Self-Assembling Protein Nanoparticles in the Design of Vaccines: 2022 Update
Abstract
:1. Introduction
2. Self-Assembling Protein Nanoparticles as Vaccine Immunogens
| Pathogen | Platform | Surface Proteins | Immunogens | Adjuvant | Disease | Model | Year | Ref. |
|---|---|---|---|---|---|---|---|---|
| Human papillomavirus | HPV-16/18 L1 | L1 | L1 y L2 | AS04 | Papillomatosis, oropharyngeal and anogenital cancer | Mice and rabbit | 2016 | [16] |
| HPV-16L1/58L2 | L1 | L1 y L2 | Alumn-MPL | Papillomatosis, anogenital and oropharyngeal cancer | Mice and rabbit | 2017 | [17] | |
| HPV-16L1 | L1 | L1 | Papillomatosis, anogenital and oropharyngeal cancer | 2016 | [18] | |||
| HPV-16L1 | L1 | L1 | Papillomatosis, anogenital and oropharyngeal cancer | Kunming mice | 2016 | [19] | ||
| Norovirus/enterovirus/rotavirus | NoV- EV-RV | VP0, VP1, VP3/ VP1/ VP6 | VP0, VP1, VP3/ VP1/ VP6 | rVP6 | Childhood gastroenteritis | Mice | 2019 | [20] |
| Norovirus | NoV-VLP | VP1 | VP1 | Severe acute gastroenteritis and diarrhoea | Mice and rats | 2020 | [21] | |
| NoV-VLP | VP1 | VP1 | Addavax | Severe acute gastroenteritis and diarrhoea | Mice and rats | 2021 | [22] | |
| P-24 nanoparticle S60-nanoparticle | VP1 | VP1 | Severe acute gastroenteritis and diarrhoea | Mice and rats | 2019 | [23] | ||
| Enterovirus 71/ Coxsackie A16/ varicella-zoster | HBc-V/1/2 | HBsAg | VP1/VP2/ VZV-gE | Alumn adjuvant | HFMD/chicken pox | Mice and rats | 2017 | [24] |
| Enterovirus 71 | EV-71 VLP’s | VP0, VP1 and VP3 | HFMD | Mice and rats | 2018 | [25] | ||
| EV-71 VLP’s | VP0, VP1 and VP3 | Alhydrogel | HFMD | Mice and rats | 2020 | [26] | ||
| EV-71 VLP’s | VP0, VP1 and VP3 | Alumn adjuvant | HFMD | Mice and rats | 2021 | [27] | ||
| MERS | MERS-CoV VLP’s | M, E and S | RBD | Alumn adjuvant | MERS | Macaques | 2017 | [28] |
| SARS-CoV2 | HCoV-NL63 | M, E and S | RBD | COVID | Cell culture | [29] | ||
| SARS-CoV-2 VLP’s | S, M, E and N | RBD | COVID | Mice | 2020 | [30] | ||
| SARS-CoV-2 VLP’s | S, M, E and N | RBD | ODN K3-CpG + Al | COVID | Mice | 2021 | [31] | |
| CuMVTT-RBM | CuMVTT | RBD | COVID | Mice and rabbits | 2021 | [32] | ||
| i301 | S | RBD | AddaVax | COVID | Mice and macaques | 2022 | [33,34] | |
| Epstein-barr | gH/gL-EBNA1/gB-LMP2 | gH/gL/gB | EBNA1/LMP2 | Mononucleosis/multiple cancers | Mice | 2016 | [35] | |
| Plasmodium | Rv21 | CSP/TRAP | PvCSP/PvTRAP | Malaria | Mice | 2018 | [36] | |
| Plasmodium falciparum | M-HBsAgS-N4 | HBsAgS | CSP | Malaria | Mice | 2019 | [37] | |
| Lutzomyia longipalpis (Leishmania) | Influenza virusomas | LJL143/Leish3/KMP11 | KMP11 and LeishF3+ | GLA-SE and TLR4 agonist | Human visceral leishmaniasis | Mice | 2017 | [38] |
| Staphylococcus aureus | LND-VLP | AP-205 | LND | Skin soft tissue infection/pneumonia | BALB/c mice | 2020 | [39] |
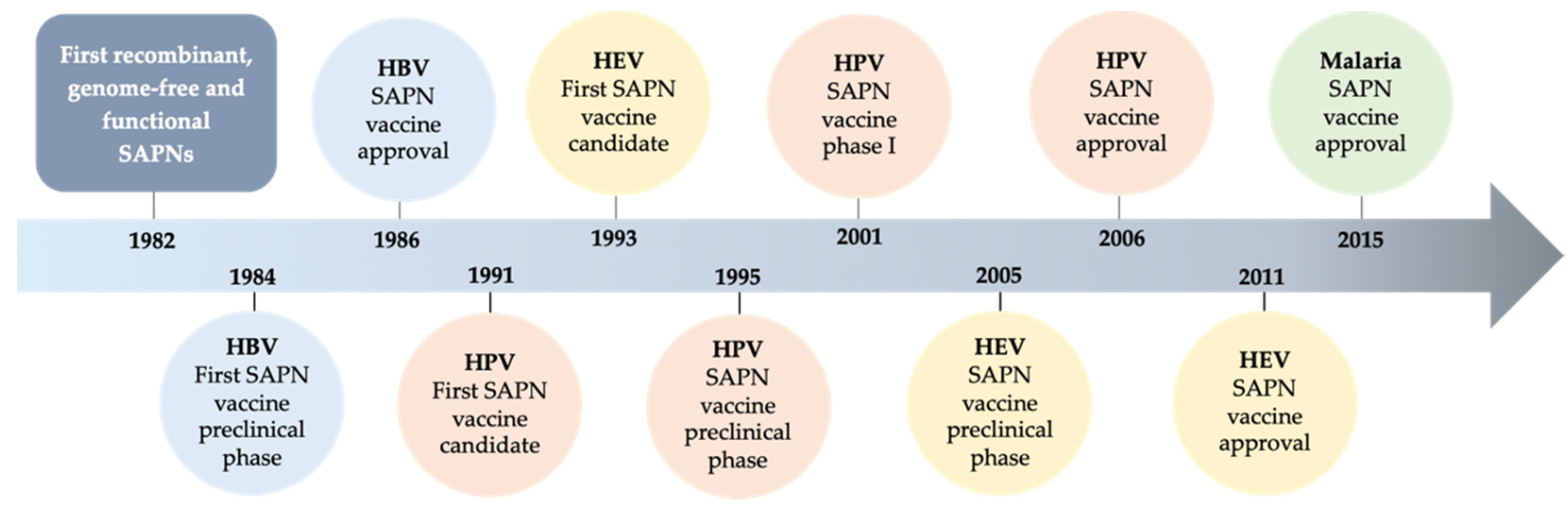
2.1. Evolution of SAPNs and Vaccines
2.1.1. Human Papillomavirus (HPV)
2.1.2. Norovirus (NoV)
2.1.3. Hand Foot Mouth Disease (HFMD) and Varicella
2.1.4. Severe Acute Respiratory Syndrome-Related Coronavirus (SARS-CoV)
2.1.5. Epstein Barr Virus (EBV)
2.1.6. Malaria
2.1.7. Leishmania
2.1.8. Bacteria
3. Messenger RNA (mRNA), Non-Replicative Adenovirus Vaccines and SAPNs
4. Future Perspectives
5. Summary
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Plotkin, S. History of Vaccination. Proc. Natl. Acad. Sci. USA 2014, 111, 12283–12287. [Google Scholar] [CrossRef] [PubMed]
- Amanna, I.J.; Slifka, M.K. Wanted, Dead or Alive: New Viral Vaccines. Antivir. Res. 2009, 84, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Ulmer, J.B.; Valley, U.; Rappuoli, R. Vaccine Manufacturing: Challenges and Solutions. Nat. Biotechnol. 2006, 24, 1377–1383. [Google Scholar] [CrossRef]
- Fuenmayor, J.; Gòdia, F.; Cervera, L. Production of Virus-like Particles for Vaccines. Nat. Biotechnol. 2017, 39, 174–180. [Google Scholar] [CrossRef]
- Schiller, J.T.; Lowy, D.R. Raising Expectations For Subunit Vaccine. J. Infect. Dis. 2015, 211, 1373–1375. [Google Scholar] [CrossRef]
- Silveira, M.M.; Moreira, G.M.S.G.; Mendonça, M. DNA Vaccines against COVID-19: Perspectives and Challenges. Life Sci. 2021, 267, 118919. [Google Scholar] [CrossRef]
- Zinkernagel, R.M. On the Role of Dendritic Cells versus Other Cells in Inducing Protective CD8+ T Cell Responses. Front. Immunol. 2014, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Kushnir, N.; Streatfield, S.J.; Yusibov, V. Virus-like Particles as a Highly Efficient Vaccine Platform: Diversity of Targets and Production Systems and Advances in Clinical Development. Vaccine 2012, 31, 58–83. [Google Scholar] [CrossRef]
- Valenzuela, P.; Medina, A.; Rutter, W.J.; Ammerer, G.; Hall, B.D. Synthesis and assembly of hepatitis B virus surface antigen particles in yeast. Nature 1982, 298, 347–350. [Google Scholar] [CrossRef]
- López-sagaseta, J.; Malito, E.; Rappuoli, R.; Bottomley, M.J. Self-Assembling Protein Nanoparticles in the Design of Vaccines. CSBJ 2016, 14, 58–68. [Google Scholar] [CrossRef] [Green Version]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like Particles: Preparation, Immunogenicity and Their Roles as Nanovaccines and Drug Nanocarriers. J. Nanobiotechnol. 2021, 19, 59. [Google Scholar] [CrossRef] [PubMed]
- Chroboczek, J.; Szurgot, I.; Szolajska, E. Virus-like Particles as Vaccine. Acta Biochim. Pol. 2014, 61, 531–539. [Google Scholar] [CrossRef]
- Abdoli, A.; Soleimanjahi, H.; Fotouhi, F.; Teimoori, A.; Pour Beiranvand, S.; Kianmehr, Z. Human Papillomavirus Type16-L1 VLP Production in Insect Cells. Iran. J. Basic Med. Sci. 2013, 16, 891. [Google Scholar]
- Lua, L.H.L.; Connors, N.K.; Sainsbury, F.; Chuan, Y.P.; Wibowo, N.; Middelberg, A.P.J. Bioengineering Virus-like Particles as Vaccines. Biotechnol. Bioeng. 2014, 111, 425–440. [Google Scholar] [CrossRef]
- Donaldson, B.; Lateef, Z.; Walker, G.F.; Young, S.L.; Ward, V.K. Virus-like Particle Vaccines: Immunology and Formulation for Clinical Translation. Expert Rev. Vaccines 2018, 17, 833–849. [Google Scholar] [CrossRef]
- Boxus, M.; Fochesato, M.; Miseur, A.; Mertens, E.; Dendouga, N.; Brendle, S.; Balogh, K.K.; Christensen, N.D.; Giannini, S.L. Broad Cross-Protection Is Induced in Preclinical Models by a Human Papillomavirus Vaccine Composed of L1/L2 Chimeric Virus-Like Particles. J. Virol. 2016, 90, 6314–6325. [Google Scholar] [CrossRef]
- Chen, X.; Liu, H.; Wang, Z.; Wang, S.; Zhang, T.; Hu, M.; Qiao, L.; Xu, X. Human Papillomavirus 16L1-58L2 Chimeric Virus-like Particles Elicit Durable Neutralizing Antibody Responses against a Broad-Spectrum of Human Papillomavirus Types. Oncotarget 2017, 8, 63333–63344. [Google Scholar] [CrossRef]
- Bang, H.B.; Lee, Y.H.; Lee, Y.J.; Jeong, K.J. High-Level Production of Human Papillomavirus (HPV) Type 16 L1 in Escherichia Coli. J. Microbiol. Biotechnol. 2015, 26, 356–363. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Zhang, G.; Wang, A.; Dong, Z.; Qi, Y.; Wang, J.; Zhao, B.; Li, N.; Jiang, M. Human Papillomavirus L1 Protein Expressed in Escherichia coli Self-Assembles into Virus-like Particles That Are Highly Immunogenic. Virus Res. 2016, 220, 97–103. [Google Scholar] [CrossRef]
- Heinimäki, S.; Hankaniemi, M.M.; Sioofy-Khojine, A.B.; Laitinen, O.H.; Hyöty, H.; Hytönen, V.P.; Vesikari, T.; Blazevic, V. Combination of Three Virus-Derived Nanoparticles as a Vaccine against Enteric Pathogens; Enterovirus, Norovirus and Rotavirus. Vaccine 2019, 37, 7509–7518. [Google Scholar] [CrossRef]
- Verardi, R.; Lindesmith, L.C.; Tsybovsky, Y.; Gorman, J.; Chuang, G.Y.; Edwards, C.E.; Brewer-Jensen, P.D.; Mallory, M.L.; Ou, L.; Schön, A.; et al. Disulfide Stabilization of Human Norovirus GI.1 Virus-like Particles Focuses Immune Response toward Blockade Epitopes. NPJ Vaccines 2020, 5, 110. [Google Scholar] [CrossRef]
- Panasiuk, M.; Zimmer, K.; Czarnota, A.; Grzyb, K.; Narajczyk, M.; Peszyńska-Sularz, G.; Żołędowska, S.; Nidzworski, D.; Hovhannisyan, L.; Gromadzka, B. Immunization with Leishmania Tarentolae-Derived Norovirus Virus-like Particles Elicits High Humoral Response and Stimulates the Production of Neutralizing Antibodies. Microb. Cell Factories 2021, 20, 186. [Google Scholar] [CrossRef]
- Tan, M.; Jiang, X. Norovirus Capsid Protein-Derived Nanoparticles and Polymers as Versatile Platforms for Antigen Presentation and Vaccine Development. Pharmaceutics 2019, 11, 472. [Google Scholar] [CrossRef]
- Wu, Y.; Zhu, R.; Xu, L.; Li, Y.; Li, S.; Yu, H.; Li, S.; Zhu, H.; Cheng, T.; Xia, N. A Novel Combined Vaccine Based on Monochimeric VLP Co-Displaying Multiple Conserved Epitopes against Enterovirus 71 and Varicella-Zoster Virus. Vaccine 2017, 35, 2728–2735. [Google Scholar] [CrossRef]
- Kim, H.J.; Son, H.S.; Lee, S.W.; Yoon, Y.; Hyeon, J.Y.; Chung, G.T.; Lee, J.W.; Yoo, J.S. Efficient Expression of Enterovirus 71 Based on Virus-like Particles Vaccine. PLoS ONE 2019, 14, e0210477. [Google Scholar] [CrossRef]
- Yang, Z.; Gao, F.; Wang, X.; Shi, L.; Zhou, Z.; Jiang, Y.; Ma, X.; Zhang, C.; Zhou, C.; Zeng, X.; et al. Development and Characterization of an Enterovirus 71 (EV71) Virus-like Particles (VLPs) Vaccine Produced in Pichia Pastoris. Hum. Vaccin. Immunother. 2020, 16, 1602–1610. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, C.; Gao, F.; Zhu, Q.; Jiang, Y.; Ma, X.; Hu, Y.; Shi, L.; Wang, X.; Zhang, C.; et al. Preclinical Evaluation of Recombinant HFMD Vaccine Based on Enterovirus 71 (EV71) Virus-like Particles (VLP): Immunogenicity, Efficacy and Toxicology. Vaccine 2021, 39, 4296–4305. [Google Scholar] [CrossRef]
- Wang, C.; Zheng, X.; Gai, W.; Zhao, Y.; Wang, H.; Wang, H.; Feng, N.; Chi, H.; Qiu, B.; Li, N.; et al. MERS-CoV Virus-like Particles Produced in Insect Cells Induce Specific Humoural and Cellular Imminity in Rhesus Macaques. Oncotarget 2017, 8, 12686–12694. [Google Scholar] [CrossRef]
- Naskalska, A.; Dabrowska, A.; Nowak, P.; Szczepanski, A.; Jasik, K.; Milewska, A.; Ochman, M.; Zeglen, S.; Rajfur, Z.; Pyrc, K. Novel Coronavirus-like Particles Targeting Cells Lining the Respiratory Tract. PLoS ONE 2018, 13, e0203489. [Google Scholar] [CrossRef]
- Xu, R.; Shi, M.; Li, J.; Song, P.; Li, N. Construction of SARS-CoV-2 Virus-Like Particles by Mammalian Expression System. Front. Bioeng. Biotechnol. 2020, 8, 862. [Google Scholar] [CrossRef]
- Yilmaz, I.C.; Ipekoglu, E.M.; Bulbul, A.; Turay, N.; Yildirim, M.; Evcili, I.; Yilmaz, N.S.; Guvencli, N.; Aydin, Y.; Gungor, B.; et al. Development and Preclinical Evaluation of Virus-like Particle Vaccine against COVID-19 Infection. Allergy Eur. J. Allergy Clin. Immunol. 2022, 77, 258–270. [Google Scholar] [CrossRef] [PubMed]
- Mohsen, M.O.; Balke, I.; Zinkhan, S.; Zeltina, V.; Liu, X.; Chang, X.; Krenger, P.S.; Plattner, K.; Gharailoo, Z.; Vogt, A.C.S.; et al. A Scalable and Highly Immunogenic Virus-like Particle-Based Vaccine against SARS-CoV-2. Allergy Eur. J. Allergy Clin. Immunol. 2022, 77, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.A.; van Doremalen, N.; Greaney, A.J.; Andersen, H.; Sharma, A.; Starr, T.N.; Keeffe, J.R.; Fan, C.; Schulz, J.E.; Gnanapragasam, P.N.P.; et al. Mosaic RBD Nanoparticles Protect against Challenge by Diverse Sarbecoviruses in Animal Models. Science 2022, 377, eabq0839. [Google Scholar] [CrossRef] [PubMed]
- Bruun, T.U.J.; Andersson, A.-M.C.; Draper, S.J.; Howarth, M. Engineering a Rugged Nanoscaffold To Enhance Plug-and-Display Vaccination. ACS Nano 2018, 12, 8855–8866. [Google Scholar] [CrossRef] [PubMed]
- Perez, E.M.; Foley, J.; Tison, T.; Silva, R.; Ogembo, J.G. Novel Epstein-Barr Virus-like Particles Incorporating GH/GLEBNA1 or GB-LMP2 Induce High Neutralizing Antibody Titers and EBV-Specific T-Cell Responses in Immunized Mice. Oncotarget 2017, 8, 19255–19273. [Google Scholar] [CrossRef] [PubMed]
- Atcheson, E.; Bauza, K.; Salman, A.M.; Alves, E.; Blight, J.; Viveros-Sandoval, M.E.; Janse, C.J.; Khan, S.M.; Hill, A.V.S.; Reyes-Sandoval, A. Tailoring a Plasmodium Vivax Vaccine to Enhance Efficacy through a Combination of a CSP Virus-like Particle and TRAP Viral Vectors. Infect. Immun. 2018, 86, e00114-18. [Google Scholar] [CrossRef]
- Kingston, N.J.; Kurtovic, L.; Walsh, R.; Joe, C.; Lovrecz, G.; Locarnini, S.; Beeson, J.G.; Netter, H.J. Hepatitis B Virus-like Particles Expressing Plasmodium Falciparum Epitopes Induce Complement-Fixing Antibodies against the Circumsporozoite Protein. Vaccine 2019, 37, 1674–1684. [Google Scholar] [CrossRef]
- Cecílio, P.; Pérez-Cabezas, B.; Fernández, L.; Moreno, J.; Carrillo, E.; Requena, J.M.; Fichera, E.; Reed, S.G.; Coler, R.N.; Kamhawi, S.; et al. Pre-Clinical Antigenicity Studies of an Innovative Multivalent Vaccine for Human Visceral Leishmaniasis. PLoS Negl. Trop. Dis. 2017, 11, e0005951. [Google Scholar] [CrossRef]
- Joyner, J.A.; Daly, S.M.; Peabody, J.; Triplett, K.D.; Pokhrel, S.; Elmore, B.O.; Adebanjo, D.; Peabody, D.S.; Chackerian, B.; Hall, P.R. Vaccination with VLPs Presenting a Linear Neutralizing Domain of S. Aureus Hla Elicits Protective Immunity. Toxins 2020, 12, 450. [Google Scholar] [CrossRef]
- Netter, H.J.; Chang, S.F.; Bruns, M. Host-Range and Pathogenicity of Hepatitis B Viruses. Futur. Virol. 2008, 3, 83–94. [Google Scholar] [CrossRef]
- Jegerlehner, A.; Storni, T.; Lipowsky, G.; Schmid, M.; Pumpens, P.; Bachmann, M.F. Regulation of IgG Antibody Responses by Epitope Density and CD21-Mediated Costimulation. Eur. J. Immunol. 2002, 32, 3305–3314. [Google Scholar] [CrossRef]
- Rosenthal, J.A.; Chen, L.; Baker, J.L.; Putnam, D.; DeLisa, M.P. Pathogen-like Particles: Biomimetic Vaccine Carriers Engineered at the Nanoscale. Curr. Opin. Biotechnol. 2014, 28, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Janeway, C.A., Jr.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2020, 20, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Quint, W.; Banks, L.; Bravo, I.G.; Stoler, M.; Broker, T.R.; Stanley, M.A. The Biology and Life-Cycle of Human Papillomaviruses. Vaccine 2012, 30, F55–F70. [Google Scholar] [CrossRef]
- Biologicals, G.; Rixensart, B.; Jenkins, D.; Harper, D.M.; Franco, E.L.; Wheeler, C.; Ferris, D.G.; Jenkins, D.; Schuind, A.; Zahaf, T.; et al. Efficacy of a Bivalent L1 Virus-like Particle Vaccine in Prevention of Infection with Human Papillomavirus Types 16 and 18 in Young Women: A Randomised Controlled Trial. Lancet 2004, 364, 1757–1765. [Google Scholar]
- Einstein, M.H.; Baron, M.; Levin, M.J.; Chatterjee, A.; Edwards, R.P.; Zepp, F.; Carletti, I.; Dessy, F.J.; Trofa, A.F.; Schuind, A.; et al. Comparison of the Immunogenicity and Safety of CervarixTM and Gardasil® Human Papillomavirus (HPV) Cervical Cancer Vaccines in Healthy Women Aged 18-45 Years. Hum. Vaccin. 2009, 5, 705–719. [Google Scholar] [CrossRef]
- Petrosky, E.; Bocchini, J.A., Jr.; Hariri, S.C.H. Use of 9-Valent Human Papillomavirus (HPV) Vaccine: Updated HPV Vaccination Recommendations of the Advisory Committee on Immunization Practices. MMWR Morb. Mortal. Wkly. Rep. 2015, 64, 300–304. [Google Scholar]
- Ahmed, S.M.; Hall, A.J.; Robinson, A.E.; Verhoef, L.; Premkumar, P.; Parashar, U.D.; Koopmans, M.; Lopman, B.A. Global Prevalence of Norovirus in Cases of Gastroenteritis: A Systematic Review and Meta-Analysis. Lancet Infect. Dis. 2014, 14, 725–730. [Google Scholar] [CrossRef]
- Noel, J.S.; Fankhauser, R.L.; Ando, T.; Monroe, S.S.; Glass, R.I. Identification of a Distinct Common Strain of “Norwalk-like Viruses” Having a Global Distribution. J. Infect. Dis. 1998, 179, 1334–1344. [Google Scholar] [CrossRef]
- Fernández, J.M.R.; Gómez, J.B. Infecciones Por Norovirus. Enferm. Infecc. Microbiol. Clín. 2010, 28, 51–55. [Google Scholar] [CrossRef]
- Hardy, M.E. Norovirus Protein Structure and Function. FEMS Microbiol. Lett. 2005, 253, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lucero, Y.; Vidal, R.; O’Ryan, G.M. Norovirus Vaccines under Development. Vaccine 2018, 36, 5435–5441. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Neill, J.D.; Estes, M.K.; Prasad, B.V.V. X-ray Structure of a Native Calicivirus: Structural Insights into Antigenic Diversity and Host Specificity. Proc. Natl. Acad. Sci. USA 2006, 103, 8048–8053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, M.; Fang, P.; Chachiyo, T.; Xia, M.; Huang, P.; Fang, Z.; Jiang, W.; Jiang, X. Noroviral P Particle: Structure, Function and Applications in Virus-Host Interaction. Virology 2008, 382, 115–123. [Google Scholar] [CrossRef]
- Xia, M.; Huang, P.; Jiang, X.; Tan, M. A Nanoparticle-Based Trivalent Vaccine Targeting the Glycan. Viruses 2021, 13, 72. [Google Scholar] [CrossRef]
- Xia, M.; Huang, P.; Sun, C.; Han, L.; Vago, F.S.; Li, K.; Zhong, W.J.; Klassen, J.S.; Tan, X.J. Bioengineered Norovirus S60 Nanoparticles as a Multifunctional Vaccine Platform. ACS Nano. 2018, 12, 10665–10682. [Google Scholar] [CrossRef]
- Xia, M.; Huang, P.; Tan, M. A Pseudovirus Nanoparticle-Based Trivalent Rotavirus Vaccine Candidate Elicits High and Cross P Type Immune Response. Pharmaceutics 2022, 14, 1597. [Google Scholar] [CrossRef]
- Tan, M.; Jiang, X. Vaccine against Norovirus. Hum. Vaccin. Immunother. 2014, 10, 1449–1456. [Google Scholar] [CrossRef]
- Du, J.; Gu, Q.; Liu, Y.; Li, Q.; Guo, T. The Endemic GII. 4 Norovirus-like-Particle Induced-Antibody Lacks of Cross-Reactivity against the Epidemic GII. 17 Strain. J. Med Virol. 2021, 93, 3974–3979. [Google Scholar] [CrossRef] [PubMed]
- Parra, G.I.; Bok, K.; Taylor, R.; Haynes, J.R.; Sosnovtsev, S.V.; Richardson, C.; Green, K.Y. Immunogenicity and Specificity of Norovirus Consensus GII.4 Virus-like Particles in Monovalent and Bivalent Vaccine Formulations. Vaccine 2012, 30, 3580–3586. [Google Scholar] [CrossRef]
- Ramani, S.; Neill, F.H.; Ferreira, J.; Treanor, J.J.; Frey, S.E.; Topham, D.J.; Goodwin, R.R.; Borkowski, A.; Baehner, F.; Mendelman, P.M.; et al. B-Cell Responses to Intramuscular Administration of a Bivalent Virus-like Particle Human Norovirus Vaccine. Clin. Vaccine Immunol. 2017, 24, e00571-16. [Google Scholar] [CrossRef] [PubMed]
- Ryu, W.S.; Kang, B.; Hong, J.; Hwang, S.; Kim, A.; Kim, J.; Cheon, D.S. Enterovirus 71 Infection with Central Nervous System Involvement, South Korea. Emerg. Infect. Dis. 2010, 16, 1764–1766. [Google Scholar] [CrossRef] [PubMed]
- Haaheim, L.R.; Pattison, J.R.; Whitley, R.J. A Practical Guide to Clinical Virology; Wiley: Hoboken, NJ, USA, 2002; ISBN 0471950971. [Google Scholar]
- Liu, W.; Wu, S.; Xiong, Y.; Li, T.; Wen, Z.; Yan, M.; Qin, K.; Liu, Y.; Wu, J. Co-Circulation and Genomic Recombination of Coxsackievirus A16 and Enterovirus 71 during a Large Outbreak of Hand, Foot, and Mouth Disease in Central China. PLoS ONE 2014, 9, e96051. [Google Scholar] [CrossRef] [Green Version]
- Brown, B.A.; Pallansch, M.A. Complete Nucleotide Sequence of Enterovirus 71 Is Distinct from Poliovirus. Virus Res. 1995, 39, 195–205. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W.; et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1953–1966. [Google Scholar] [CrossRef]
- Yang, Y.; Peng, F.; Wang, R.; Guan, K.; Jiang, T.; Xu, G.; Sun, J.; Chang, C. The Deadly Coronaviruses: The 2003 SARS Pandemic and the 2020 Novel Coronavirus Epidemic in China, the Company’ s Public News and Information. J. Autoimmun. 2020, 109, 102487. [Google Scholar] [CrossRef]
- Drosten, C.; Gunther, S.; Preiser, W.; van der Werf, S.; Brodt, H.R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.; et al. Identification of a Novel Coronavirus in Patients with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348, 1967–1976. [Google Scholar] [CrossRef]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Ho, Y.; Lin, P.H.; Liu, C.Y.Y.; Lee, S.P.; Chao, Y.C. Assembly of Human Severe Acute Respiratory Syndrome Coronavirus-like Particles. Biochem. Biophys. Res. Commun. 2004, 318, 833–838. [Google Scholar] [CrossRef]
- Cao, Y.; Xie, L.; Shi, F.; Tang, M.; Li, Y.; Hu, J.; Zhao, L.; Zhao, L.; Yu, X.; Luo, X.; et al. Targeting the Signaling in Epstein–Barr Virus-Associated Diseases: Mechanism, Regulation, and Clinical Study. Signal Transduct. Target Ther. 2021, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Gething, P.W.; Elyazar, I.R.F.; Moyes, C.L.; Smith, D.L.; Battle, K.E.; Guerra, C.A.; Patil, A.P.; Tatem, A.J.; Howes, R.E.; Myers, M.F.; et al. A Long Neglected World Malaria Map: Plasmodium Vivax Endemicity in 2010. PLoS Negl. Trop. Dis. 2012, 6, e1814. [Google Scholar] [CrossRef] [PubMed]
- Markus, M.B. Malaria: Origin of the Term “Hypnozoite”. J. Hist. Biol. 2011, 44, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, N. European Medicines Agency Approves First Malaria Vaccine. BMJ 2015, 351, h4067. [Google Scholar] [CrossRef]
- Salman, A.M.; Montoya-DIáz, E.; West, H.; Lall, A.; Atcheson, E.; Lopez-Camacho, C.; Ramesar, J.; Bauza, K.; Collins, K.A.; Brod, F.; et al. Rational Development of a Protective P. Vivax Vaccine Evaluated with Transgenic Rodent Parasite Challenge Models. Sci. Rep. 2017, 7, srep46482. [Google Scholar] [CrossRef]
- Kaye, P.; Scott, P. Leishmaniasis: Complexity at the Host-Pathogen Interface. Nat. Rev. Microbiol. 2011, 9, 604–615. [Google Scholar] [CrossRef]
- Alvar, J.; Vélez, I.D.; Bern, C.; Herrero, M.; Desjeux, P.; Cano, J.; Jannin, J.; de Boer, M. Leishmaniasis Worldwide and Global Estimates of Its Incidence. PLoS ONE 2012, 7, e35671. [Google Scholar] [CrossRef]
- Abdeladhim, M.; Kamhawi, S.; Valenzuela, J.G. What’s behind a Sand Fly Bite? The Profound Effect of Sand Fly Saliva on Host Hemostasis, Inflammation and Immunity. Infect. Genet. Evol. 2014, 28, 691–703. [Google Scholar] [CrossRef]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus Aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and Virulence of Staphylococcus Aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef]
- Oscherwitz, J.; Cease, K.B. Identification and Validation of a Linear Protective Neutralizing Epitope in the β-Pore Domain of Alpha Toxin. PLoS ONE 2015, 10, e0116882. [Google Scholar] [CrossRef]
- Fiolet, T.; Kherabi, Y.; Macdonald, C.; Ghosn, J.; Peiffer-smadja, N. Comparing COVID-19 Vaccines for Their Characteristics, Efficacy and Effectiveness against SARS-CoV-2 and Variants of Concern: A Narrative Review. Clin. Microbiol. Infect. 2020, 28, 202–221. [Google Scholar] [CrossRef] [PubMed]
- Pollard, A.J.; Bijker, E.M. A Guide to Vaccinology: From Basic Principles to New Developments. Nat. Rev. Immunol. 2021, 21, 83–100. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.A.; Bauer, M.; Manolova, V.; Muntwiler, S.; Saudan, P.; Bachmann, M.F. Cutting Edge: Limited Specialization of Dendritic Cell Subsets for MHC Class II-Associated Presentation of Viral Particles. J. Immunol. 2010, 184, 26–29. [Google Scholar] [CrossRef]
- Leen, A.M.; Christin, A.; Khalil, M.; Weiss, H.; Gee, A.P.; Brenner, M.K.; Heslop, H.E.; Rooney, C.M.; Bollard, C.M. Identification of Hexon-Specific CD4 and CD8 T-Cell Epitopes for Vaccine and Immunotherapy. J. Virol. 2008, 82, 546–554. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. MRNA Vaccines-a New Era in Vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef]
- Logunov, D.Y.; Dolzhikova, I.V.; Shcheblyakov, D.V.; Tukhvatulin, A.I.; Zubkova, O.V.; Dzharullaeva, A.S.; Kovyrshina, A.V.; Lubenets, N.L.; Grousova, D.M.; Erokhova, A.S.; et al. Safety and Efficacy of an RAd26 and RAd5 Vector-Based Heterologous Prime-Boost COVID-19 Vaccine: An Interim Analysis of a Randomised Controlled Phase 3 Trial in Russia. Lancet 2021, 397, 671–681. [Google Scholar] [CrossRef]
- Wilson, J.M.; Engelhardt, J. Adenovirus Vectors for Gene Therapy. Biotechnol. Adv. 1997, 15, 769. [Google Scholar] [CrossRef]
- Kurokawa, N.; Robinson, M.K.; Bernard, C.; Kawaguchi, Y.; Koujin, Y.; Koen, A.; Madhi, S.; Polasek, T.M.; McNeal, M.; Dargis, M.; et al. Safety and Immunogenicity of a Plant-Derived Rotavirus-like Particle Vaccine in Adults, Toddlers and Infants. Vaccine 2021, 39, 5513–5523. [Google Scholar] [CrossRef]
- Ward, B.J.; Séguin, A.; Couillard, J.; Trépanier, S.; Landry, N. Phase III: Randomized Observer-Blind Trial to Evaluate Lot-to-Lot Consistency of a New Plant-Derived Quadrivalent Virus like Particle Influenza Vaccine in Adults 18–49 Years of Age. Vaccine 2021, 39, 1528–1533. [Google Scholar] [CrossRef]
- Pillet, S.; Arunachalam, P.S.; Andreani, G.; Golden, N.; Fontenot, J.; Aye, P.P.; Röltgen, K.; Lehmicke, G.; Gobeil, P.; Dubé, C.; et al. Safety, Immunogenicity, and Protection Provided by Unadjuvanted and Adjuvanted Formulations of a Recombinant Plant-Derived Virus-like Particle Vaccine Candidate for COVID-19 in Nonhuman Primates. Cell Mol. Immunol. 2022, 19, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Lai, H.; Sun, H.; Chen, Q. Virus-like Particles That Display Zika Virus Envelope Protein Domain III Induce Potent Neutralizing Immune Responses in Mice. Sci. Rep. 2017, 7, 7679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
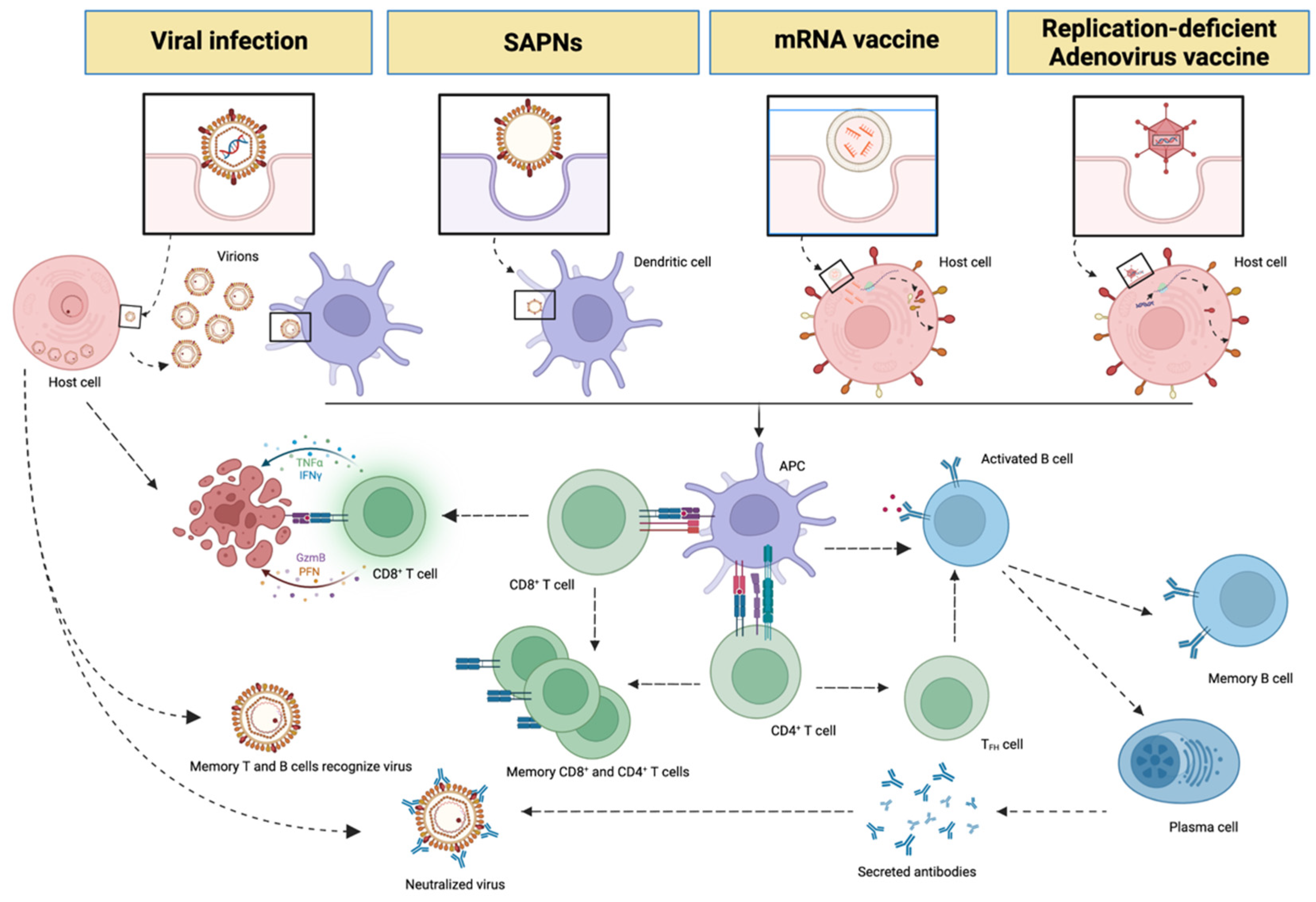


| SAPNs Vaccines | mRNA Vaccines | Non-Replicating Adenovirus Vaccines | |
|---|---|---|---|
| Advantages |
|
|
|
| Disadvantages |
|
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales-Hernández, S.; Ugidos-Damboriena, N.; López-Sagaseta, J. Self-Assembling Protein Nanoparticles in the Design of Vaccines: 2022 Update. Vaccines 2022, 10, 1447. https://doi.org/10.3390/vaccines10091447
Morales-Hernández S, Ugidos-Damboriena N, López-Sagaseta J. Self-Assembling Protein Nanoparticles in the Design of Vaccines: 2022 Update. Vaccines. 2022; 10(9):1447. https://doi.org/10.3390/vaccines10091447
Chicago/Turabian StyleMorales-Hernández, Sergio, Nerea Ugidos-Damboriena, and Jacinto López-Sagaseta. 2022. "Self-Assembling Protein Nanoparticles in the Design of Vaccines: 2022 Update" Vaccines 10, no. 9: 1447. https://doi.org/10.3390/vaccines10091447
APA StyleMorales-Hernández, S., Ugidos-Damboriena, N., & López-Sagaseta, J. (2022). Self-Assembling Protein Nanoparticles in the Design of Vaccines: 2022 Update. Vaccines, 10(9), 1447. https://doi.org/10.3390/vaccines10091447







