Meta-Data Analysis to Explore the Hub of the Hub-Genes That Influence SARS-CoV-2 Infections Highlighting Their Pathogenetic Processes and Drugs Repurposing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Metadata Sources and Descriptions
2.1.1. Collection of Hub-DEGs to Explore Drug Targets
2.1.2. Collection of Drug Agents
2.2. Methods
2.2.1. Protein–Protein Interaction (PPI) Network Analysis of Hub-DEGs
2.2.2. Functional and Pathway Enrichment Analysis of hHub-DEGs
2.2.3. Regulatory Network Analysis of hHub-DEGs
2.2.4. Association of hHub-DEGs with Comorbidities
2.2.5. Drug Repurposing by Molecular Docking Simulation
3. Results
3.1. Basic Characteristics of the Selected Studies
3.2. Identification of Hub of Hub-Proteins (hHub-Proteins)
3.3. Functional and Pathway Enrichment Analysis of hHub-DEGs
3.4. Transcriptional and Post-Transcriptional Regulatory Factors
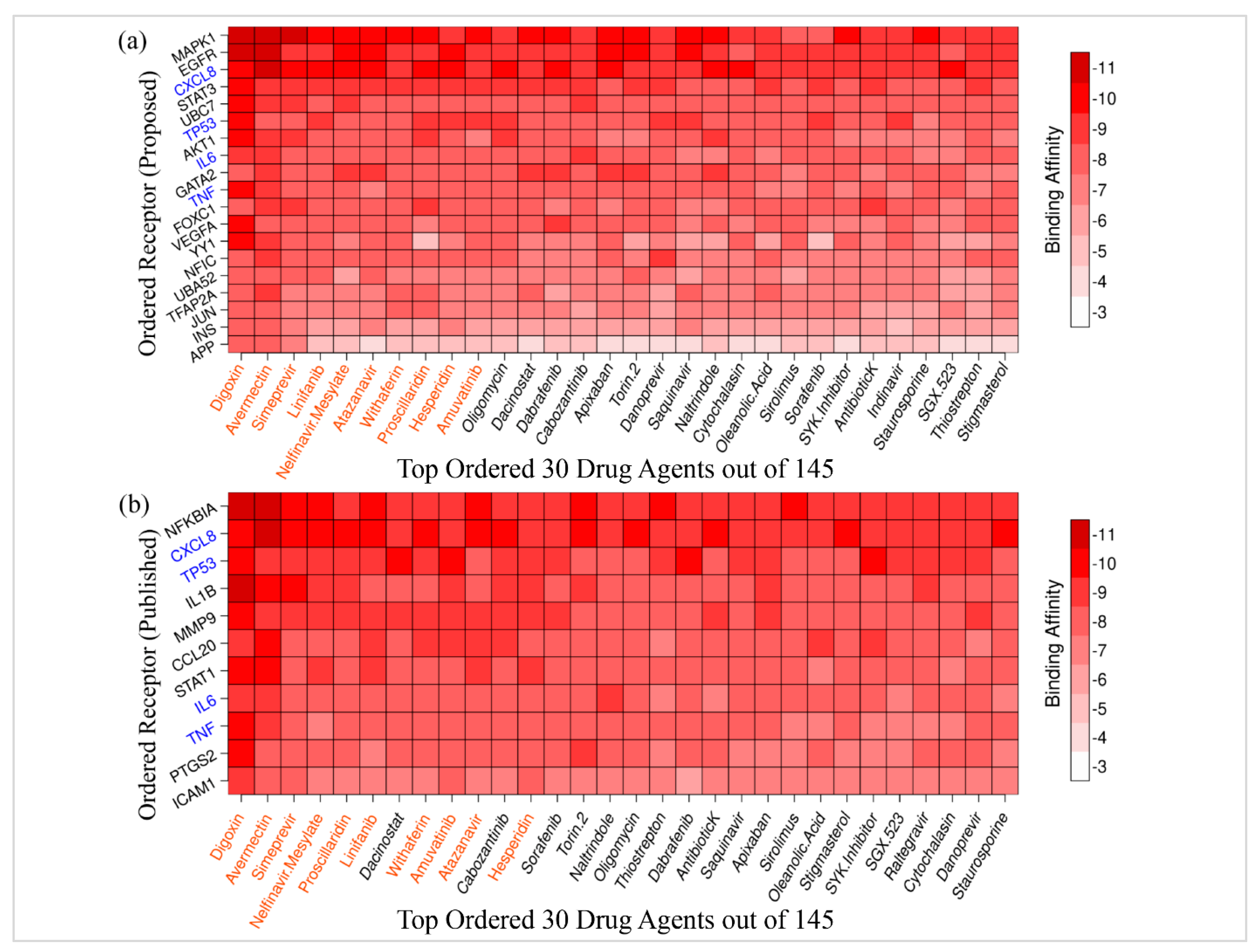
3.5. Drug Repurposing by Molecular Docking
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, R.F.; Gao, Y.L.; Robert, S.H.; Gao, J.P.; Yang, S.G.; Zhu, C.T. Systematic Review of the Registered Clinical Trials for Coronavirus Disease 2019 (COVID-19). J. Transl. Med. 2020, 18, 274. [Google Scholar] [CrossRef] [PubMed]
- Treatments and Vaccines for COVID-19|European Medicines Agency. Available online: https://www.ema.europa.eu/en/human-regulatory/overview/public-health-threats/coronavirus-disease-covid-19/treatments-vaccines-covid-19#authorised-medicines-section (accessed on 11 January 2021).
- Santiago, J.A.; Bottero, V.; Potashkin, J.A. Dissecting the Molecular Mechanisms of Neurodegenerative Diseases through Network Biology. Front. Aging Neurosci. 2017, 9, 166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, M.R.; Islam, T.; Turanli, B.; Zaman, T.; Faruquee, H.M.; Rahman, M.M.; Mollah, M.N.H.; Nanda, R.K.; Arga, K.Y.; Gov, E.; et al. Network-Based Approach to Identify Molecular Signatures and Therapeutic Agents in Alzheimer’s Disease. Comput. Biol. Chem. 2019, 78, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Islam, T.; Rahman, R.; Gov, E.; Turanli, B.; Gulfidan, G.; Haque, A.; Arga, K.Y.; Haque Mollah, N. Drug Targeting and Biomarkers in Head and Neck Cancers: Insights from Systems Biology Analyses. Omi. A J. Integr. Biol. 2018, 22. [Google Scholar] [CrossRef] [PubMed]
- Shahjaman, M.; Rezanur Rahman, M.; Shahinul Islam, S.M.; Nurul Haque Mollah, M. A Robust Approach for Identification of Cancer Biomarkers and Candidate Drugs. Medicina 2019, 55, 269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moni, M.A.; Islam, M.B.; Rahman, M.R.; Rashed-Al-Mahfuz, M.; Awal, M.A.; Islam, S.M.S.; Mollah, M.N.H.; Quinn, J.M.W. Network-Based Computational Approach to Identify Delineating Common Cell Pathways Influencing Type 2 Diabetes and Diseases of Bone and Joints. IEEE Access 2020, 8, 1486–1497. [Google Scholar] [CrossRef]
- Xie, T.A.; Han, M.Y.; Su, X.R.; Li, H.H.; Chen, J.C.; Guo, X.G. Identification of Hub Genes Associated with Infection of Three Lung Cell Lines by SARS-CoV-2 with Integrated Bioinformatics Analysis. J. Cell. Mol. Med. 2020, 24, 12225–12230. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.H.; Tannenbaum, A.; Deasy, J.O. Identification of Biological Correlates Associated with Respiratory Failure in COVID-19. BMC Med. Genom. 2020, 13, 186. [Google Scholar] [CrossRef]
- Ge, C.; He, Y. In Silico Prediction of Molecular Targets of Astragaloside IV for Alleviation of COVID-19 Hyperinflammation by Systems Network Pharmacology and Bioinformatic Gene Expression Analysis. Front. Pharmacol. 2020, 11. [Google Scholar] [CrossRef]
- Aishwarya, S.; Gunasekaran, K.; Margret, A.A. Computational Gene Expression Profiling in the Exploration of Biomarkers, Non-Coding Functional RNAs and Drug Perturbagens for COVID-19. J. Biomol. Struct. Dyn. 2022, 40, 3681–3696. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A.; Chaudhary, U.; Bharadwaj, A.; Wahi, N.; Kalli, J.R.; Gupta, S.; Kumar, S.; Gupta, S.; Raj, U. A Lung Transcriptomic Analysis for Exploring Host Response in COVID-19. J. Pure Appl. Microbiol. 2020, 14, 1077–1081. [Google Scholar] [CrossRef]
- Tao, Q.; Du, J.; Li, X.; Zeng, J.; Tan, B.; Xu, J.; Lin, W.; Chen, X. lin Network Pharmacology and Molecular Docking Analysis on Molecular Targets and Mechanisms of Huashi Baidu Formula in the Treatment of COVID-19. Drug Dev. Ind. Pharm. 2020, 46, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhao, Y.D.; Wang, X.M. CXCL10 an Important Chemokine Associated with Cytokine Storm in COVID-19 Infected Patients. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7497–7505. [Google Scholar] [CrossRef]
- Han, L.; Wei, X.X.; Zheng, Y.J.; Zhang, L.L.; Wang, X.M.; Yang, H.Y.; Ma, X.; Zhao, L.H.; Tong, X.L. Potential Mechanism Prediction of Cold-Damp Plague Formula against COVID-19 via Network Pharmacology Analysis and Molecular Docking. Chin. Med. 2020, 15, 78. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, C.; Zhang, X.; Zhang, Y.; Ren, Y. Identication of Key Genes and Pathways in SARS-CoV-2 Infection Using Bioinformatics Analysis. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Gu, H.; Yuan, G. Identification of Potential Key Genes for SARS-CoV-2 Infected Human Bronchial Organoids Based on Bioinformatics Analysis. bioRxiv 2020. [Google Scholar] [CrossRef]
- Soon Nan, K.; Karuppanan, K.; Kumar, S.; Alam, S. Identification of Common Key Genes and Pathways between COVID-19 and Lung Cancer by Using Protein-Protein Interaction Network Analysis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Sardar, R.; Satish, D.; Gupta, D. Identification of Novel SARS-CoV-2 Drug Targets by Host MicroRNAs and Transcription Factors Co-Regulatory Interaction Network Analysis. Front. Genet. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Vastrad, B.; Vastrad, C.; Tengli, A. Identification of Potential MRNA Panels for Severe Acute Respiratory Syndrome Coronavirus 2 (COVID-19) Diagnosis and Treatment Using Microarray Dataset and Bioinformatics Methods. 3 Biotech 2020, 10, 422. [Google Scholar] [CrossRef] [PubMed]
- Chong, X.; Peng, R.; Sun, Y.; Zhang, L.; Zhang, Z. Identification of Key Genes in Gastric Cancer by Bioinformatics Analysis. Biomed Res. Int. 2020, 2020, 7658230. [Google Scholar] [CrossRef] [PubMed]
- Prasad, K.; Khatoon, F.; Rashid, S.; Ali, N.; AlAsmari, A.F.; Ahmed, M.Z.; Alqahtani, A.S.; Alqahtani, M.S.; Kumar, V. Targeting Hub Genes and Pathways of Innate Immune Response in COVID-19: A Network Biology Perspective. Int. J. Biol. Macromol. 2020, 163, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Satu, S.; Khan, I.; Rahman, R.; Howlader, K.C.; Roy, S.; Roy, S.S.; Quinn, J.M.W.; Moni, M.A. Diseasome and Comorbidities Complexities of SARS-CoV-2 Infection with Common Malignant Diseases. Brief. Bioinform. 2021, 22, 1415–1429. [Google Scholar] [CrossRef] [PubMed]
- Taz, T.A.; Ahmed, K.; Paul, B.K.; Kawsar, M.; Aktar, N.; Mahmud, S.M.H.; Moni, M.A. Network-Based Identification Genetic Effect of SARS-CoV-2 Infections to Idiopathic Pulmonary Fibrosis (IPF) Patients. Brief. Bioinform. 2021, 22, 1254–1266. [Google Scholar] [CrossRef]
- Moni, M.A.; Quinn, J.M.W.; Sinmaz, N.; Summers, M.A. Gene Expression Profiling of SARS-CoV-2 Infections Reveal Distinct Primary Lung Cell and Systemic Immune Infection Responses That Identify Pathways Relevant in COVID-19 Disease. Brief. Bioinform. 2020, 2020, 1324–1337. [Google Scholar] [CrossRef]
- Islam, T.; Rahman, M.R.; Aydin, B.; Beklen, H.; Arga, K.Y.; Shahjaman, M. Integrative Transcriptomics Analysis of Lung Epithelial Cells and Identification of Repurposable Drug Candidates for COVID-19. Eur. J. Pharmacol. 2020, 887, 173594. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Hou, Y.; Shen, J.; Huang, Y.; Martin, W.; Cheng, F. Network-Based Drug Repurposing for Novel Coronavirus 2019-NCoV/SARS-CoV-2. Cell Discov. 2020, 6, 14. [Google Scholar] [CrossRef] [Green Version]
- Caradonna, A.; Patel, T.; Toleska, M.; Alabed, S.; Chang, S.L. Meta-Analysis of APP Expression Modulated by SARS-CoV-2 Infection via the ACE2 Receptor. Int. J. Mol. Sci. 2022, 23, 1182. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Yuan, G. Identification of Key Genes in SARS-CoV-2 Patients on Bioinformatics Analysis. bioRxiv 2020. [Google Scholar] [CrossRef]
- Gu, H.; Yuan, G. Identification of Key Genes and Pathways in the HPSC-Derived Lungs Infected by the SARS-CoV-2. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Selvaraj, G.; Kaliamurthi, S.; Peslherbe, G.H.; Wei, D.Q. Identifying Potential Drug Targets and Candidate Drugs for COVID-19: Biological Networks and Structural Modeling Approaches. F1000Research 2021, 10, 127. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Sun, D.; Xie, Y.; Liu, K.; Ma, Y. Network Pharmacology-Based Study of the Molecular Mechanisms of Qixuekang in Treating COVID-19 during the Recovery Period. Int. J. Clin. Exp. Pathol. 2020, 13, 2677–2690. [Google Scholar]
- Jha, P.K.; Vijay, A.; Halu, A.; Uchida, S.; Aikawa, M. Gene Expression Profiling Reveals the Shared and Distinct Transcriptional Signatures in Human Lung Epithelial Cells Infected With SARS-CoV-2, MERS-CoV, or SARS-CoV: Potential Implications in Cardiovascular Complications of COVID-19. Front. Cardiovasc. Med. 2021, 7, 1–15. [Google Scholar] [CrossRef]
- Ramesh, P.; Veerappapillai, S.; Karuppasamy, R. Gene Expression Profiling of Corona Virus Microarray Datasets to Identify Crucial Targets in COVID-19 Patients. Gene Rep. 2021, 22, 100980. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yang, L. Underlying Mechanisms and Candidate Drugs for COVID-19 Based on the Connectivity Map Database. Front. Genet. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; He, X.; Zhang, L.; Ran, Q.; Wang, J.; Xiong, A.; Wu, D.; Chen, F.; Sun, J.; Chang, C. Assessing ACE2 Expression Patterns in Lung Tissues in the Pathogenesis of COVID-19. J. Autoimmun. 2020, 112, 102463. [Google Scholar] [CrossRef] [PubMed]
- Prasad, K.; AlOmar, S.Y.; Alqahtani, S.A.M.; Malik, M.Z.; Kumar, V. Brain Disease Network Analysis to Elucidate the Neurological Manifestations of COVID-19. Mol. Neurobiol. 2021, 58, 1875–1893. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Li, Y.; Yang, Y.; Li, M.; Du, Y.; Zhang, Y.; Wang, J.; Shi, Y. Study on Mechanism of Matrine in Treatment of COVID-19 Combined with Liver Injury by Network Pharmacology and Molecular Docking Technology. Drug Deliv. 2021, 28, 325. [Google Scholar] [CrossRef] [PubMed]
- Nain, Z.; Rana, H.K.; Liò, P.; Islam, S.M.S.; Summers, M.A.; Moni, M.A. Pathogenetic Profiling of COVID-19 and SARS-like Viruses. Brief. Bioinform. 2021, 22, 1175–1196. [Google Scholar] [CrossRef]
- Fang, K.Y.; Cao, W.C.; Xie, T.A.; Lv, J.; Chen, J.X.; Cao, X.J.; Li, Z.W.; Deng, S.T.; Guo, X.G. Exploration and Validation of Related Hub Gene Expression during SARS-CoV-2 Infection of Human Bronchial Organoids. Hum. Genom. 2021, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Rezaei-Tavirani, M.; Nejad, M.R.; Arjmand, B.; Tavirani, S.R.; Razzaghi, M.; Mansouri, V. Fibrinogen Dysregulation Is a Prominent Process in Fatal Conditions of COVID-19 Infection; a Proteomic Analysis. Arch. Acad. Emerg. Med. 2021, 9, 1–5. [Google Scholar] [CrossRef]
- Li, S.; Wang, W.; Li, T.; Han, X.; Hu, C.; Wang, Y.; Shen, M.; Du, L.; Nai, Y.; Wang, J.; et al. Immune Characteristics Analysis Reveals Two Key Inflammatory Factors Correlated to the Expressions of SARS-CoV-2 S1-Specific Antibodies. Genes Dis. 2022, 9, 522. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S. COVID-19: A Drug Repurposing and Biomarker Identification by Using Comprehensive Gene-Disease Associations through Protein-Protein Interaction Network Analysis. Preprints 2020. [Google Scholar] [CrossRef]
- Zhu, Y.W.; Yan, X.F.; Ye, T.J.; Hu, J.; Wang, X.L.; Qiu, F.J.; Liu, C.H.; Hu, X.D. Analyzing the Potential Therapeutic Mechanism of Huashi Baidu Decoction on Severe COVID-19 through Integrating Network Pharmacological Methods. J. Tradit. Complement. Med. 2021, 11, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Wang, L.J.; He, K.; Lin, X.; Yu, T.; Li, J.; Gong, J. Xiang High Expression of Ace2 in the Human Lung Leads to the Release of Il6 by Suppressing Cellular Immunity: Il6 Plays a Key Role in COVID-19. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Z.; Li, K.; Maskey, A.R.; Huang, W.; Toutov, A.A.; Yang, N.; Srivastava, K.; Geliebter, J.; Tiwari, R.; Miao, M.; et al. A Small Molecule Compound Berberine as an Orally Active Therapeutic Candidate against COVID-19 and SARS: A Computational and Mechanistic Study. FASEB J. 2021, 35, 1–19. [Google Scholar] [CrossRef]
- Auwul, M.R.; Rahman, M.R.; Gov, E.; Shahjaman, M.; Moni, M.A. Bioinformatics and Machine Learning Approach Identifies Potential Drug Targets and Pathways in COVID-19. Brief. Bioinform. 2021, 22, bbab120. [Google Scholar] [CrossRef] [PubMed]
- Mosharaf, M.P.; Reza, M.S.; Kibria, M.K.; Ahmed, F.F.; Kabir, M.H.; Hasan, S.; Mollah, M.N.H. Computational Identification of Host Genomic Biomarkers Highlighting Their Functions, Pathways and Regulators That Influence SARS-CoV-2 Infections and Drug Repurposing. Sci. Rep. 2022, 12, 1–22. [Google Scholar] [CrossRef]
- Lee, H.; Park, J.; Im, H.J.; Na, K.J.; Choi, H. Discovery of Potential Imaging and Therapeutic Targets for Severe Inflammation in COVID-19 Patients. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef]
- Alanazi, K.M.; Farah, M.A.; Hor, Y.Y. Multi-Targeted Approaches and Drug Repurposing Reveal Possible SARS-CoV-2 Inhibitors. Vaccines 2022, 10, 24. [Google Scholar] [CrossRef]
- Beck, B.R.; Shin, B.; Choi, Y.; Park, S.; Kang, K. Predicting Commercially Available Antiviral Drugs That May Act on the Novel Coronavirus (SARS-CoV-2) through a Drug-Target Interaction Deep Learning Model. Comput. Struct. Biotechnol. J. 2020, 18, 784–790. [Google Scholar] [CrossRef]
- Xia, J.; Gill, E.E.; Hancock, R.E.W. NetworkAnalyst for Statistical, Visual and Network-Based Meta-Analysis of Gene Expression Data. Nat. Protoc. 2015, 10, 823–844. [Google Scholar] [CrossRef] [PubMed]
- Boyle, E.I.; Weng, S.; Gollub, J.; Jin, H.; Botstein, D.; Cherry, J.M.; Sherlock, G. GO::TermFinder-Open Source Software for Accessing Gene Ontology Information and Finding Significantly Enriched Gene Ontology Terms Associated with a List of Genes INTRODUCTION: MOTIVATION AND DESIGN. Bioinforma. Appl. NOTE 2004, 20, 3710–3715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING Database in 2017: Quality-Controlled Protein-Protein Association Networks, Made Broadly Accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Khan, A.; Fornes, O.; Stigliani, A.; Gheorghe, M.; Castro-Mondragon, J.A.; Van Der Lee, R.; Bessy, A.; Chèneby, J.; Kulkarni, S.R.; Tan, G.; et al. JASPAR 2018: Update of the Open-Access Database of Transcription Factor Binding Profiles and Its Web Framework. Nucleic Acids Res. 2018, 46, D260–D266. [Google Scholar] [CrossRef] [Green Version]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A Decade-Long Collection of Experimentally Supported MiRNA-Gene Interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguirre-Gamboa, R.; Gomez-Rueda, H.; Martínez-Ledesma, E.; Martínez-Torteya, A.; Chacolla-Huaringa, R.; Rodriguez-Barrientos, A.; Tamez-Peña, J.G.; Treviño, V. SurvExpress: An Online Biomarker Validation Tool and Database for Cancer Gene Expression Data Using Survival Analysis. PLoS ONE 2013, 8, e74250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 Update: Improved Access to Chemical Data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [Green Version]
- Discovery Studio. Discovery Studio Visualizer. Discovery 2014, 3–5. [Google Scholar]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Software News and Updates AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeerschd, T.; Zurek, E.; Hutchison, G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delano, W.L.; Bromberg, S. PyMOL User’s Guide; DeLano Scientific LLC: San Francisco, CA, USA, 2004. [Google Scholar]
- The UniProt Consortium. UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Tanimoto, T. An Elementary Mathematical Theory of Classification and Prediction; International Business Machines Corp.: New York, NY, USA, 1958; pp. 1–10. [Google Scholar]
- Willett, P.; Barnard, J.M.; Downs, G.M. Chemical Similarity Searching. J. Chem. Inf. Comput. Sci. 1998, 38, 983–996. [Google Scholar] [CrossRef] [Green Version]
- Carpten, J.D.; Faber, A.L.; Horn, C.; Donoho, G.P.; Briggs, S.L.; Robbins, C.M.; Hostetter, G.; Boguslawski, S.; Moses, T.Y.; Savage, S.; et al. A Transforming Mutation in the Pleckstrin Homology Domain of AKT1 in Cancer. Nature 2007, 448, 439–444. [Google Scholar] [CrossRef]
- Emamian, E.S. AKT/GSK3 Signaling Pathway and Schizophrenia. Front. Mol. Neurosci. 2012, 5, 33. [Google Scholar] [CrossRef] [Green Version]
- Lindhurst, M.J.; Sapp, J.C.; Teer, J.K.; Johnston, J.J.; Finn, E.M.; Peters, K.; Turner, J.; Cannons, J.L.; Bick, D.; Blakemore, L.; et al. A Mosaic Activating Mutation in AKT1 Associated with the Proteus Syndrome. N. Engl. J. Med. 2011, 365, 611–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwab, S.G.; Hoefgen, B.; Hanses, C.; Hassenbach, M.B.; Albus, M.; Lerer, B.; Trixler, M.; Maier, W.; Wildenauer, D.B. Further Evidence for Association of Variants in the AKT1 Gene with Schizophrenia in a Sample of European Sib-Pair Families. Biol. Psychiatry 2005, 58. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Tang, Y.; Li, Q.; Chen, Y.; Liu, D.; Liu, G.; Fan, X.; Ma, R.; Wang, S.; Li, L.; et al. Network Pharmacology Integrated Molecular Docking Reveals the Bioactive Components and Potential Targets of Morinda Officinalis–Lycium Barbarum Coupled-Herbs against Oligoasthenozoospermia. Sci. Rep. 2021, 11, 2220. [Google Scholar] [CrossRef] [PubMed]
- Gassen, N.C.; Niemeyer, D.; Muth, D.; Corman, V.M.; Martinelli, S.; Gassen, A.; Hafner, K.; Papies, J.; Mösbauer, K.; Zellner, A.; et al. SKP2 Attenuates Autophagy through Beclin1-Ubiquitination and Its Inhibition Reduces MERS-Coronavirus Infection. Nat. Commun. 2019, 10, 5770. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Khan, Z. System Biological Investigations of Hydroxychloroquine and Azithromycin Targets and Their Implications in QT Interval Prolongation. Chem. Biol. Interact. 2020, 332, 109299. [Google Scholar] [CrossRef] [PubMed]
- Fraiman, P.; Freire, M.; Moreira-Neto, M.; Godeiro-Junior, C. Hemorrhagic Stroke and COVID-19 Infection: Coincidence or Causality? eNeurologicalSci 2020, 21, 100274. [Google Scholar] [CrossRef]
- XQ, C.; Z, X.; QD, C.; RJ, S.; X, Z.; B, T.; WC, M.; YE, Y. Mechanistic Analysis of Age-Related Clinical Manifestations in Down Syndrome. Front. Aging Neurosci. 2021, 13, 700280. [Google Scholar] [CrossRef]
- Bie, Y.; Ge, W.; Yang, Z.; Cheng, X.; Zhao, Z.; Li, S.; Wang, W.; Wang, Y.; Zhao, X.; Yin, Z.; et al. The Crucial Role of CXCL8 and Its Receptors in Colorectal Liver Metastasis. Dis. Markers 2019, 2019, 8023460. [Google Scholar] [CrossRef] [PubMed]
- Metzemaekers, M.; Vandendriessche, S.; Berghmans, N.; Gouwy, M.; Proost, P. Truncation of CXCL8 to CXCL8(9-77) Enhances Actin Polymerization and in Vivo Migration of Neutrophils. J. Leukoc. Biol. 2020, 107, 1167–1173. [Google Scholar] [CrossRef]
- Bethune, G.; Bethune, D.; Ridgway, N.; Xu, Z. Epidermal Growth Factor Receptor (EGFR) in Lung Cancer: An Overview and Update. J. Thorac. Dis. 2010, 2, 48–51. [Google Scholar] [PubMed]
- Purcaru, O.S.; Artene, S.A.; Barcan, E.; Silosi, C.A.; Stanciu, I.; Danoiu, S.; Tudorache, S.; Tataranu, L.G.; Dricu, A. The Interference between SARS-CoV-2 and Tyrosine Kinase Receptor Signaling in Cancer. Int. J. Mol. Sci. 2021, 22, 4830. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Zhang, S.; Shen, Y.; Yang, Q. Epidermal Growth Factor Receptor Is a Co-Factor for Transmissible Gastroenteritis Virus Entry. Virology 2018, 521, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Zhu, L.; Yang, X.; Lin, J.; Yang, Q. The Epidermal Growth Factor Receptor Regulates Cofilin Activity and Promotes Transmissible Gastroenteritis Virus Entry into Intestinal Epithelial Cells. Oncotarget 2016, 7, 12206–12221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balmanno, K.; Cook, S.J. Tumour Cell Survival Signalling by the ERK1/2 Pathway. Cell Death Differ. 2009, 16, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Vagapova, E.R.; Lebedev, T.D.; Prassolov, V.S. Viral Fibrotic Scoring and Drug Screen Based on MAPK Activity Uncovers EGFR as a Key Regulator of COVID-19 Fibrosis. Sci. Rep. 2021, 11, 11234. [Google Scholar] [CrossRef] [PubMed]
- Channappanavar, R.; Zhao, J.; Perlman, S. T Cell-Mediated Immune Response to Respiratory Coronaviruses. Immunol. Res. 2014, 59, 118–128. [Google Scholar] [CrossRef] [Green Version]
- Wen, J.; Wang, Y.T.; Valentine, K.M.; dos Santos Alves, R.P.; Xu, Z.; Regla-Nava, J.A.; Ngono, A.E.; Young, M.P.; Ferreira, L.C.S.; Shresta, S. CD4+ T Cells Cross-Reactive with Dengue and Zika Viruses Protect against Zika Virus Infection. Cell Rep. 2020, 31, 107566. [Google Scholar] [CrossRef]
- Maes, B.; Bosteels, C.; De Leeuw, E.; Declercq, J.; Van Damme, K.; Delporte, A.; Demeyere, B.; Vermeersch, S.; Vuylsteke, M.; Willaert, J.; et al. Treatment of Severely Ill COVID-19 Patients with Anti-Interleukin Drugs (COV-AID): A Structured Summary of a Study Protocol for a Randomised Controlled Trial. Trials 2020, 21, 468. [Google Scholar] [CrossRef] [PubMed]
- Sinaei, R.; Pezeshki, S.; Parvaresh, S.; Sinaei, R. Why COVID-19 Is Less Frequent and Severe in Children: A Narrative Review. World J. Pediatr. 2021, 17, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Fajgenbaum, D.C. Novel Insights and Therapeutic Approaches in Idiopathic Multicentric Castleman Disease. Blood 2018, 132, 2323–2330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Miller, E.B.; Goswami, M.; Zhang, P.; Ronning, K.E.; Karlen, S.J.; Zawadzki, R.J.; Pugh, E.N.; Burns, M.E. Rapid Monocyte Infiltration Following Retinal Detachment Is Dependent on Non-Canonical IL6 Signaling through Gp130. J. Neuroinflammation 2017, 14, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Li, M.; Zhou, Z.; Guan, X.; Xiang, Y. Can We Use Interleukin-6 (IL-6) Blockade for Coronavirus Disease 2019 (COVID-19)-Induced Cytokine Release Syndrome (CRS)? J. Autoimmun. 2020, 111, 102452. [Google Scholar] [CrossRef] [PubMed]
- Gubernatorova, E.O.; Gorshkova, E.A.; Polinova, A.I.; Drutskaya, M.S. IL-6: Relevance for Immunopathology of SARS-CoV-2. Cytokine Growth Factor Rev. 2020, 53, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Ma, Q.; Li, C.; Liu, R.; Zhao, L.; Wang, W.; Zhang, P.; Liu, X.; Gao, G.; Liu, F.; et al. Profiling Serum Cytokines in COVID-19 Patients Reveals IL-6 and IL-10 Are Disease Severity Predictors. Emerg. Microbes Infect. 2020, 9, 1123–1130. [Google Scholar] [CrossRef]
- Saghazadeh, A.; Rezaei, N. Towards Treatment Planning of COVID-19: Rationale and Hypothesis for the Use of Multiple Immunosuppressive Agents: Anti-Antibodies, Immunoglobulins, and Corticosteroids. Int. Immunopharmacol. 2020, 84, 106560. [Google Scholar] [CrossRef]
- Kaur, S.; Bansal, Y.; Kumar, R.; Bansal, G. A Panoramic Review of IL-6: Structure, Pathophysiological Roles and Inhibitors. Bioorganic Med. Chem. 2020, 28, 115327. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhao, B.; Qu, Y.; Chen, Y.; Xiong, J.; Feng, Y.; Men, D.; Huang, Q.; Liu, Y.; Yang, B.; et al. Detectable Serum SARS-CoV-2 Viral Load (RNAaemia) Is Closely Correlated with Drastically Elevated Interleukin 6 (IL-6) Level in Critically Ill COVID-19 Patients. Clin. Infect. Dis. 2020, 71, 1937–1942. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.S.; Liu, D.X. Activation of the C-Jun NH2-Terminal Kinase Pathway by Coronavirus Infectious Bronchitis Virus Promotes Apoptosis Independently of c-Jun Article. Cell Death Dis. 2017, 8, 3215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiwari, R.; Mishra, A.R.; Gupta, A.; Nayak, D. Structural Similarity-Based Prediction of Host Factors Associated with SARS-CoV-2 Infection and Pathogenesis. J. Biomol. Struct. Dyn. 2022, 40, 5868–5879. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Geng, Y.; Luo, J.; Shen, W.; Zhu, W.; Meng, C.; Li, M.; Zhou, X.; Zhang, S.; Cao, J. Downregulation of Ubiquitin Inhibits the Proliferation and Radioresistance of Non-Small Cell Lung Cancer Cells In Vitro and In Vivo. Sci. Rep. 2015, 5, 9476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, B.; Qu, Y.; Jin, Y.; Yu, Y.; Deng, N.; Wawrowsky, K.; Zhang, X.; Li, N.; Bose, S.; Wang, Q.; et al. FOXC1 Activates Smoothened-Independent Hedgehog Signaling in Basal-like Breast Cancer. Cell Rep. 2015, 13, P1046–P1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haffner, M.C.; Weier, C.; Xu, M.M.; Vaghasia, A.; Gürel, B.; Gümüşkaya, B.; Esopi, D.M.; Fedor, H.; Tan, H.L.; Kulac, I.; et al. Molecular Evidence That Invasive Adenocarcinoma Can Mimic Prostatic Intraepithelial Neoplasia (PIN) and Intraductal Carcinoma through Retrograde Glandular Colonization. J. Pathol. 2016, 238, 31–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, S.; Jiang, X.; Li, J.; Li, C.; Guo, M.; Ye, F.; Zhang, M.; Jiao, Y.; Guo, B. Comprehensive Analysis of the GATA Transcription Factor Gene Family in Breast Carcinoma Using Gene Microarrays, Online Databases and Integrated Bioinformatics. Sci. Rep. 2019, 9, 4467. [Google Scholar] [CrossRef] [PubMed]
- Tessema, M.; Yingling, C.M.; Snider, A.M.; Do, K.; Juri, D.E.; Picchi, M.A.; Zhang, X.; Liu, Y.; Leng, S.; Tellez, C.S.; et al. GATA2 Is Epigenetically Repressed in Human and Mouse Lung Tumors and Is Not Requisite for Survival of KRAS Mutant Lung Cancer. J. Thorac. Oncol. 2014, 9, P784–P793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, T.; Wang, G.; Yang, L.; Peng, B.; Wen, Y.; Ding, G.; Wang, Z. Transcription Factor YY1 Modulates Lung Cancer Progression by Activating LncRNA-PVT1. DNA Cell Biol. 2017, 36, 947–958. [Google Scholar] [CrossRef]
- Zhang, S.; Li, M.; Ji, H.; Fang, Z. Landscape of Transcriptional Deregulation in Lung Cancer. BMC Genom. 2018, 19, 435. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Lee, D.S.; Park, J.C. Nuclear Factor I-C Regulates E-Cadherin via Control of KLF4 in Breast Cancer. BMC Cancer 2015, 15, 113. [Google Scholar] [CrossRef] [Green Version]
- Marimuthu, A.; Jacob, H.K.C.; Jakharia, A.; Subbannayya, Y.; Keerthikumar, S.; Kashyap, M.K.; Goel, R.; Balakrishnan, L.; Dwivedi, S.; Pathare, S.; et al. Gene Expression Profiling of Gastric Cancer. J. Proteom. Bioinforma. 2011, 4, 4. [Google Scholar] [CrossRef] [Green Version]
- Brun, M.; Coles, J.E.; Monckton, E.A.; Glubrecht, D.D.; Bisgrove, D.; Godbout, R. Nuclear Factor I Regulates Brain Fatty Acid-Binding Protein and Glial Fibrillary Acidic Protein Gene Expression in Malignant Glioma Cell Lines. J. Mol. Biol. 2009, 391, 282–300. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.R.; Islam, T.; Gov, E.; Turanli, B.; Gulfidan, G.; Shahjaman, M.; Banu, N.A.; Mollah, M.N.H.; Arga, K.Y.; Moni, M.A. Identification of Prognostic Biomarker Signatures and Candidate Drugs in Colorectal Cancer: Insights from Systems Biology Analysis. Medicina 2019, 55, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Wang, W.; Azhati, B.; Wang, Y.; Tusong, H. MiR-106a-5p Functions as a Tumor Suppressor by Targeting VEGFA in Renal Cell Carcinoma. Dis. Markers 2020, 2020, 8837941. [Google Scholar] [CrossRef] [PubMed]
- Majd, M.; Hosseini, A.; Ghaedi, K.; Kiani-Esfahani, A.; Tanhaei, S.; Shiralian-Esfahani, H.; Rahnamaee, S.Y.; Mowla, S.J.; Nasr-Esfahani, M.H. MiR-9-5p and MiR-106a-5p Dysregulated in CD4+ T-Cells of Multiple Sclerosis Patients and Targeted Essential Factors of T Helper17/Regulatory T-Cells Differentiation. Iran. J. Basic Med. Sci. 2018, 21, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; He, J.; Disoma, C.; Hu, Y.; Li, J.; Chen, G.; Sheng, Y.; Cai, X.; Li, C.; Cheng, K.; et al. Hsa_circ_0107593 Suppresses the Progression of Cervical Cancer via Sponging Hsa-MiR-20a-5p/93-5p/106b-5p. Front. Oncol. 2021, 10, 590627. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Wu, Q.; Zhou, L.; Mu, S.; Fu, Q. MiR-106b-5p and MiR-17-5p Suppress Osteogenic Differentiation by Targeting Smad5 and Inhibit Bone Formation. Exp. Cell Res. 2016, 347, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, H.E.; Song, Y.S.; Cho, E.Y.; Lee, A. MiR-106b-5p and MiR-17-5p Could Predict Recurrence and Progression in Breast Ductal Carcinoma in Situ Based on the Transforming Growth Factor-Beta Pathway. Breast Cancer Res. Treat. 2019, 176, 119–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Dou, R.; Yin, T.; Ding, J. MiRNA-106b-5p in Human Cancers: Diverse Functions and Promising Biomarker. Biomed. Pharmacother. 2020, 127, 110211. [Google Scholar] [CrossRef]
- Kelleci Cakir, B.; Bayraktar-Ekincioglu, A.; Demirkan, K. Benefit versus Toxicity Risk of Digoxin in Patients with COVID-19. Eur. J. Hosp. Pharm. 2021. [Google Scholar] [CrossRef] [PubMed]
- Peltzer, B.; Lerman, B.B.; Goyal, P.; Cheung, J.W. Role for Digoxin in Patients Hospitalized with COVID-19 and Atrial Arrhythmias. J. Cardiovasc. Electrophysiol. 2021, 32, 880. [Google Scholar] [CrossRef] [PubMed]
- Siniorakis, E.; Arvanitakis, S.; Katsianis, A.; Elkouris, M. Atrial Fibrillation and Flutter in Patients Hospitalized for COVID-19: The Challenging Role of Digoxin. J. Cardiovasc. Electrophysiol. 2021, 32, 878. [Google Scholar] [CrossRef] [PubMed]
- Mezaal, M.H.; Farhan, H.A.; Dakhil, Z.A. COVID-19 Pandemic Impact on Physicians’ Decision-Making: Digoxin Toxicity in View of Combination of Hydroxychloroquine and Azithromycin: A Case Report. Open Access Maced. J. Med. Sci. 2020, 8, 150–153. [Google Scholar] [CrossRef]
- Pandey, S.; Pathak, S.K.; Pandey, A.; Salunke, A.A.; Chawla, J.; Ortho, M.S.; Sharma, A.; Sharma, S.; Thivari, P.; Ratna, H.V.K. Ivermectin in COVID-19: What Do We Know? Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 1921. [Google Scholar] [CrossRef] [PubMed]
- Heidary, F.; Gharebaghi, R. Ivermectin: A Systematic Review from Antiviral Effects to COVID-19 Complementary Regimen. J. Antibiot. 2020, 73, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.; Sahoo, A.K.; Singh, A. Ivermectin: Potential Candidate for the Treatment of COVID 19. Braz. J. Infect. Dis. 2020, 24, 369–371. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.S.; Hsueh, P.R. Old and Re-Purposed Drugs for the Treatment of COVID-19. Expert Rev. Anti. Infect. Ther. 2020, 18, 843–847. [Google Scholar] [CrossRef] [PubMed]
- Hossen, M.S.; Barek, M.A.; Jahan, N.; Safiqul Islam, M. A Review on Current Repurposing Drugs for the Treatment of COVID-19: Reality and Challenges. SN Compr. Clin. Med. 2020, 2, 1777–1789. [Google Scholar] [CrossRef] [PubMed]
- Rosenquist, Å.; Samuelsson, B.; Johansson, P.O.; Cummings, M.D.; Lenz, O.; Raboisson, P.; Simmen, K.; Vendeville, S.; De Kock, H.; Nilsson, M.; et al. Discovery and Development of Simeprevir (TMC435), a HCV NS3/4A Protease Inhibitor. J. Med. Chem. 2014, 57, 1673–1693. [Google Scholar] [CrossRef]
- Muturi, E.; Hong, W.; Li, J.; Yang, W.; He, J.; Wei, H.; Yang, H. Effects of Simeprevir on the Replication of SARS-CoV-2 in Vitro and in Transgenic HACE2 Mice. Int. J. Antimicrob. Agents 2022, 59, 106499. [Google Scholar] [CrossRef]
- Hosseini, F.S.; Amanlou, M. Anti-HCV and Anti-Malaria Agent, Potential Candidates to Repurpose for Coronavirus Infection: Virtual Screening, Molecular Docking, and Molecular Dynamics Simulation Study. Life Sci. 2020, 258, 118205. [Google Scholar] [CrossRef] [PubMed]
- Gurung, A.B.; Ali, M.A.; Lee, J.; Farah, M.A.; Al-Anazi, K.M. The Potential of Paritaprevir and Emetine as Inhibitors of SARS-CoV-2 RdRp. Saudi J. Biol. Sci. 2021, 28, 1426–1432. [Google Scholar] [CrossRef] [PubMed]
- Gurung, A.B. In Silico Structure Modelling of SARS-CoV-2 Nsp13 Helicase and Nsp14 and Repurposing of FDA Approved Antiviral Drugs as Dual Inhibitors. Gene Rep. 2020, 21, 100860. [Google Scholar] [CrossRef] [PubMed]

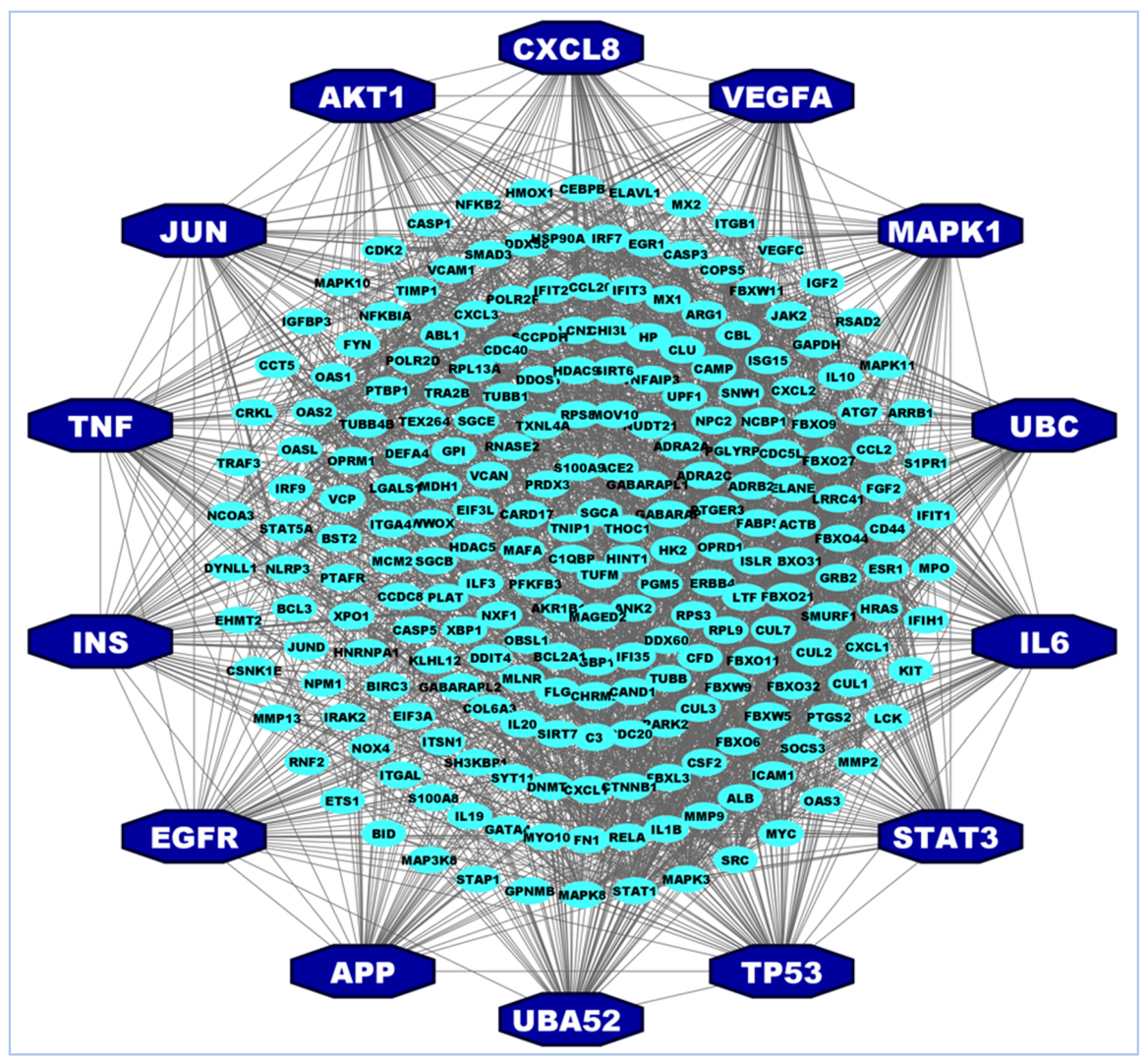



| SL | Articles & Datasets | Hub-Genes/Proteins | Number of Proteins |
|---|---|---|---|
| 1 | Caradonna, A et al., 2022 [29] | ACE2, APP | 2 |
| 2 | Hanming Gu et al., 2020 [30] | NFKBIA, C3, CCL20, BCL2A1, BID | 5 |
| 3 | Kang Soon Nan et al., 2021 [18] | ALB, CXCL8, FGF2, IL6, INS, MMP2, MMP9, PTGS2, STAT3, VEGFA | 10 |
| 4 | Hanming Gu et al., 2020 [31] | CDC20, NCBP1, POLR2D, DYNLL1, FBXW5, LRRC41, FBXO21, FBXW9, FBXO44, FBXO6 | 10 |
| 5 | Rahila Sardar et al., 2020 [19] | HMOX1, DNMT1, PLAT, GDF1, ITGB1 | 5 |
| 6 | Hanming Gu et al., 2020 [21] | FLOC, DYNLL1, FBXL3, FBXW11, FBXO27, FBXO44, FBXO32, FBXO31, FBXO9, CUL2 | 10 |
| 7 | Tian-Ao Xie et al., 2020 [8] | CXCL1, CXCL2, TNF, NFKBIA, CSF2, TNFAIP3, IL6, CXCL3, CCL20, ICAM1 | 10 |
| 8 | Jung Hun Oh et al., 2020 [9] | GATA4, ID2, MAFA, NOX4, PTBP1, SMAD3, TUBB1, WWOX | 8 |
| 9 | Basavaraj Vastrad et al., 2020 [20] | TP53, HRAS, CTNNB1, FYN, ABL1, STAT3, STAT1, JAK2, C1QBP, XBP1, BST2, CD99, IFI35, MAPK11, RELA, LCK, KIT, EGR1, IL20, ILF3, CASP3, IL19, ATG7, GPI, S1PR1 | 25 |
| 10 | Kartikay Prasad et al., 2020 [22] | STAT1, IRF7, IFIH1, MX1, ISG15, IFIT3, OAS2, DDX58, IRF9, IFIT1, OAS1, OAS3, DDX60, OASL, IFIT2 | 15 |
| 11 | Gurudeeban Selvaraj et al., 2021 [32] | MYC, HDAC9, NCOA3, CEBPB, VEGFA, BCL3, SMAD3, SMURF1, KLHL12, CBL, ERBB4, CRKL | 12 |
| 12 | Md. Shahriare Satu et al., 2021 [24] | MARCO, VCAN, ACTB, LGALS1, HMOX1, TIMP1, OAS2, GAPDH, MSH3, FN1, NPC2, JUND, CHI3L1, GPNMB, SYTL2, CASP1, S100A8, MYO10, IGFBP3, APCDD1, COL6A3, FABP5, PRDX3, CLEC1B, DDIT4, CXCL10, CXCL8 | 27 |
| 13 | Tasnimul Alam Taz et al., 2020 [25] | VEGFA, AKT1, MMP9, ICAM1, CD44 | 5 |
| 14 | Mohammad Ali Moni et al., 2020 [26] | MX1, IRF7, BST2 | 3 |
| 15 | Tania Islam et al., 2020 [27] | BIRC3, ICAM1, IRAK2, MAP3K8, S100A8, SOCS3, STAT5A, TNF, TNFAIP3, TNIP1 | 10 |
| 16 | Yadi Zhou et al., 2020 [28] | JUN, XPO1, NPM1, HNRNPA1 | 4 |
| 17 | Ge C et al., 2020 [10] | MMP13, NLRP3, GBP1, ADORA2A, PTAFR, TNF, MLNR, IL1B, NFKBIA, ADRB2, IL6 | 11 |
| 18 | Aishwarya et al., 2020 [11] | IGF2, HINT1, MAPK10, SGCE, HDAC5, SGCA, SGCB, CFD, ITSN1, EHMT2, CLU, ISLR, PGM5, ANK2, HDAC9, SYT11, MDH1, SCCPDH, SIRT6, DTNA, FN1, ARRB1, MAGED2, TEX264, VEGFC, HK2, TXNL4A, SLC16A3, NUDT21, TRA2B, HNRNPA1, CDC40, THOC1, PFKFB3 | 34 |
| 19 | Saxena, A. et al., 2020 [12] | STAP1, CASP5, FDCSP, CARD17, ST20, AKR1B10, CLC, KCNJ2-AS1, RNASE2, FLG | 10 |
| 20 | Tao Q et al., 2020 [13] | MAPK3, MAPK1, MAPK8, IL10, TNF, CXCL8, IL6, PTGS2, TP53, CCL2, CASP3, IL1B | 12 |
| 21 | Zhang N et al., 2020 [14] | CXCL10, ISG15, DDX58, MX2, OASL, STAT1, RSAD2, MX1, IRF7, OAS1 | 10 |
| 22 | Han L et al., 2020 [15] | IL6, TNF, IL10, MAPK8, MAPK3, CXCL8, CASP3, PTGS2, TP53, MAPK1 | 10 |
| 23 | Tian J et al., 2020 [33] | CXCL8, CXCL2, CXCL10, ADRA2A, ADRA2C, CHRM2, PTGER3, OPRM1, OPRD1, JUN. | 10 |
| 24 | Jha PK et al., 2021 [34] | SMAD3, STAT1, SH3KBP1, HDGF, TUBB, NFKB2, ETS1, UBC, TUFM, TRAF3, CCT5, RPL9, TUBB4B, CSNK1E, S100A9 | 15 |
| 25 | Ramesh P et al., 2020 [35] | ELANE, MPO, ARG1, DEFA4,CAMP, MMP9, LTF, LCN2,PGLYRP1,HP | 10 |
| 26 | Li Zhonglin et al., 2020 [36] | DDOST, UPF1, HIST2H2A, ITGAL, EGFR, CXCL1, DYNLL1, POLR2F, RPL13A, FBXO11, CSNK1E | 11 |
| 27 | Li G et al., 2020 [37] | RPS3, RPS8, PRS9, VCP, LARP1, UBA52, PRKN, EIF3A, EIF3L, SRC, CASP1, RIPK, ACE2 | 13 |
| 28 | Prasad K et al., 2021 [38] | MOV10, NXF1, APP, ELAVL1, CUL3, XPO, TP53, EGFR, MCM2, MYC, COPS5, ESR1, UBC, FN1, CUL7, VCAM1, RNF2, CUL1, SIRT7, CAND1, OBSL1, HSP90AA1, CDK2, NPM1, GRB2, FBXO6, CDC5L, GABARAPL2, VCP, CCDC8, GABARAPL1, CUL2, SNW1, ITGA4, GABARAP | 35 |
| 29 | Fangzhou Liu et al., 2021 [39] | AKT1, TP53, TNF, IL6, BCL2L1, ATM | 6 |
| 30 | Zulkar Nain et al., 2020 [40] | NFKBIA, BUB3, EIF2S3, GADD45A, MET, MCL1, SOCS3 | 7 |
| 31 | Ke-Ying Fang et al., 2021 [41] | IL6, FN1, CXCL1, CCL5, CCL2, CXCL10, EGF, FGF2, ICAM1, CXCL8, IL1B, MMP9 | 12 |
| 32 | Mostafa Rezaei-Tavirani et al., 2021 [42] | FGA, FGG, FGF, ORM1, ORM2, PPBP, PF4, CRP, APOA2, SAA1, ACTB, CFB, LCAT, CETP, TLN1, SAA2, FGL1, CFI, YWHAZ, YWHAE, AZGP1, S100A8, CFHR1, CFHR3, PON3, PRDX6, ARHGDIB, TAGLN2, TRIM33, TUBB1, SH3BGRL3 | 31 |
| 33 | Shenglong Li et al., 2020 [43] | IL1b and IL6 | 2 |
| 34 | Suresh Kumar et al., 2020 [44] | VEGFA, TNF, IL-6, CXCL8, IL-10, CCL2, IL1B, TLR4, ICAM1, MMP9 | 10 |
| 35 | Yi-Wei Zhu et al., 2020 [45] | RELA, TNF, IL6, IL1B, MAPK14, TP53, CXCL8, MAPK3, MAPK1, IL4, MAPK8, CASP8 and STAT1 | 13 |
| 36 | Z. Bao et al., 2021 [46] | CCL11, TNFAIP6, AGTR2, FGA, CRM2, HBB, IRF1, IL1RN, IDO1, ATF3, CRM1, CCL4L1, CD163, FGG, CCL21, CCL3, SELE, CCL19, HSP90AA1, CX3CL1, SERPINA1, CSF3, THBS1, HP, SERPNE1, VCAM1, CXCL9, CCL4, PTGS2, CXCL10, CCL2, CXCL8, ALB, IL6 | 34 |
| 37 | Zhen-Zhen Wang et al., 2021 [47] | TNF, EGFR, CASP9, EGFA, NFKB1, TP53, IL6, CASP3, MAPK8, PTGS2, GAPDH, CCL2, NFKBIA, MMP9, MMP2, CCND1, MCL1, MAPK1, MYC, CXCL8, JUN, CASP8, PPARG, IL1B | 24 |
| 38 | Auwul et al., 2021 [48] | PLK1, AURKB, AURKA, CDK1, CDC20, KIF11, CCNB1, KIF2C, DTL and CDC6 | 10 |
| 39 | Mosharaf et al., 2022 [49] | TLR2, USP53, GUCY1A2, SNRPD2, NEDD9, IGF2, CXCL2, KLF6, PAG1 and ZFP36 | 10 |
| 40 | Lee H et al., 2021 [50] | SLC3A2, SLC2A3, FOLR2, CCR1, FPR1, GPR183, CD68, FCGR3B, KLRD1, CD3D, KRT7, TPPP3, CD6, HBB, PPBP and MS4A1 | 16 |
| 41 | Alanazi et al., 2022 [51] | NSP1, NSP3, NSP5, NSP9, NSP12, NSP13, NSP15, 3a, S, E, M, 6, 7a and N | 14 |
| Common genes in at least 5 articles | CXCL8, IL6, TNF, TP53, IL1B, MMP9, NFKBIA, PTGS2, ICAM1, STAT1, CCL2 | 11 | |
| Common genes in at least 6 articles | CXCL8, IL6, TNF, TP53, IL1B, MMP9 | 6 | |
| Common genes in at least 7 articles | CXCL8, IL6, TNF, TP53, IL1B | 5 | |
| Common genes in at least 9 articles | CXCL8, IL6, TNF | 3 | |
| Common genes in at least 11 articles | CXCL8, IL6 | 2 | |
| Source | GO Term ID | Description | Padj-Value | Gene Count | Enriched Genes |
|---|---|---|---|---|---|
| GO:MF | GO:0019899 | enzyme binding | 0.00000000 | 11 | AKT1, APP, EGFR, INS, JUN, MAPK1, STAT3, TNF, TP53, UBA52, UBC |
| GO:0098772 | molecular function regulator activity | 0.00000000 | 10 | AKT1, APP, CXCL8, EGFR, IL6, INS, JUN, TNF, TP53, VEGFA | |
| GO:0042802 | identical protein binding | 0.00000000 | 10 | AKT1, APP, EGFR, INS, JUN, MAPK1, STAT3, TNF, TP53, VEGFA | |
| GO:0005102 | signaling receptor binding | 0.00000000 | 9 | APP, CXCL8, EGFR, IL6, INS, STAT3, TNF, TP53, VEGFA | |
| GO:0019902 | phosphatase binding | 0.00000003 | 5 | AKT1, EGFR, MAPK1, STAT3, TP53 | |
| GO:0030546 | signaling receptor activator activity | 0.00000003 | 6 | APP, CXCL8, IL6, INS, TNF, VEGFA | |
| GO:0005515 | protein binding | 0.00000008 | 14 | AKT1, APP, CXCL8, EGFR, IL6, INS, JUN, MAPK1, STAT3, TNF, TP53, UBA52, UBC, VEGFA | |
| GO:0005126 | cytokine receptor binding | 0.00000010 | 5 | CXCL8, IL6, STAT3, TNF, VEGFA | |
| GO:0031625 | ubiquitin protein ligase binding | 0.00000013 | 5 | EGFR, JUN, TP53, UBA52, UBC | |
| GO:0044389 | ubiquitin-like protein ligase binding | 0.00000015 | 5 | EGFR, JUN, TP53, UBA52, UBC | |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 0.00000000 | 14 | AKT1, APP, CXCL8, EGFR, IL6, INS, JUN, MAPK1, STAT3, TNF, TP53, UBA52, UBC, VEGFA |
| GO:0051090 | regulation of DNA-binding transcription factor activity | 0.00000000 | 11 | AKT1, APP, IL6, INS, JUN, MAPK1, STAT3, TNF, UBA52, UBC, VEGFA | |
| GO:0009891 | positive regulation of biosynthetic process | 0.00000000 | 14 | AKT1, APP, CXCL8, EGFR, IL6, INS, JUN, MAPK1, STAT3, TNF, TP53, UBA52, UBC, VEGFA | |
| GO:0001934 | positive regulation of protein phosphorylation | 0.00000000 | 12 | AKT1, APP, EGFR, IL6, INS, MAPK1, STAT3, TNF, TP53, UBA52, UBC, VEGFA | |
| GO:0042327 | positive regulation of phosphorylation | 0.00000000 | 12 | AKT1, APP, EGFR, IL6, INS, MAPK1, STAT3, TNF, TP53, UBA52, UBC, VEGFA | |
| GO:0010562 | positive regulation of phosphorus metabolic process | 0.00000000 | 12 | AKT1, APP, EGFR, IL6, INS, MAPK1, STAT3, TNF, TP53, UBA52, UBC, VEGFA | |
| GO:0045937 | positive regulation of phosphate metabolic process | 0.00000000 | 12 | AKT1, APP, EGFR, IL6, INS, MAPK1, STAT3, TNF, TP53, UBA52, UBC, VEGFA | |
| GO:0031401 | positive regulation of protein modification process | 0.00000000 | 12 | AKT1, APP, EGFR, IL6, INS, MAPK1, STAT3, TNF, TP53, UBA52, UBC, VEGFA | |
| GO:0009719 | response to endogenous stimulus | 0.00000000 | 13 | AKT1, APP, CXCL8, EGFR, IL6, INS, JUN, MAPK1, STAT3, TNF, TP53, UBA52, UBC | |
| GO:0071310 | cellular response to organic substance | 0.00000000 | 14 | AKT1, APP, CXCL8, EGFR, IL6, INS, JUN, MAPK1, STAT3, TNF, TP53, UBA52, UBC, VEGFA | |
| GO:CC | GO:0043233 | organelle lumen | 0.00000000 | 12 | AKT1, APP, EGFR, IL6, INS, JUN, MAPK1, STAT3, TP53, UBA52, UBC, VEGFA |
| GO:0070013 | intracellular organelle lumen | 0.00000000 | 12 | AKT1, APP, EGFR, IL6, INS, JUN, MAPK1, STAT3, TP53, UBA52, UBC, VEGFA | |
| GO:0031974 | membrane-enclosed lumen | 0.00000000 | 12 | AKT1, APP, EGFR, IL6, INS, JUN, MAPK1, STAT3, TP53, UBA52, UBC, VEGFA | |
| GO:0016020 | membrane | 0.00000004 | 13 | AKT1, APP, EGFR, IL6, INS, JUN, MAPK1, STAT3, TNF, TP53, UBA52, UBC, VEGFA | |
| GO:0005768 | endosome | 0.00000005 | 7 | APP, EGFR, INS, MAPK1, TNF, UBA52, UBC | |
| GO:0005783 | endoplasmic reticulum | 0.00000012 | 8 | APP, EGFR, IL6, INS, MAPK1, TP53, UBA52, UBC | |
| GO:0071944 | cell periphery | 0.00000012 | 11 | AKT1, APP, EGFR, IL6, JUN, MAPK1, STAT3,TNF, UBA52, UBC, VEGFA | |
| GO:0005576 | extracellular region | 0.00000013 | 10 | APP, CXCL8, EGFR, IL6, INS, MAPK1, TNF, UBA52, UBC, VEGFA | |
| GO:0012505 | endomembrane system | 0.00000014 | 10 | APP, EGFR, IL6, INS, MAPK1, TNF, TP53, UBA52, UBC, VEGFA | |
| GO:0031983 | vesicle lumen | 0.00000017 | 5 | APP, EGFR, INS, MAPK1, VEGFA |
| Potential Targets | Structure of Ligand | Binding Affinity (kCal/mol) | Surface View of Complex | Pose View of Complex | Target Ligand Interaction | Interacting Amino Acid | Bond Type | Distance (A0) |
|---|---|---|---|---|---|---|---|---|
 MAPK1 MAPK1 | 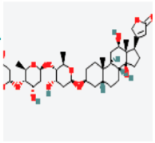 Digoxin Digoxin | −11.0 | 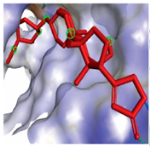 |  |  | ARG191 TRP192 ALA52 LUE170 VAL39 LUE56 ILE84 | CH CH A A A A A | 2.55 2.783 3.544 4.820 5.029 4.883 4.404 |
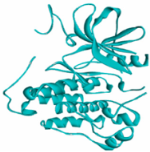 EGFR EGFR |  Avermectin Avermectin | −10.8 | 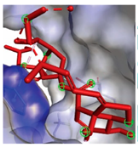 |  |  | VAL876 ASP855 VAL726 LYS745 ILE878 LYS878 ARG858 PHE723 | CH CHB A A A A A PA | 2.068 3.581 4.181 4.063 5.221 4.334 5.086 4.743 |
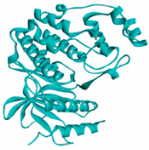 MAPK1 MAPK1 | 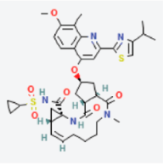 Simprevir Simprevir | −10.3 |  |  | 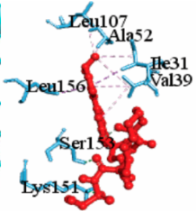 | LYS151 SER153 ILE31 LEU156 ALA52 VAL39 LYS54 LEU107 LEU156 | CH CH PS PS A A A A A | 2.176 2.985 3.804 3.638 3.738 4.700 4.539 4.940 4.662 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mosharaf, M.P.; Kibria, M.K.; Hossen, M.B.; Islam, M.A.; Reza, M.S.; Mahumud, R.A.; Alam, K.; Gow, J.; Mollah, M.N.H. Meta-Data Analysis to Explore the Hub of the Hub-Genes That Influence SARS-CoV-2 Infections Highlighting Their Pathogenetic Processes and Drugs Repurposing. Vaccines 2022, 10, 1248. https://doi.org/10.3390/vaccines10081248
Mosharaf MP, Kibria MK, Hossen MB, Islam MA, Reza MS, Mahumud RA, Alam K, Gow J, Mollah MNH. Meta-Data Analysis to Explore the Hub of the Hub-Genes That Influence SARS-CoV-2 Infections Highlighting Their Pathogenetic Processes and Drugs Repurposing. Vaccines. 2022; 10(8):1248. https://doi.org/10.3390/vaccines10081248
Chicago/Turabian StyleMosharaf, Md. Parvez, Md. Kaderi Kibria, Md. Bayazid Hossen, Md. Ariful Islam, Md. Selim Reza, Rashidul Alam Mahumud, Khorshed Alam, Jeff Gow, and Md. Nurul Haque Mollah. 2022. "Meta-Data Analysis to Explore the Hub of the Hub-Genes That Influence SARS-CoV-2 Infections Highlighting Their Pathogenetic Processes and Drugs Repurposing" Vaccines 10, no. 8: 1248. https://doi.org/10.3390/vaccines10081248
APA StyleMosharaf, M. P., Kibria, M. K., Hossen, M. B., Islam, M. A., Reza, M. S., Mahumud, R. A., Alam, K., Gow, J., & Mollah, M. N. H. (2022). Meta-Data Analysis to Explore the Hub of the Hub-Genes That Influence SARS-CoV-2 Infections Highlighting Their Pathogenetic Processes and Drugs Repurposing. Vaccines, 10(8), 1248. https://doi.org/10.3390/vaccines10081248






