with COVID-19. J. Clin. Microbiol. 2020, 58, e00760-20. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Odiwuor, N.; Xiong, J.; Sun, L.; Nyaruaba, R.O.; Wei, H.; Tanner, N.A. Rapid Molecular Detection of SARS-CoV-2 (COVID-19) Virus RNA Using Colorimetric LAMP. medRxiv 2020. [Google Scholar] [CrossRef]
- El-Tholoth, M.; Bau, H.H.; Song, J. A Single and Two-Stage, Closed-Tube, Molecular Test for the 2019 Novel Coronavirus (COVID-19) at Home, Clinic, and Points of Entry. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Jhou, Y.-R.; Wang, C.-H.; Tsai, H.-P.; Shan, Y.-S.; Lee, G.-B. An integrated microfluidic platform featuring real-time reverse transcription loop-mediated isothermal amplification for detection of COVID-19. Sens. Actuators B Chem. 2022, 358, 131447. [Google Scholar] [CrossRef]
- de Oliveira, K.G.; Estrela, P.F.N.; Mendes, G.d.M.; dos Santos, C.A.; Silveira-Lacerda, E.d.P.; Duarte, G.R.M. Rapid molecular diagnostics of COVID-19 by RT-LAMP in a centrifugal polystyrene-toner based microdevice with end-point visual detection. Analyst 2021, 146, 1178–1187. [Google Scholar] [CrossRef]
- Monis, P.T.; Giglio, S. Nucleic acid amplification-based techniques for pathogen detection and identification. Infect. Genet. Evol. 2006, 6, 2–12. [Google Scholar] [CrossRef]
- Gorzalski, A.J.; Tian, H.; Laverdure, C.; Morzunov, S.; Verma, S.C.; VanHooser, S.; Pandori, M.W. High-Throughput Transcription-mediated amplification on the Hologic Panther is a highly sensitive method of detection for SARS-CoV-2. J. Clin. Virol. 2020, 129, 104501. [Google Scholar] [CrossRef]
- Newsom, K.; Zhang, Y.; Chamala, S.; Martinez, K.; Clare-Salzler, M.; Starostik, P. The Hologic Aptima SARS-CoV-2 assay enables high ratio pooling saving reagents and improving turnaround time. J. Clin. Lab. Anal. 2021, 35, e23888. [Google Scholar] [CrossRef]
- Goo, N.-I.; Kim, D.-E. Rolling circle amplification as isothermal gene amplification in molecular diagnostics. BioChip J. 2016, 10, 262–271. [Google Scholar] [CrossRef]
- Huang, W.; Ting, J.; Fang, M.; Hsu, H.; Su, J.; Misaki, T.; Chan, D.; Yang, J.; Yeh, T.-Y.; Yang, K.; et al. Room Temperature Isothermal Colorimetric Padlock Probe Rolling Circle Amplification for Viral DNA and RNA Detection. bioRxiv 2021. [Google Scholar] [CrossRef]
- Jain, S.; Dandy, D.S.; Geiss, B.J.; Henry, C.S. Padlock probe-based rolling circle amplification lateral flow assay for point-of-need nucleic acid detection. Analyst 2021, 146, 4340–4347. [Google Scholar] [CrossRef] [PubMed]
- Hille, F.; Charpentier, E. CRISPR-Cas: Biology, mechanisms and relevance. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150496. [Google Scholar] [CrossRef]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Yu, L.; Liu, C.; Ye, S.; Chen, W.; Li, D.; Huang, W. One-tube SARS-CoV-2 detection platform based on RT-RPA and CRISPR/Cas12a. J. Transl. Med. 2021, 19, 74. [Google Scholar] [CrossRef]
- Sheridan, C. COVID-19 spurs wave of innovative diagnostics. Nat. Biotechnol. 2020, 38, 769–772. [Google Scholar] [CrossRef]
- Hajian, R.; Balderston, S.; Tran, T.; deBoer, T.; Etienne, J.; Sandhu, M.; Wauford, N.A.; Chung, J.-Y.; Nokes, J.; Athaiya, M.; et al. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019, 3, 427–437. [Google Scholar] [CrossRef]
- Bruch, R.; Baaske, J.; Chatelle, C.; Meirich, M.; Madlener, S.; Weber, W.; Dincer, C.; Urban, G.A. CRISPR/Cas13a-Powered Electrochemical Microfluidic Biosensor for Nucleic Acid Amplification-Free miRNA Diagnostics. Adv. Mater. 2019, 31, 1905311. [Google Scholar] [CrossRef] [Green Version]
- Fozouni, P.; Son, S.; Díaz de León Derby, M.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2021, 184, 323–333.e329. [Google Scholar] [CrossRef] [PubMed]
- Long, W.-H.; Xiao, H.-S.; Gu, X.-M.; Zhang, Q.-H.; Yang, H.-J.; Zhao, G.-P.; Liu, J.-H. A universal microarray for detection of SARS coronavirus. J. Virol. Methods 2004, 121, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Li, J.; Deng, Z.; Xiong, W.; Wang, Q.; Hu, Y. Comprehensive Detection and Identification of Seven Animal Coronaviruses and Human Respiratory Coronavirus 229E with a Microarray Hybridization Assay. Intervirology 2010, 53, 95–104. [Google Scholar] [CrossRef]
- Shaffaf, T.; Ghafar-Zadeh, E. COVID-19 Diagnostic Strategies. Part I: Nucleic Acid-Based Technologies. Bioengineering 2021, 8, 49. [Google Scholar] [CrossRef]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.-S.F.; Kehe, J.; et al. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef]
- Damin, F.; Galbiati, S.; Gagliardi, S.; Cereda, C.; Dragoni, F.; Fenizia, C.; Savasi, V.; Sola, L.; Chiari, M. CovidArray: A Microarray-Based Assay with High Sensitivity for the Detection of Sars-Cov-2 in Nasopharyngeal Swabs. Sensors 2021, 21, 2490. [Google Scholar] [CrossRef]
- Moore, S.C.; Penrice-Randal, R.; Alruwaili, M.; Dong, X.; Pullan, S.T.; Carter, D.P.; Bewley, K.; Zhao, Q.; Sun, Y.; Hartley, C.; et al. Amplicon based MinION sequencing of SARS-CoV-2 and metagenomic characterisation of nasopharyngeal swabs from patients with COVID-19. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Togun, T.; Kampmann, B.; Stoker, N.G.; Lipman, M. Anticipating the impact of the COVID-19 pandemic on TB patients and TB control programmes. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 21. [Google Scholar] [CrossRef]
- Dhar, D.; Mohanty, A. Gut microbiota and Covid-19- possible link and implications. Virus Res. 2020, 285, 198018. [Google Scholar] [CrossRef] [PubMed]
- Freed, N.E.; Vlková, M.; Faisal, M.B.; Silander, O.K. Rapid and inexpensive whole-genome sequencing of SARS-CoV-2 using 1200 bp tiled amplicons and Oxford Nanopore Rapid Barcoding. Biol. Methods Protoc. 2020, 5, bpaa014. [Google Scholar] [CrossRef] [PubMed]
- Yermanos, A.; Hong, K.-L.; Agrafiotis, A.; Han, J.; Nadeau, S.; Valenzuela, C.; Azizoglu, A.; Ehling, R.; Gao, B.; Spahr, M.; et al. DeepSARS: Simultaneous diagnostic detection and genomic surveillance of SARS-CoV-2. BMC Genom. 2022, 23, 289. [Google Scholar] [CrossRef] [PubMed]
- Bechtold, P.; Wagner, P.; Hosch, S.; Siegrist, D.; Ruiz-Serrano, A.; Gregorini, M.; Mpina, M.; Ondó, F.A.; Obama, J.; Ayekaba, M.O.o.; et al. Rapid Identification of SARS-CoV-2 Variants of Concern Using a Portable peakPCR Platform. Anal. Chem. 2021, 93, 16350–16359. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.A.; Williams, C.L.; Penrod, Y.; McCloskey, C.; Carpenter-Azevedo, K.; Huard, R.C.; King, E.; Terence Dunn, S. A pyrosequencing protocol for rapid identification of SARS-CoV-2 variants. J. Med. Virol. 2022, 94, 3661–3668. [Google Scholar] [CrossRef]
- Ghaffari, A.; Meurant, R.; Ardakani, A. COVID-19 Serological Tests: How Well Do They Actually Perform? Diagnostics 2020, 10, 453. [Google Scholar] [CrossRef]
- Carvalho, T.; Krammer, F.; Iwasaki, A. The first 12 months of COVID-19: A timeline of immunological insights. Nat. Rev. Immunol. 2021, 21, 245–256. [Google Scholar] [CrossRef]
- Jacofsky, D.; Jacofsky, E.M.; Jacofsky, M. Understanding Antibody Testing for COVID-19. J. Arthroplast. 2020, 35, S74–S81. [Google Scholar] [CrossRef]
- Lee, C.Y.-P.; Lin, R.T.P.; Renia, L.; Ng, L.F.P. Serological Approaches for COVID-19: Epidemiologic Perspective on Surveillance and Control. Front. Immunol. 2020, 11, 879. [Google Scholar] [CrossRef]
- Premkumar, L.; Segovia-Chumbez, B.; Jadi, R.; Martinez, D.R.; Raut, R.; Markmann, A.J.; Cornaby, C.; Bartelt, L.; Weiss, S.; Park, Y.; et al. The receptor-binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020, 5, eabc8413. [Google Scholar] [CrossRef]
- Ge, J.; Wang, R.; Ju, B.; Zhang, Q.; Sun, J.; Chen, P.; Zhang, S.; Tian, Y.; Shan, S.; Cheng, L.; et al. Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry. Nat. Commun. 2021, 12, 250. [Google Scholar] [CrossRef]
- Mehdi, F.; Chattopadhyay, S.; Thiruvengadam, R.; Yadav, S.; Kumar, M.; Sinha, S.K.; Goswami, S.; Kshetrapal, P.; Wadhwa, N.; Chandramouli Natchu, U.; et al. Development of a Fast SARS-CoV-2 IgG ELISA, Based on Receptor-Binding Domain, and Its Comparative Evaluation Using Temporally Segregated Samples From RT-PCR Positive Individuals. Front. Microbiol. 2021, 11, 618097. [Google Scholar] [CrossRef]
- Bendavid, E.; Mulaney, B.; Sood, N.; Shah, S.; Bromley-Dulfano, R.; Lai, C.; Weissberg, Z.; Saavedra-Walker, R.; Tedrow, J.; Bogan, A.; et al. COVID-19 antibody seroprevalence in Santa Clara County, California. Int. J. Epidemiol. 2021, 50, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Michel, M.; Bouam, A.; Edouard, S.; Fenollar, F.; Di Pinto, F.; Mège, J.-L.; Drancourt, M.; Vitte, J. Evaluating ELISA, Immunofluorescence, and Lateral Flow Assay for SARS-CoV-2 Serologic Assays. Front. Microbiol. 2020, 11, 597529. [Google Scholar] [CrossRef]
- Espejo, A.P.; Akgun, Y.; Al Mana, A.F.; Tjendra, Y.; Millan, N.C.; Gomez-Fernandez, C.; Cray, C. Review of Current Advances in Serologic Testing for COVID-19. Am. J. Clin. Pathol. 2020, 154, 293–304. [Google Scholar] [CrossRef]
- Song, Y.; Song, J.; Wei, X.; Huang, M.; Sun, M.; Zhu, L.; Lin, B.; Shen, H.; Zhu, Z.; Yang, C. Discovery of Aptamers Targeting the Receptor-Binding Domain of the SARS-CoV-2 Spike Glycoprotein. Anal. Chem. 2020, 92, 9895–9900. [Google Scholar] [CrossRef]
- Zhang, G.; Pomplun, S.; Loftis, A.R.; Tan, X.; Loas, A.; Pentelute, B.L. Investigation of ACE2 N-terminal fragments binding to SARS-CoV-2 Spike RBD. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Huergo, L.F.; Selim, K.A.; Conzentino, M.S.; Gerhardt, E.C.M.; Santos, A.R.S.; Wagner, B.; Alford, J.T.; Deobald, N.; Pedrosa, F.O.; de Souza, E.M.; et al. Magnetic Bead-Based Immunoassay Allows Rapid, Inexpensive, and Quantitative Detection of Human SARS-CoV-2 Antibodies. ACS Sens. 2021, 6, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Yamayoshi, S.; Sakai-Tagawa, Y.; Koga, M.; Akasaka, O.; Nakachi, I.; Koh, H.; Maeda, K.; Adachi, E.; Saito, M.; Nagai, H.; et al. Comparison of Rapid Antigen Tests for COVID-19. Viruses 2020, 12, 1420. [Google Scholar] [CrossRef]
- Vogl, T.; Leviatan, S.; Segal, E. SARS-CoV-2 antibody testing for estimating COVID-19 prevalence in the population. Cell Rep. Med. 2021, 2, 100191. [Google Scholar] [CrossRef] [PubMed]
- Srivastav, S.; Dankov, A.; Adanalic, M.; Grzeschik, R.; Tran, V.; Pagel-Wieder, S.; Gessler, F.; Spreitzer, I.; Scholz, T.; Schnierle, B.; et al. Rapid and Sensitive SERS-Based Lateral Flow Test for SARS-CoV2-Specific IgM/IgG Antibodies. Anal. Chem. 2021, 93, 12391–12399. [Google Scholar] [CrossRef]
- Lijia, S.; Lihong, S.; Huabin, W.; Xiaoping, X.; Xiaodong, L.; Yixuan, Z.; Pin, H.; Yina, X.; Xiaoyun, S.; Junqi, W. Serological chemiluminescence immunoassay for the diagnosis of SARS-CoV-2 infection. J. Clin. Lab. Anal. 2020, 34, e23466. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Wen, D.; Wu, J.; Liu, L.; Wu, W.; Fang, X.; Kong, J. Microfluidic Immunoassays for Sensitive and Simultaneous Detection of IgG/IgM/Antigen of SARS-CoV-2 within 15 min. Anal. Chem. 2020, 92, 9454–9458. [Google Scholar] [CrossRef]
- Jiang, H.-W.; Li, Y.; Zhang, H.-N.; Wang, W.; Yang, X.; Qi, H.; Li, H.; Men, D.; Zhou, J.; Tao, S.-C. SARS-CoV-2 proteome microarray for global profiling of COVID-19 specific IgG and IgM responses. Nat. Commun. 2020, 11, 3581. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Chaudhary, V.K. Whole-Blood Agglutination Assay for On-Site Detection of Human Immunodeficiency Virus Infection. J. Clin. Microbiol. 2003, 41, 2814–2821. [Google Scholar] [CrossRef] [Green Version]
- Alves, D.; Curvello, R.; Henderson, E.; Kesarwani, V.; Walker, J.A.; Leguizamon, S.C.; McLiesh, H.; Raghuwanshi, V.S.; Samadian, H.; Wood, E.M.; et al. Rapid Gel Card Agglutination Assays for Serological Analysis Following SARS-CoV-2 Infection in Humans. ACS Sens. 2020, 5, 2596–2603. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7001–7003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyakawa, K.; Jeremiah, S.S.; Ohtake, N.; Matsunaga, S.; Yamaoka, Y.; Nishi, M.; Morita, T.; Saji, R.; Nishii, M.; Kimura, H.; et al. Rapid quantitative screening assay for SARS-CoV-2 neutralizing antibodies using HiBiT-tagged virus-like particles. J. Mol. Cell Biol. 2020, 12, 987–990. [Google Scholar] [CrossRef] [PubMed]
- Sholukh, A.M.; Fiore-Gartland, A.; Ford, E.S.; Hou, Y.J.; Tse, V.; Kaiser, H.; Park, A.; Lempp, F.A.; Germain, R.S.; Bossard, E.; et al. Evaluation of SARS-CoV-2 neutralization assays for antibody monitoring in natural infection and vaccine trials. medRxiv 2020. [Google Scholar] [CrossRef]
- Hufsky, F.; Lamkiewicz, K.; Almeida, A.; Aouacheria, A.; Arighi, C.; Bateman, A.; Baumbach, J.; Beerenwinkel, N.; Brandt, C.; Cacciabue, M.; et al. Computational strategies to combat COVID-19: Useful tools to accelerate SARS-CoV-2 and coronavirus research. Brief. Bioinform. 2021, 22, 642–663. [Google Scholar] [CrossRef]
- Sarki, R.; Ahmed, K.; Wang, H.; Zhang, Y.; Wang, K. Automated detection of COVID-19 through convolutional neural network using chest x-ray images. PLoS ONE 2022, 17, e0262052. [Google Scholar] [CrossRef]
- Chakraborty, S.; Murali, B.; Mitra, A.K. An Efficient Deep Learning Model to Detect COVID-19 Using Chest X-ray Images. Int. J. Environ. Res. Public Health 2022, 19, 2013. [Google Scholar] [CrossRef]
- Pata, D.; Valentini, P.; De Rose, C.; De Santis, R.; Morello, R.; Buonsenso, D. Chest Computed Tomography and Lung Ultrasound Findings in COVID-19 Pneumonia: A Pocket Review for Non-radiologists. Front. Med. 2020, 7, 375. [Google Scholar] [CrossRef] [PubMed]
- Allinovi, M.; Parise, A.; Giacalone, M.; Amerio, A.; Delsante, M.; Odone, A.; Franci, A.; Gigliotti, F.; Amadasi, S.; Delmonte, D.; et al. Lung Ultrasound May Support Diagnosis and Monitoring of COVID-19 Pneumonia. Ultrasound Med. Biol. 2020, 46, 2908–2917. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, S.; Savinelli, C.; Paolucci, E.; Pelagatti, L.; Sibona, E.; Fersini, N.; Buggea, M.; Tozzi, C.; Allescia, G.; Paolini, D.; et al. Point-of-care ultrasound (PoCUS) in the early diagnosis of novel coronavirus 2019 disease (COVID-19) in a first-level emergency department during a SARS-CoV-2 outbreak in Italy: A real-life analysis. Intern. Emerg. Med. 2022, 17, 193–204. [Google Scholar] [CrossRef]
- Di Gioia, C.C.; Artusi, N.; Xotta, G.; Bonsano, M.; Sisto, U.G.; Tecchiolli, M.; Orso, D.; Cominotto, F.; Amore, G.; Meduri, S.; et al. Lung ultrasound in ruling out COVID-19 pneumonia in the ED: A multicentre prospective sensitivity study. Emerg. Med. J. 2022, 39, 199. [Google Scholar] [CrossRef] [PubMed]
- Copetti, R.; Amore, G.; Giudice, C.A.; Orso, D.; Cola, S.; Pillinini, P.; Rocco, C.; Cappello, D.; Dibenedetto, A.G.; Meduri, S. Lung Ultrasonography in Ruling Out COVID-19 Among Health Care Workers in Two Italian Emergency Departments: A Multicenter Study. J. Diagn. Med. Sonogr. 2021, 38, 45–51. [Google Scholar] [CrossRef]
- Zhao, V.X.T.; Wong, T.I.; Zheng, X.T.; Tan, Y.N.; Zhou, X. Colorimetric biosensors for point-of-care virus detections. Mater. Sci. Energy Technol. 2020, 3, 237–249. [Google Scholar] [CrossRef]
- Singh, B.; Datta, B.; Ashish, A.; Dutta, G. A comprehensive review on current COVID-19 detection methods: From lab care to point of care diagnosis. Sens. Int. 2021, 2, 100119. [Google Scholar] [CrossRef] [PubMed]
- Ardekani, L.S.; Thulstrup, P.W. Gold Nanoparticle-Mediated Lateral Flow Assays for Detection of Host Antibodies and COVID-19 Proteins. Nanomaterials 2022, 12, 1456. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Shrivastava, S.; Kausley, S.B.; Rai, B.; Pandit, A.B. Coronavirus: A comparative analysis of detection technologies in the wake of emerging variants. Infection 2022. Online ahead of print. [Google Scholar] [CrossRef]
- Alberti, G.; Zanoni, C.; Magnaghi, L.R.; Biesuz, R. Gold and Silver Nanoparticle-Based Colorimetric Sensors: New Trends and Applications. Chemosensors 2021, 9, 305. [Google Scholar] [CrossRef]
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moitra, P.; Alafeef, M.; Dighe, K.; Frieman, M.B.; Pan, D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14, 7617–7627. [Google Scholar] [CrossRef] [PubMed]
- Murugan, D.; Bhatia, H.; Sai, V.V.R.; Satija, J. P-FAB: A Fiber-Optic Biosensor Device for Rapid Detection of COVID-19. Trans. Indian Natl. Acad. Eng. 2020, 5, 211–215. [Google Scholar] [CrossRef]
- Calvo-Lozano, O.; Sierra, M.; Soler, M.; Estévez, M.C.; Chiscano-Camón, L.; Ruiz-Sanmartin, A.; Ruiz-Rodriguez, J.C.; Ferrer, R.; González-López, J.J.; Esperalba, J.; et al. Label-Free Plasmonic Biosensor for Rapid, Quantitative, and Highly Sensitive COVID-19 Serology: Implementation and Clinical Validation. Anal. Chem. 2022, 94, 975–984. [Google Scholar] [CrossRef]
- Basso, C.R.; Malossi, C.D.; Haisi, A.; de Albuquerque Pedrosa, V.; Barbosa, A.N.; Grotto, R.T.; Araujo Junior, J.P. Fast and reliable detection of SARS-CoV-2 antibodies based on surface plasmon resonance. Anal. Methods 2021, 13, 3297–3306. [Google Scholar] [CrossRef]
- Arduini, F.; Cinti, S.; Scognamiglio, V.; Moscone, D.; Palleschi, G. How cutting-edge technologies impact the design of electrochemical (bio)sensors for environmental analysis. A review. Anal. Chim. Acta 2017, 959, 15–42. [Google Scholar] [CrossRef]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef] [Green Version]
- Mavrikou, S.; Moschopoulou, G.; Tsekouras, V.; Kintzios, S. Development of a Portable, Ultra-Rapid and Ultra-Sensitive Cell-Based Biosensor for the Direct Detection of the SARS-CoV-2 S1 Spike Protein Antigen. Sensors 2020, 20, 3121. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-I.; Subramanian, A.; Mueller, S.; Levon, K.; Nam, C.-Y.; Rafailovich, M.H. Potentiometric Biosensors Based on Molecular-Imprinted Self-Assembled Monolayer Films for Rapid Detection of Influenza A Virus and SARS-CoV-2 Spike Protein. ACS Appl. Nano Mater. 2022, 5, 5045–5055. [Google Scholar] [CrossRef]
- Tam, C.C.; Flannery, A.R.; Cheng, L.W. A Rapid, Sensitive, and Portable Biosensor Assay for the Detection of Botulinum Neurotoxin Serotype A in Complex Food Matrices. Toxins 2018, 10, 476. [Google Scholar] [CrossRef] [Green Version]
- Kashefi-Kheyrabadi, L.; Nguyen, H.V.; Go, A.; Baek, C.; Jang, N.; Lee, J.M.; Cho, N.-H.; Min, J.; Lee, M.-H. Rapid, multiplexed, and nucleic acid amplification-free detection of SARS-CoV-2 RNA using an electrochemical biosensor. Biosens. Bioelectron. 2022, 195, 113649. [Google Scholar] [CrossRef] [PubMed]
- Carr, A.R.; Dopp, J.L.; Wu, K.; Sadat Mousavi, P.; Jo, Y.R.; McNeley, C.E.; Lynch, Z.T.; Pardee, K.; Green, A.A.; Reuel, N.F. Toward Mail-in-Sensors for SARS-CoV-2 Detection: Interfacing Gel Switch Resonators with Cell-Free Toehold Switches. ACS Sens. 2022, 7, 806–815. [Google Scholar] [CrossRef] [PubMed]
- Snitz, K.; Andelman-Gur, M.; Pinchover, L.; Weissgross, R.; Weissbrod, A.; Mishor, E.; Zoller, R.; Linetsky, V.; Medhanie, A.; Shushan, S.; et al. Proof of concept for real-time detection of SARS CoV-2 infection with an electronic nose. PLoS ONE 2021, 16, e0252121. [Google Scholar] [CrossRef]
- Shan, B.; Broza, Y.Y.; Li, W.; Wang, Y.; Wu, S.; Liu, Z.; Wang, J.; Gui, S.; Wang, L.; Zhang, Z.; et al. Multiplexed Nanomaterial-Based Sensor Array for Detection of COVID-19 in Exhaled Breath. ACS Nano 2020, 14, 12125–12132. [Google Scholar] [CrossRef]
- Wintjens, A.G.W.E.; Hintzen, K.F.H.; Engelen, S.M.E.; Lubbers, T.; Savelkoul, P.H.M.; Wesseling, G.; van der Palen, J.A.M.; Bouvy, N.D. Applying the electronic nose for pre-operative SARS-CoV-2 screening. Surg. Endosc. 2021, 35, 6671–6678. [Google Scholar] [CrossRef]
- Heo, W.; Lee, K.; Park, S.; Hyun, K.-A.; Jung, H.-I. Electrochemical biosensor for nucleic acid amplification-free and sensitive detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA via CRISPR/Cas13a trans-cleavage reaction. Biosens. Bioelectron. 2022, 201, 113960. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, F.; Stewart Aitchison, J.; Mojahedi, M. Multimode Spectroscopy in Optical Biosensors. In Biomedical Optical Sensors: Differentiators for Winning Technologies; De La Rue, R., Herzig, H.P., Gerken, M., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 57–79. [Google Scholar]
- Carlomagno, C.; Bertazioli, D.; Gualerzi, A.; Picciolini, S.; Banfi, P.I.; Lax, A.; Messina, E.; Navarro, J.; Bianchi, L.; Caronni, A.; et al. COVID-19 salivary Raman fingerprint: Innovative approach for the detection of current and past SARS-CoV-2 infections. Sci. Rep. 2021, 11, 4943. [Google Scholar] [CrossRef]
- Yeh, Y.-T.; Gulino, K.; Zhang, Y.; Sabestien, A.; Chou, T.-W.; Zhou, B.; Lin, Z.; Albert, I.; Lu, H.; Swaminathan, V.; et al. A rapid and label-free platform for virus capture and identification from clinical samples. Proc. Natl. Acad. Sci. USA 2020, 117, 895–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ember, K.; Daoust, F.; Mahfoud, M.; Dallaire, F.; Zamani, E.; Tran, T.; Plante, A.; Diop, M.-K.; Nguyen, T.; St-Georges-Robillard, A.; et al. Saliva-based detection of COVID-19 infection in a real-world setting using reagent-free Raman spectroscopy and machine learning. J. Biomed. Opt. 2022, 27, 025002. [Google Scholar] [CrossRef]
- Stanborough, T.; Given, F.M.; Koch, B.; Sheen, C.R.; Stowers-Hull, A.B.; Waterland, M.R.; Crittenden, D.L. Optical Detection of CoV-SARS-2 Viral Proteins to Sub-Picomolar Concentrations. ACS Omega 2021, 6, 6404–6413. [Google Scholar] [CrossRef]
- Finlayson, D.; Rinaldi, C.; Baker, M.J. Is Infrared Spectroscopy Ready for the Clinic? Anal. Chem. 2019, 91, 12117–12128. [Google Scholar] [CrossRef] [Green Version]
- Su, K.-Y.; Lee, W.-L. Fourier Transform Infrared Spectroscopy as a Cancer Screening and Diagnostic Tool: A Review and Prospects. Cancers 2020, 12, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakudo, A. Near-infrared spectroscopy for medical applications: Current status and future perspectives. Clin. Chim. Acta 2016, 455, 181–188. [Google Scholar] [CrossRef]
- Nogueira, M.S.; Leal, L.B.; Marcarini, W.D.; Pimentel, R.L.; Muller, M.; Vassallo, P.F.; Campos, L.C.G.; dos Santos, L.; Luiz, W.B.; Mill, J.G.; et al. Rapid diagnosis of COVID-19 using FT-IR ATR spectroscopy and machine learning. Sci. Rep. 2021, 11, 15409. [Google Scholar] [CrossRef] [PubMed]
- Kitane, D.L.; Loukman, S.; Marchoudi, N.; Fernandez-Galiana, A.; El Ansari, F.Z.; Jouali, F.; Badir, J.; Gala, J.-L.; Bertsimas, D.; Azami, N.; et al. A simple and fast spectroscopy-based technique for Covid-19 diagnosis. Sci. Rep. 2021, 11, 16740. [Google Scholar] [CrossRef] [PubMed]
- Narita, F.; Wang, Z.; Kurita, H.; Li, Z.; Shi, Y.; Jia, Y.; Soutis, C. A Review of Piezoelectric and Magnetostrictive Biosensor Materials for Detection of COVID-19 and Other Viruses. Adv. Mater. 2021, 33, 2005448. [Google Scholar] [CrossRef] [PubMed]
- Pohanka, M. Overview of Piezoelectric Biosensors, Immunosensors and DNA Sensors and Their Applications. Materials 2018, 11, 448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabir, H.; Merati, M.; Abdekhodaie, M.J. Design of an effective piezoelectric microcantilever biosensor for rapid detection of COVID-19. J. Med. Eng. Technol. 2021, 45, 423–433. [Google Scholar] [CrossRef]
- Mandal, D.; Indaleeb, M.M.; Younan, A.; Banerjee, S. Piezoelectric point-of-care biosensor for the detection of SARS-COV-2 (COVID-19) antibodies. Sens. Bio-Sens. Res. 2022, 37, 100510. [Google Scholar] [CrossRef] [PubMed]
- Berkenbrock, J.A.; Grecco-Machado, R.; Achenbach, S. Microfluidic devices for the detection of viruses: Aspects of emergency fabrication during the COVID-19 pandemic and other outbreaks. Proc. R. Soc. A Math. Phys. Eng. Sci. 2020, 476, 20200398. [Google Scholar] [CrossRef]
- Flores-Contreras, E.A.; González-González, R.B.; Rodríguez-Sánchez, I.P.; Yee-de León, J.F.; Iqbal, H.M.N.; González-González, E. Microfluidics-Based Biosensing Platforms: Emerging Frontiers in Point-of-Care Testing SARS-CoV-2 and Seroprevalence. Biosensors 2022, 12, 179. [Google Scholar] [CrossRef]
- Fabiani, L.; Caratelli, V.; Fiore, L.; Scognamiglio, V.; Antonacci, A.; Fillo, S.; De Santis, R.; Monte, A.; Bortone, M.; Moscone, D.; et al. State of the Art on the SARS-CoV-2 Toolkit for Antigen Detection: One Year Later. Biosensors 2021, 11, 310. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lillehoj, P.B. Microfluidic Magneto Immunosensor for Rapid, High Sensitivity Measurements of SARS-CoV-2 Nucleocapsid Protein in Serum. ACS Sens. 2021, 6, 1270–1278. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-S.; Abbas, N.; Shin, S. A rapid diagnosis of SARS-CoV-2 using DNA hydrogel formation on microfluidic pores. Biosens. Bioelectron. 2021, 177, 113005. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandey, S.K.; Mohanta, G.C.; Kumar, V.; Gupta, K. Diagnostic Tools for Rapid Screening and Detection of SARS-CoV-2 Infection. Vaccines 2022, 10, 1200. https://doi.org/10.3390/vaccines10081200
Pandey SK, Mohanta GC, Kumar V, Gupta K. Diagnostic Tools for Rapid Screening and Detection of SARS-CoV-2 Infection. Vaccines. 2022; 10(8):1200. https://doi.org/10.3390/vaccines10081200
Chicago/Turabian StylePandey, Satish Kumar, Girish C. Mohanta, Vinod Kumar, and Kuldeep Gupta. 2022. "Diagnostic Tools for Rapid Screening and Detection of SARS-CoV-2 Infection" Vaccines 10, no. 8: 1200. https://doi.org/10.3390/vaccines10081200
APA StylePandey, S. K., Mohanta, G. C., Kumar, V., & Gupta, K. (2022). Diagnostic Tools for Rapid Screening and Detection of SARS-CoV-2 Infection. Vaccines, 10(8), 1200. https://doi.org/10.3390/vaccines10081200






