Identification and Characterization of Epithelial Cell-Derived Dense Bodies Produced upon Cytomegalovirus Infection
Abstract
:1. Introduction
2. Material and Methods
2.1. Cells, Viruses and Viral Infections
2.2. Dense Bodies’Purifications and Protein Analysis
2.3. Analysis of Virus Replication
2.4. Ultrastructural Analysis of CMV-Infected Cells
2.5. Negative Staining
2.6. Particle Size and Abundance Analysis
3. Results
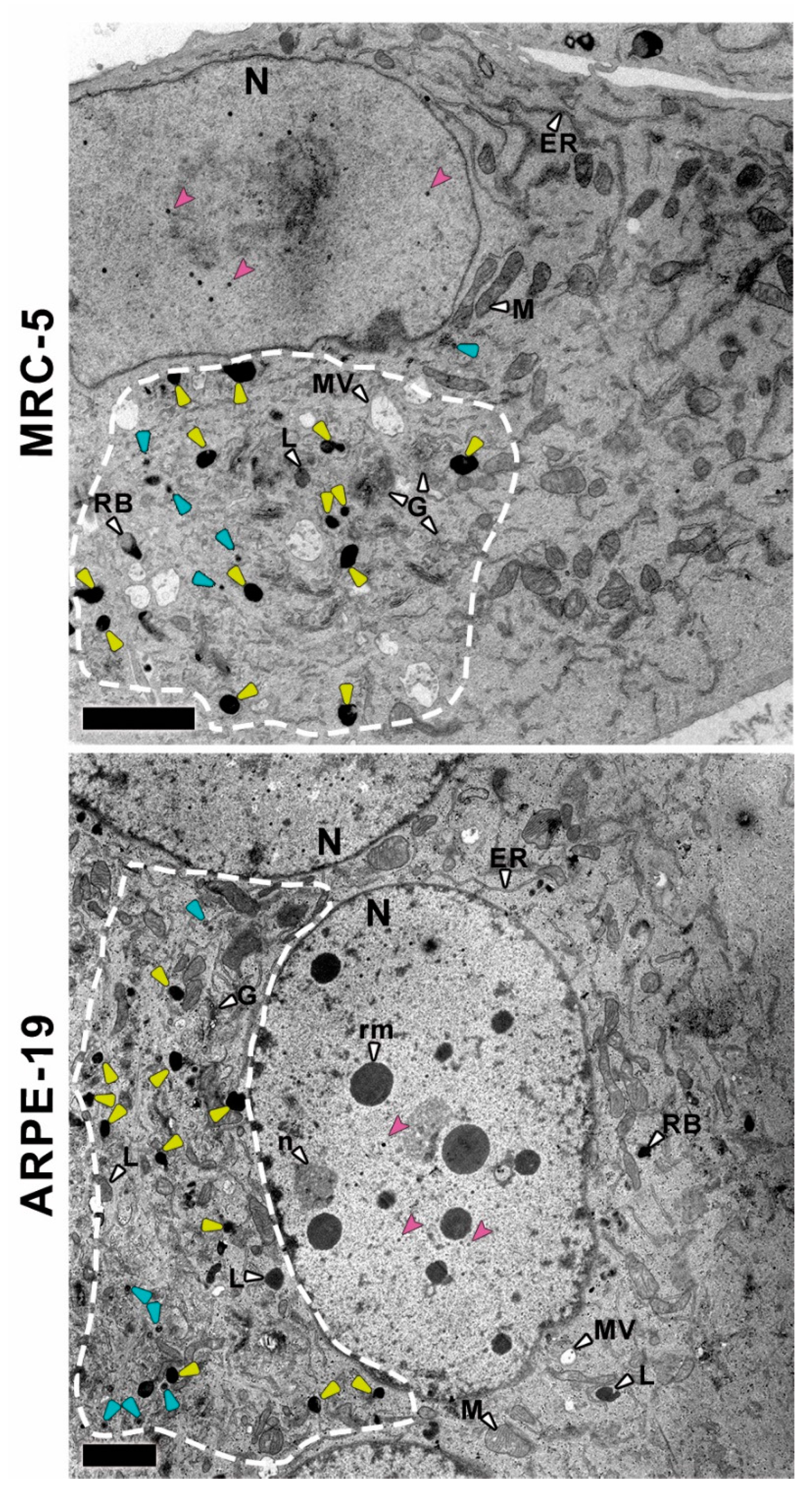
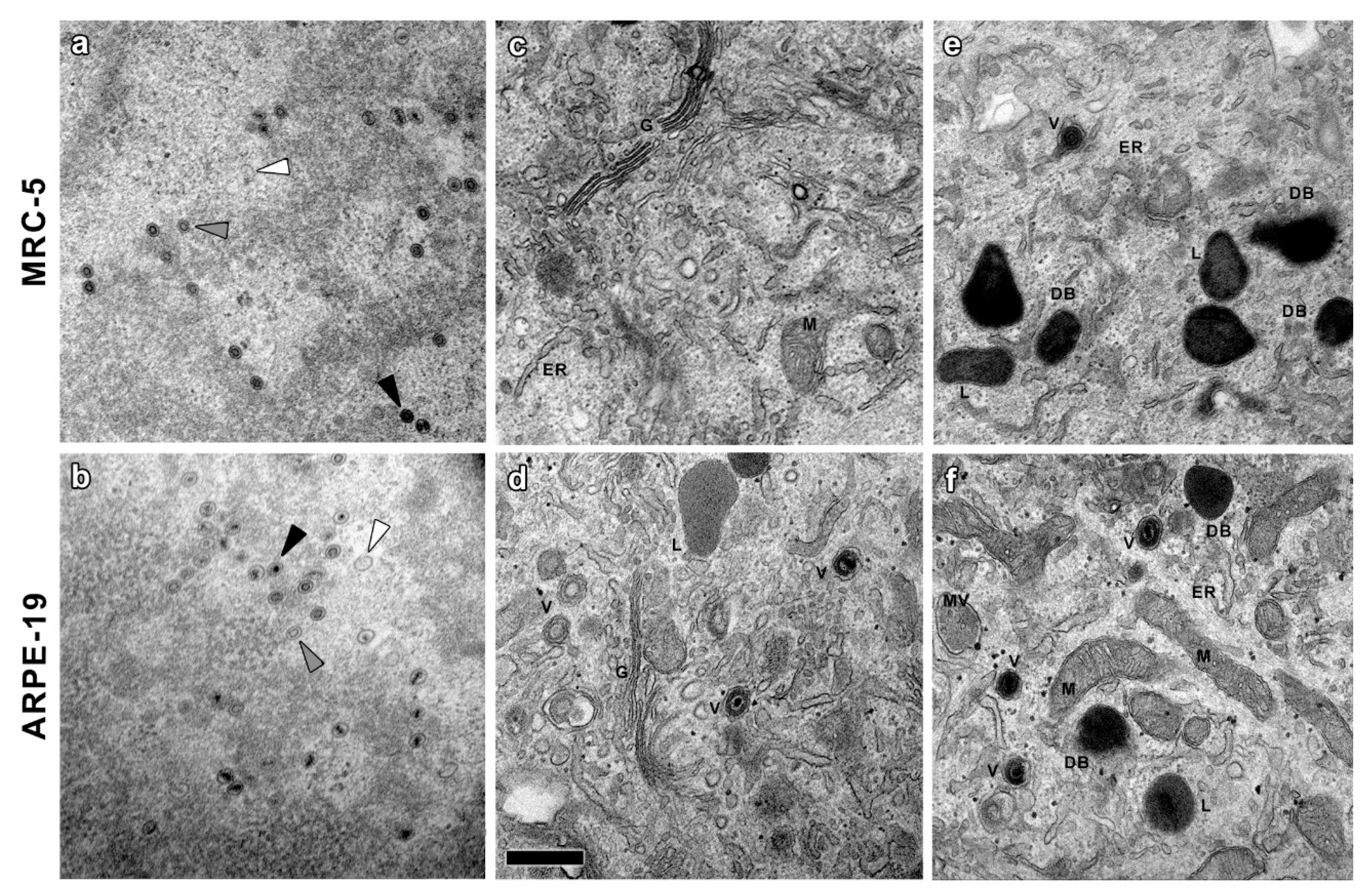
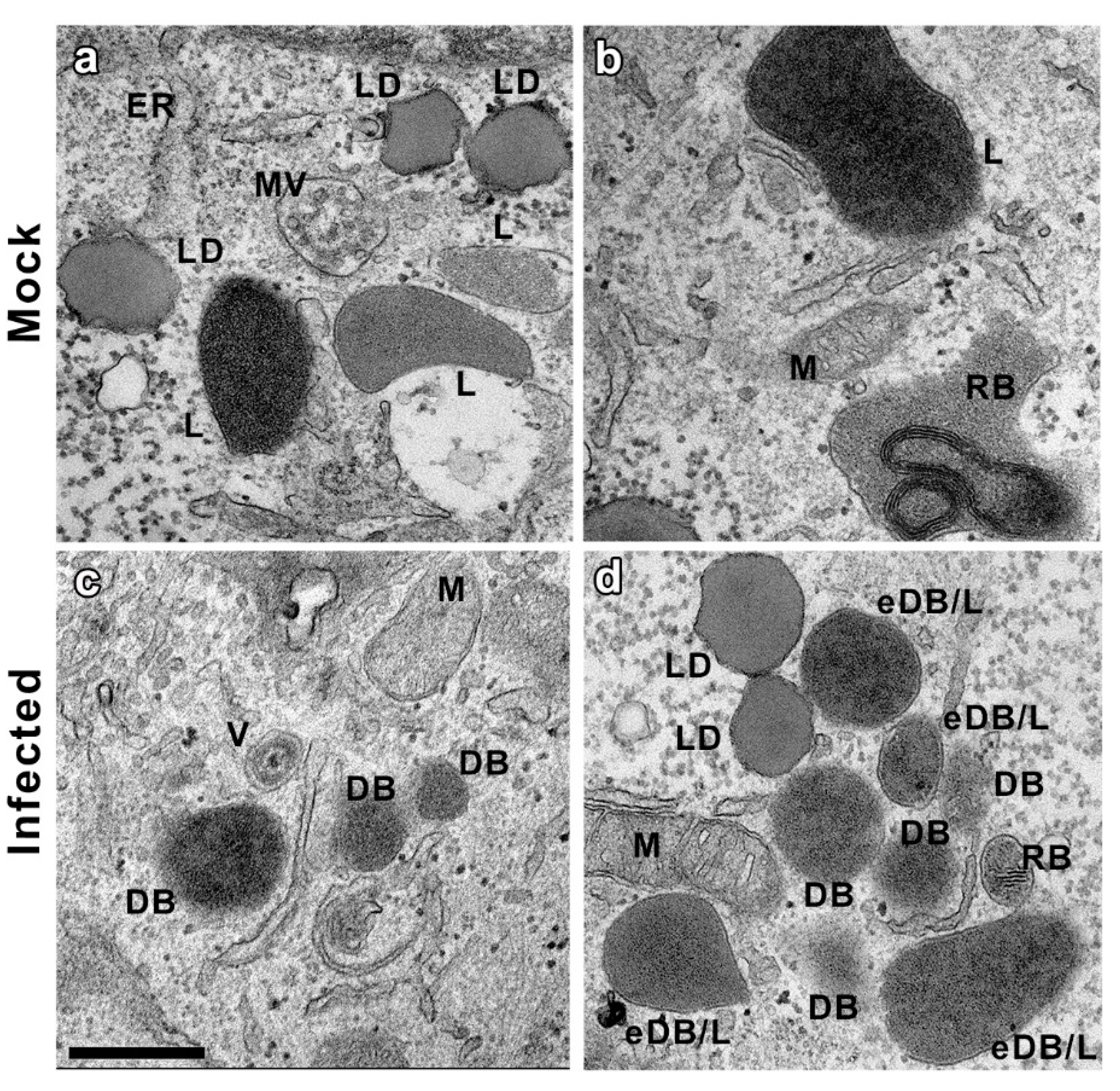
3.1. Ultrastructural Analysis of CMV-Infected MRC-5 and ARPE-19 Cells
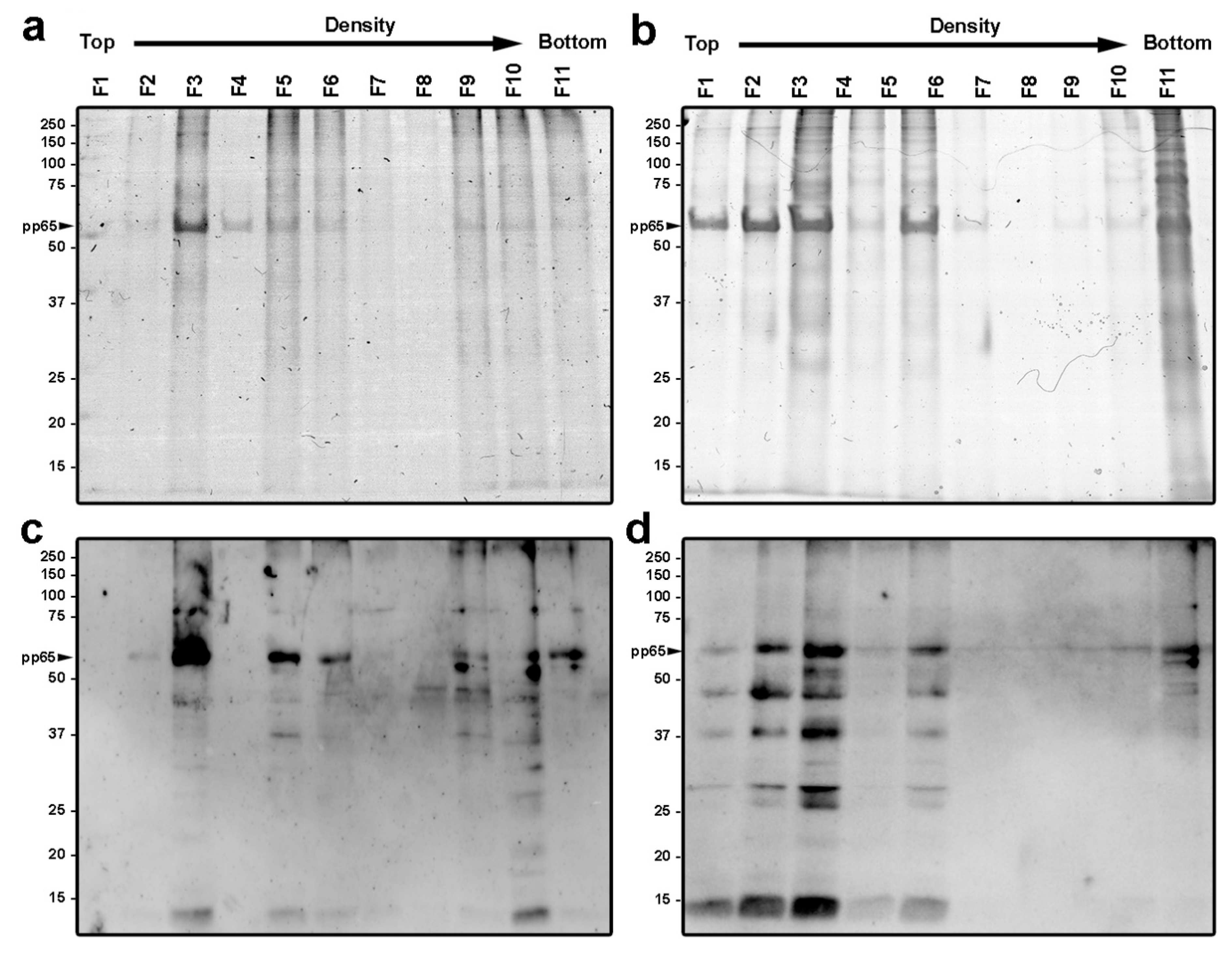
3.2. Purification and Characterization of Viral Particles
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Murphy, E.; Shenk, T.E. Human Cytomegalovirus Genome; Shenk, T.E., Stinski, M.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; Volume 325, ISBN 9783540773481. [Google Scholar]
- Boeckh, M.; Geballe, A.P. Science in medicine Cytomegalovirus: Pathogen, paradigm, and puzzle. J. Clin. Investig. 2011, 121, 1673–1680. [Google Scholar] [CrossRef]
- Seitz, R. Human Cytomegalovirus (HCMV)-Revised. Transfus. Med. Hemotherapy 2010, 37, 365–375. [Google Scholar] [CrossRef]
- Manandhar, T.; Hò, G.G.T.; Pump, W.C.; Blasczyk, R.; Bade-Doeding, C. Battle between host immune cellular responses and HCMV immune evasion. Int. J. Mol. Sci. 2019, 20, 3626. [Google Scholar] [CrossRef]
- Boppana, S.B.; Ross, S.A.; Fowler, K.B. Congenital Cytomegalovirus Infection: Clinical Outcome. Clin. Infect. Dis. 2013, 57, S178–S181. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Shah, S.; Lee, M.; Dai, W.; Lo, P.; Britt, W.; Zhu, H.; Liu, F.; Zhou, H.Z. Biochemical and Structural Characterization of the Capsid- Bound Tegument Proteins of Human Cytomegalovirus. J. Struct. Biol. 2011, 174, 451–456. [Google Scholar] [CrossRef]
- Pepperl, S.; Munster, J.; Mach, M.; Harris, J.R.; Plachter, B. Dense Bodies of Human Cytomegalovirus Induce both Humoral and Cellular Immune Responses in the Absence of Viral Gene Expression. J. Virol. 2000, 74, 6132–6146. [Google Scholar] [CrossRef] [PubMed]
- Büscher, N.; Paulus, C.; Nevels, M.; Tenzer, S.; Plachter, B. The proteome of human cytomegalovirus virions and dense bodies is conserved across different strains. Med. Microbiol. Immunol. 2015, 204, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Varnum, S.M.; Nelson, J.A.; Wiley, S.; Britt, W.; Shenk, T.; Smith, R.D. Identification of Proteins in Human Cytomegalovirus (HCMV) Particles: The HCMV Proteome. J. Virol. 2004, 78, 10960–10966. [Google Scholar] [CrossRef]
- Gogesch, P.; Penner, I.; Krauter, S.; Büscher, N.; Grode, L.; Aydin, I.; Plachter, B. Production strategies for pentamer-positive subviral dense bodies as a safe human cytomegalovirus vaccine. Vaccines 2019, 7, 104. [Google Scholar] [CrossRef]
- Pepperl-Klindworth, S.; Frankenberg, N.; Plachter, B. Development of novel vaccine strategies against human cytomegalovirus infection based on subviral particles. J. Clin. Virol. 2002, 25, 75–85. [Google Scholar] [CrossRef]
- Schneider-Ohrum, K.; Cayatte, C.; Liu, Y.; Wang, Z.; Irrinki, A.; Cataniag, F.; Nguyen, N.; Lambert, S.; Liu, H.; Aslam, S.; et al. Production of Cytomegalovirus Dense Bodies by Scalable Bioprocess Methods Maintains Immunogenicity and Improves Neutralizing Antibody Titers. J. Virol. 2016, 90, 10133–10144. [Google Scholar] [CrossRef] [PubMed]
- Cayatte, C.; Schneider-Ohrum, K.; Wang, Z.; Irrinki, A.; Nguyen, N.; Lu, J.; Nelson, C.; Servat, E.; Gemmell, L.; Citkowicz, A.; et al. Cytomegalovirus Vaccine Strain Towne-Derived Dense Bodies Induce Broad Cellular Immune Responses and Neutralizing Antibodies That Prevent Infection of Fibroblasts and Epithelial Cells. J. Virol. 2013, 87, 11107–11120. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, C.; Falk, J.J.; Büscher, N.; Penner, I.; Zimmermann, C.; Gogesch, P.; Sinzger, C.; Plachter, B. Dense Bodies of a gH/gL/UL128/UL130/UL131 Pentamer-Repaired Towne Strain of Human Cytomegalovirus Induce an Enhanced Neutralizing Antibody Response. J. Virol. 2019, 93, e00931-19. [Google Scholar] [CrossRef] [PubMed]
- Sauer, C.; Klobuch, S.; Herr, W.; Thomas, S.; Plachter, B. Subviral Dense Bodies of Human Cytomegalovirus Stimulate Maturation and Activation of Monocyte-Derived Immature Dendritic Cells. J. Virol. 2013, 87, 11287–11291. [Google Scholar] [CrossRef] [PubMed]
- Ahlqvist, J.; Mocarski, E. Cytomegalovirus UL103 Controls Virion and Dense Body Egress. J. Virol. 2011, 85, 5125–5135. [Google Scholar] [CrossRef]
- Gerna, G.; Kabanova, A.; Lilleri, D. Human Cytomegalovirus Cell Tropism and Host Cell Receptors. Vaccines 2019, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Krause, P.R.; Bialek, S.R.; Boppana, S.B.; Griffiths, P.D.; Laughlin, C.A.; Ljungdahl, P.O.; Mocarski, E.S.; Pass, R.F.; Read, J.S.; Schleiss, M.R.; et al. Priorities for CMV vaccine development. Vaccine 2013, 32, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, S.A.; Boppana, S.B. Vaccination against the human cytomegalovirus. Vaccine 2019, 37, 7437–7442. [Google Scholar] [CrossRef] [PubMed]
- Anderholm, K.; Bierle, C.; Schleiss, M. Cytomegalovirus Vaccines: Current Status and Future Prospects. Drugs 2016, 76, 1625–1645. [Google Scholar] [CrossRef] [PubMed]
- Gerna, G.; Lilleri, D. Human cytomegalovirus (HCMV) infection/re-infection: Development of a protective HCMV vaccine. New Microbiol. 2019, 42, 1–20. [Google Scholar] [PubMed]
- Ferri, C.; Giuggioli, D.; Raimondo, V.; L’Andolina, M.; Tavoni, A.; Cecchetti, R.; Guiducci, S.; Ursini, F.; Caminiti, M.; Varcasia, G.; et al. COVID-19 and rheumatic autoimmune systemic diseases: Report of a large Italian patients series. Clin. Rheumatol. 2020, 39, 3195–3204. [Google Scholar] [CrossRef] [PubMed]
- Hall, V.; Ferreira, V.; Kumar, D.; Humal, A. Impact of Immunosuppression on the Immune Response to SARS-CoV-2 Infection: A Mechanistic Study. Transpl. Infect. Dis. 2021, 23, e13743. [Google Scholar] [CrossRef] [PubMed]
- Ferri, C.; Giuggioli, D.; Raimondo, V.; Dagna, L.; Riccieri, V.; Zanatta, E.; Guiducci, S.; Tavoni, A.; Foti, R.; Cuomo, G.; et al. COVID-19 and systemic sclerosis: Clinicopathological implications from Italian nationwide survey study. Lancet Rheumatol. 2021, 3, e166–e168. [Google Scholar] [CrossRef]
- Fouts, A.E.; Chan, P.; Stephan, J.-P.; Vandlen, R.; Feierbach, B. Antibodies against the gH/gL/UL128/UL130/UL131 Complex Comprise the Majority of the Anti-Cytomegalovirus (Anti-CMV) Neutralizing Antibody Response in CMV Hyperimmune Globulin. J. Virol. 2012, 86, 7444–7447. [Google Scholar] [CrossRef] [PubMed]
- Marshall, G.S.; Rabalais, G.P.; Stout, G.G.; Waldeyer, S.L. Antibodies to recombinant-derived glycoprotein b after natural human cytomegalovirus infection correlate with neutralizing activity. J. Infect. Dis. 1992, 165, 381–384. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.; Klein, M.; Britt, W.J.; Haßfurther, E.; Mach, M. Glycoprotein H of human cytomegalovirus is a major antigen for the neutralizing humoral immune response. J. Gen. Virol. 1996, 77, 1537–1547. [Google Scholar] [CrossRef]
- Sandonís, V.; García-Ríos, E.; McConnell, M.J.; Pérez-Romero, P. Role of Neutralizing Antibodies in CMV Infection: Implications for New Therapeutic Approaches. Trends Microbiol. 2020, 28, 900–912. [Google Scholar] [CrossRef] [PubMed]
- Sinzger, C.; Grefte, A.; Plachter, B.; Gouw, A.S.H.; The, T.H.; Jahn, G. Fibroblasts, epithelial cells, endothelial cells and smooth muscle cells are major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J. Gen. Virol. 1995, 76, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Ryckman, B.J.; Rainish, B.L.; Chase, M.C.; Borton, J.A.; Nelson, J.A.; Jarvis, M.A.; Johnson, D.C. Characterization of the Human Cytomegalovirus gH/gL/UL128-131 Complex That Mediates Entry into Epithelial and Endothelial Cells. J. Virol. 2008, 82, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Freed, D.C.; He, X.; Li, F.; Tang, A.; Cox, K.S.; Dubey, S.A.; Cole, S.; Medi, M.B.; Liu, Y.; et al. A replication-defective human cytomegalovirus vaccine for prevention of congenital infection. Sci. Transl. Med. 2016, 8, 362ra145. [Google Scholar] [CrossRef]
- Irmiere, A.; Gibson, W. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 1983, 130, 118–133. [Google Scholar] [CrossRef]
- Talbot, P.; Almeida, J.D. Human cytomegalovirus: Purification of enveloped virions and dense bodies. J. Gen. Virol. 1977, 36, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Tandon, R.; Mocarski, E.S. Viral and host control of cytomegalovirus maturation. Trends Microbiol. 2012, 20, 392–401. [Google Scholar] [CrossRef]
- Becke, S.; Aue, S.; Thomas, D.; Schader, S.; Podlech, J.; Bopp, T.; Sedmak, T.; Wolfrum, U.; Plachter, B.; Reyda, S. Optimized recombinant dense bodies of human cytomegalovirus efficiently prime virus specific lymphocytes and neutralizing antibodies without the addition of adjuvant. Vaccine 2010, 28, 6191–6198. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Smith, G.A.; Enquist, L.W.; Shenk, T. Construction of a Self-Excisable Bacterial Artificial Chromosome Containing the Human Cytomegalovirus Genome and Mutagenesis of the Diploid TRL/IRL13 Gene. J. Virol. 2002, 76, 2316–2328. [Google Scholar] [CrossRef] [PubMed]
- Giménez, E.; Muñoz-Cobo, B.; Solano, C.; Amat, P.; Navarro, D. Early kinetics of plasma cytomegalovirus DNA load in allogeneic stem cell transplant recipients in the era of highly sensitive real-time PCR assays: Does it have any clinical value? J. Clin. Microbiol. 2014, 52, 654–656. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Mocarski, E.S.; Shenk, T.E.; Pass, R.F. Cytomegaloviruses. In Fields Virology; Fields, B.N., Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; ISBN 9780781760607. [Google Scholar]
- Kerr, J.F.R.; Wyllie, A.H.; Currie, A.R. Apoptosis: A Basic Biological Phenomenon With Wide-ranging implications in tissue kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Niquet, J.; Baldwin, R.A.; Allen, S.G.; Fujikawa, D.G.; Wasterlain, C.G. Hypoxic neuronal necrosis: Protein synthesis-independent activation of a cell death program. Proc. Natl. Acad. Sci. USA 2003, 100, 2825–2830. [Google Scholar] [CrossRef] [PubMed]
- Buser, C.; Walther, P.; Mertens, T.; Michel, D. Cytomegalovirus Primary Envelopment Occurs at Large Infoldings of the Inner Nuclear Membrane. J. Virol. 2007, 81, 3042–3048. [Google Scholar] [CrossRef]
- Das, S.; Vasanji, A.; Pellett, P.E. Three-Dimensional Structure of the Human Cytomegalovirus Cytoplasmic Virion Assembly Complex Includes a Reoriented Secretory Apparatus. J. Virol. 2007, 81, 11861–11869. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.D.; de Harven, E. Herpes simplex virus and human cytomegalovirus replication in WI-38 cells. III. Cytochemical localization of lysosomal enzymes in infected cells. J. Virol. 1978, 26, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Craighead, J.E.; Kanich, R.E.; Almeida, J.D. Nonviral Microbodies with Viral Antigenicity Produced in Cytomegalovirus-Infected Cells. J. Virol. 1972, 10, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Roby, C.; Gibson, W. Characterization of phosphoproteins and protein kinase activity of virions, noninfectious enveloped particles, and dense bodies of human cytomegalovirus. J. Virol. 1986, 59, 714–727. [Google Scholar] [CrossRef] [PubMed]
- Krömmelbein, N.; Wiebusch, L.; Schiedner, G.; Büscher, N.; Sauer, C.; Florin, L.; Sehn, E.; Wolfrum, U.; Plachter, B. Adenovirus E1A/E1B transformed amniotic fluid cells support human cytomegalovirus replication. Viruses 2016, 8, 37. [Google Scholar] [CrossRef]
- Schampera, M.S.; Arellano-Galindo, J.; Kagan, K.O.; Adler, S.P.; Jahn, G.; Hamprecht, K. Role of pentamer complex-specific and IgG subclass 3 antibodies in HCMV hyperimmunoglobulin and standard intravenous IgG preparations. Med. Microbiol. Immunol. 2019, 208, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Scrivano, L.; Sinzger, C.; Nitschko, H.; Koszinowski, U.H.; Adler, B. HCMV spread and cell tropism are determined by distinct virus populations. PLoSPathog. 2011, 7, e1001256. [Google Scholar] [CrossRef]
- Barnes, S.; Schilizzi, O.; Audsley, K.M.; Newnes, H.V.; Foley, B. Deciphering the immunological phenomenon of adaptive natural killer (NK) cells and cytomegalovirus (CMV). Int. J. Mol. Sci. 2020, 21, 8864. [Google Scholar] [CrossRef]
- White, M.J.; Nielsen, C.M.; Mcgregor, R.H.C.; Riley, E.M.; Goodier, M.R. Differential activation of CD57-defined natural killer cell subsets during recall responses to vaccine antigens. Immunology 2014, 142, 140–150. [Google Scholar] [CrossRef]
- Horowitz, A.; Hafalla, J.C.R.; King, E.; Lusingu, J.; Dekker, D.; Leach, A.; Moris, P.; Cohen, J.; Vekemans, J.; Villafana, T.; et al. Antigen-Specific IL-2 Secretion Correlates with NK Cell Responses after Immunization of Tanzanian Children with the RTS,S/AS01 Malaria Vaccine. J. Immunol. 2012, 188, 5054–5062. [Google Scholar] [CrossRef]
- Costa-Garcia, M.; Vera, A.; Moraru, M.; Vilches, C.; López-Botet, M.; Muntasell, A. Antibody-Mediated Response of NKG2C bright NK Cells against Human Cytomegalovirus. J. Immunol. 2015, 194, 2715–2724. [Google Scholar] [CrossRef] [PubMed]
- Costa-García, M.; Ataya, M.; Moraru, M.; Vilches, C.; López-Botet, M.; Muntasell, A. Human cytomegalovirus antigen presentation by HLA-DR+ NKG2C+ adaptive NK cells specifically activates polyfunctional effector memory CD4+ T lymphocytes. Front. Immunol. 2019, 10, 687. [Google Scholar] [CrossRef] [PubMed]
- Pupuleku, A.; Costa-García, M.; Farré, D.; Hengel, H.; Angulo, A.; Muntasell, A.; López-Botet, M. Elusive role of the CD94/NKG2C NK cell receptor in the response to cytomegalovirus: Novel experimental observations in a reporter cell system. Front. Immunol. 2017, 8, 1317. [Google Scholar] [CrossRef] [PubMed]
- Beninga, J.; Kropff, B.; Mach, N. Comparative analysis of fourteen individual human cytomegalovirus proteins for helper T cell response. J. Gen. Virol. 1995, 76, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Gaffen, S.L.; Jain, R.; Garg, A.V.; Cua, D.J. IL-23-IL-17 immune axis: Discovery, Mechanistic Understanding, and Clinical Testing. Nat. Rev. Immunol. 2014, 14, 585–600. [Google Scholar] [CrossRef] [PubMed]
- Paiva, I.A.; Badolato-Corrêa, J.; Familiar-Macedo, D.; De-Oliveira-pinto, L.M. Th17 cells in viral infections—Friend or foe? Cells 2021, 10, 1159. [Google Scholar] [CrossRef] [PubMed]
- Murdaca, G.; Colombo, B.M.; Puppo, F. The role of Th17 lymphocytes in the autoimmune and chronic inflammatory diseases. Intern. Emerg. Med. 2011, 6, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Wacleche, V.S.; Landay, A.; Routy, J.P.; Ancuta, P. The Th17 lineage: From barrier surfaces homeostasis to autoimmunity, cancer, and HIV-1 pathogenesis. Viruses 2017, 9, 303. [Google Scholar] [CrossRef]
- Alwine, J.C. The Human Cytomegalovirus Assembly Compartment: A Masterpiece of Viral Manipulation of Cellular Processes That Facilitates Assembly and Egress. PLoS Pathog. 2012, 8, e1002878. [Google Scholar] [CrossRef]
- Smith, J.D.; De Harven, E. Herpes simplex virus and human cytomegalovirus replication in WI-38 cells. I. Sequence of viral replication. J. Virol. 1973, 12, 919–930. [Google Scholar] [CrossRef]
- Sanchez, V.; Greis, K.D.; Sztul, E.; Britt, W.J. Accumulation of Virion Tegument and Envelope Proteins in a Stable Cytoplasmic Compartment during Human Cytomegalovirus Replication: Characterization of a Potential Site of Virus Assembly. J. Virol. 2000, 74, 975–986. [Google Scholar] [CrossRef] [PubMed]
- Mersseman, V.; Besold, K.; Reddehase, M.J.; Wolfrum, U.; Strand, D.; Plachter, B.; Reyda, S. Exogenous introduction of an immunodominant peptide from the non-structural IE1 protein of human cytomegalovirus into the MHC class I presentation pathway by recombinant dense bodies. J. Gen. Virol. 2008, 89, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Dunn, K.C.; Aotaki-Keen, A.E.; Putkey, F.R.; Hjelmeland, L.M. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp. Eye Res. 1996, 62, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Topilko, A.; Michelson, S. Hyperimmediate Entry of Human Cytomegalovirus Virions and Dense Bodies Into Human Fibroblasts. Res. Virol. 1994, 145, 75–82. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Ríos, E.; Rodríguez, M.J.; Terrón, M.C.; Luque, D.; Pérez-Romero, P. Identification and Characterization of Epithelial Cell-Derived Dense Bodies Produced upon Cytomegalovirus Infection. Vaccines 2022, 10, 1308. https://doi.org/10.3390/vaccines10081308
García-Ríos E, Rodríguez MJ, Terrón MC, Luque D, Pérez-Romero P. Identification and Characterization of Epithelial Cell-Derived Dense Bodies Produced upon Cytomegalovirus Infection. Vaccines. 2022; 10(8):1308. https://doi.org/10.3390/vaccines10081308
Chicago/Turabian StyleGarcía-Ríos, Estéfani, María Josefa Rodríguez, María Carmen Terrón, Daniel Luque, and Pilar Pérez-Romero. 2022. "Identification and Characterization of Epithelial Cell-Derived Dense Bodies Produced upon Cytomegalovirus Infection" Vaccines 10, no. 8: 1308. https://doi.org/10.3390/vaccines10081308





