Abstract
The etiological agent of some anogenital tract cancers is infection with the high-risk human papillomavirus (HPV). Currently, prophylactic vaccines against HPV have been validated, but the presence of drug treatment directed against the infection and its oncogenic effects remain essential. Among the best drug targets, viral oncoprotein E6 has been identified as a key factor in cell immortalization and tumor progression in HPV-positive cells. E6, through interaction with the cellular ubiquitin ligase E6AP, can promote the degradation of p53, a tumor suppressor protein. Therefore, suppression of the creation of the E6-E6AP complex is one of the essential strategies to inhibit the survival and proliferation of infected cells. In the present study, we proposed an in-silico approach for the discovery of small molecules with inhibitory activity on the E6-E6AP interaction. The first three compounds (F0679-0355, F33774-0275, and F3345-0326) were selected on the basis of virtual screening and prediction of the molecules’ ADMET properties and docking with E6 protein, these molecules were selected for further study by investigating their stability in the E6 complex and their inhibitory effect on the E6-E6AP interaction by molecular dynamics (MD) simulation. The identified molecules thus represent a good starting point for the development of anti-HPV drugs.
1. Introduction
Cervical cancer is the second most common cancer in women. More than “500,000” new cases are registered annually with an overall rate of 18.8 per 100,000 women. In 2020, it was an estimated 604,000 new cases and 342,000 deaths worldwide [1]. Currently, it is widely accepted that Human papillomavirus is the etiological agent of cervical cancer [2] and that high-risk (HR) HPVs are responsible for the occurrence and development of approximately 5% of all cancers and is associated with 30% of all pathogen-related cancers [3]. Human papillomaviruses (HPV) belong to the Papillomaviridae family. There are more than 200 distinct HPV genotypes known for their ability to infect mucous membranes and human skin epithelial cells HPVs are the etiological agent of the majority of the most common sexually transmitted viral infections in men and women and recent studies have shown that HPV can also affect fertility, pregnancy rates, and other health outcomes [4,5,6]. The viral genome consists of seven genes classified as early genes (E1, E2, E3, E4, E5, E6, and E7), which regulate viral transcription and genome replication, and two Late genes (L1 and L2) encode the structural proteins involved in capsid formation [7]. The HR-HPV16 is the causative agent of most cervical cancer cases. Its high oncogenic potential is mainly due to the oncoproteins E6 and E7 [8,9]. Scientific evidence has shown that p53 is the main target of the E6 oncoprotein. However, several studies have reported that E6 binds to a diverse range of cellular proteins playing a key role in the cell cycle control and regulation [10,11]. The E6 contains two zinc-binding domains with a conserved fold that are connected to each other by a helical linker The E6 amino-terminal zinc-binding domain and the carboxy-terminal zinc-binding domain have a globally conserved fold in the crystal [12]. The two zinc domains, together with an alpha helix tube connecting them, form a deep pocket in which the LXXLL peptide makes close contact [13]. During HPV-induced carcinogenesis, HR E6 oncoproteins induce p53 degradation. During E6-mediated p53 degradation, HR-HPV E6 proteins interact with the LxxLL motif of E6AP, LxxLL a 20-amino acid peptide in E6AP, resulting in the recruitment and polyubiquitination of p53. The LxxLL peptide isolated from E6AP is sufficient to render E6 susceptible to interaction with p53. [14]. the central pocket of E6 binds directly to the LxxLL motif of the ubiquitin ligase E6AP [15,16], which induce a conformational change in HR E6 proteins allowing the formation of a complex with p53. In this ternary complex, called E6/E6AP/p53, p53 is polyubiquitinated by E6AP and then degraded by a proteasome, thereby directly inhibiting apoptosis [17]. Of particular interest, HR E6 oncoproteins were reported to be involved in regulation of p53 gene transactivation and are able to abolish p53 transcriptional transactivation activity [18]. HR E6 oncoproteins can also interact with p300/CBP co-activators to control p53-dependent gene regulation [19].
Currently, three prophylactic vaccines have been approved and used effectively to prevent persistent viral infections and HPV-associated cervical lesions: Cervarix®2, which is effective for HPV genotypes 16 and 18 [20]; Gardasil®4, effective for HPV [21] genotypes 6, 11, and 16; and Gardasil®9 is effective for HPV genotypes 6, 11, 16, 18, 31, 33, 45, 52, and 58 [22]. Nevertheless, the vaccination program is not well established and, except in some developed countries, the vaccination is not provided in most countries. Thus, management of cervical cancer and precancerous lesions is still limited to the use of chemotherapeutic agents and/or the implementation of surgical and ablative techniques to remove the developed tumors. Both of these treatments are invasive, non-specific, and tend to be expensive, making their accessibility limited to millions of patients, especially in developing countries [23]. There is, therefore, an urgent need to develop accessible drug-based therapies targeting the onco-virus for specific treatment of HPV-associated diseases and better management of cervical cancer and precancerous lesions.
Thus, the present study was planned to identify small molecules from 6346 chemicals available in the Life Chemicals database (http://www.lifechemicals.com Format SDF accessed on 11 February 2020) with potential inhibitory activity against HPV E6. In this study, an integrated bioinformatics approach was used to identify lead compounds that could serve as inhibitors for the HPV E6 oncoproteins.
We identified three drug-like compounds, resulting in the discovery of several chemical entities that offer novel scaffolds that could be used as the core of new families of E6 HPV16 inhibitors. All three compounds were used in the 50 ns MDS study. Based on various parameters such as RMSD, and RMSF, we report (F0679-0355, F33774-0275, and F3345-0326) that they have the same backbone as lead compounds, which could serve as E6 HPV16 inhibitors. However, further in vitro and/or in vivo research is needed to validate the in-silico results
2. Material and Methods
2.1. Collection and Curation of the Chemical Library
Chemical structures of 6340 compounds with potential anti-tumor activity, targeting various types of cancer, such as prostate, breast cancers, leukemia, lymphoma, carcinoma, etc., were downloaded as 3D sdf files from the Life Chemicals Anti-Cancer Screening Library (https://lifechemicals.com Format SDF accessed on 11 February 2022) and prepared with Open babel [24].Lipinski’s rule, with 300 < Mw < 700 g/mol and 5 < Number of rotatable bonds < 12), was applied to filter out non-drug-like molecules [25].
2.2. Structure-Based Virtual Screening
Drug-like molecules were considered as a database to identify new compounds with high binding affinity and chemical complementarity to the E6/E6AP/p53 tetramer active site, using a virtual screen. The 3D structures of E6/E6AP/p53 (PDB code: 4xR8) were downloaded from the Protein Data Bank [26]. In order to prepare the selected protein for molecular docking, water and co-crystallized small molecules were removed. The protein contains eight chains; seven chains were removed, and the H-chain was particularly retained to make the calculations shorter and simpler, the non-polar hydrogens were added using discovery studio [27]. A grid box with a size of (x = 40; y = 80; z = 60) and a center of (x = 16.0; y = −32.0; z = −19.0) was defined to cover the ligand-binding site at 4xr8.
2.3. Molecular Docking
Selected molecules from the virtual screening were molecularly docked to the PDB target (4xr8). The Autodockvina [28], and MGL Tools [29] programs were run with their default settings.
2.4. Post Docking Analysis
Structures built on molecular docking results (E6/E6AP/p53) were examined separately. The molecules were prioritized by taking into account the binding energy of their highest-scoring conformation and then the top 10 candidates were visually inspected for their interactions with the active site. Their interactions with the active site residues were visually examined. Finally, the selected molecules were analyzed in more detail (ADMET prediction and molecular dynamics). Visual inspection of docking poses and analysis of protein–ligand interactions were performed in Biovia Discovery Studio Visualizer version 2016 (Yashoda Technical Campus, Satara, India) [27]. Visualization images were rendered by PyMOL 2.3 (Warren Lyford DeLano, 1 August 2006) [30].
2.5. In silico Pharmacokinetics (PK)/Pharmacodynamics (PD) Evaluation
Drug development requires many phases, starting with target identification and ending with ADMET prediction. Early detection of these characteristics is crucial to reduce the cost and time of the drug development process. To define the passage of this drug in the body, the measurement of the ADMET parameters of pharmacokinetics (Adsorption, Distribution, Metabolism, Excretion, and Toxicity) was performed. To this end, selected molecules based on their energy score were exploited to determine these in silico pharmacokinetic parameters using http://admetsar.com/ (Format SMILES accessed on 9 March 2020) to prevent the failure of these compounds in clinical trials and to increase their potential to reach the stage of drug candidates in the future.
2.6. Molecular Dynamics
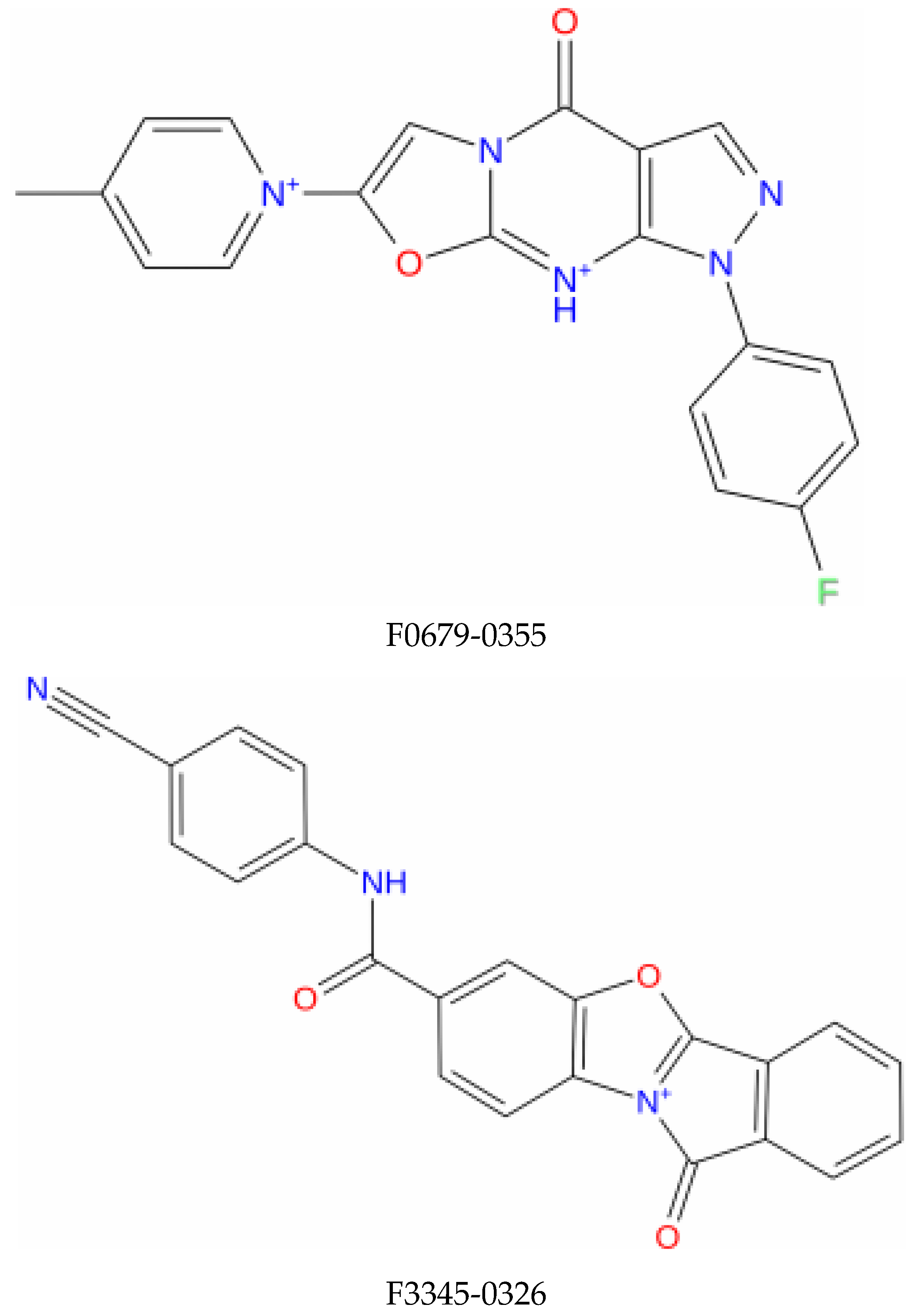
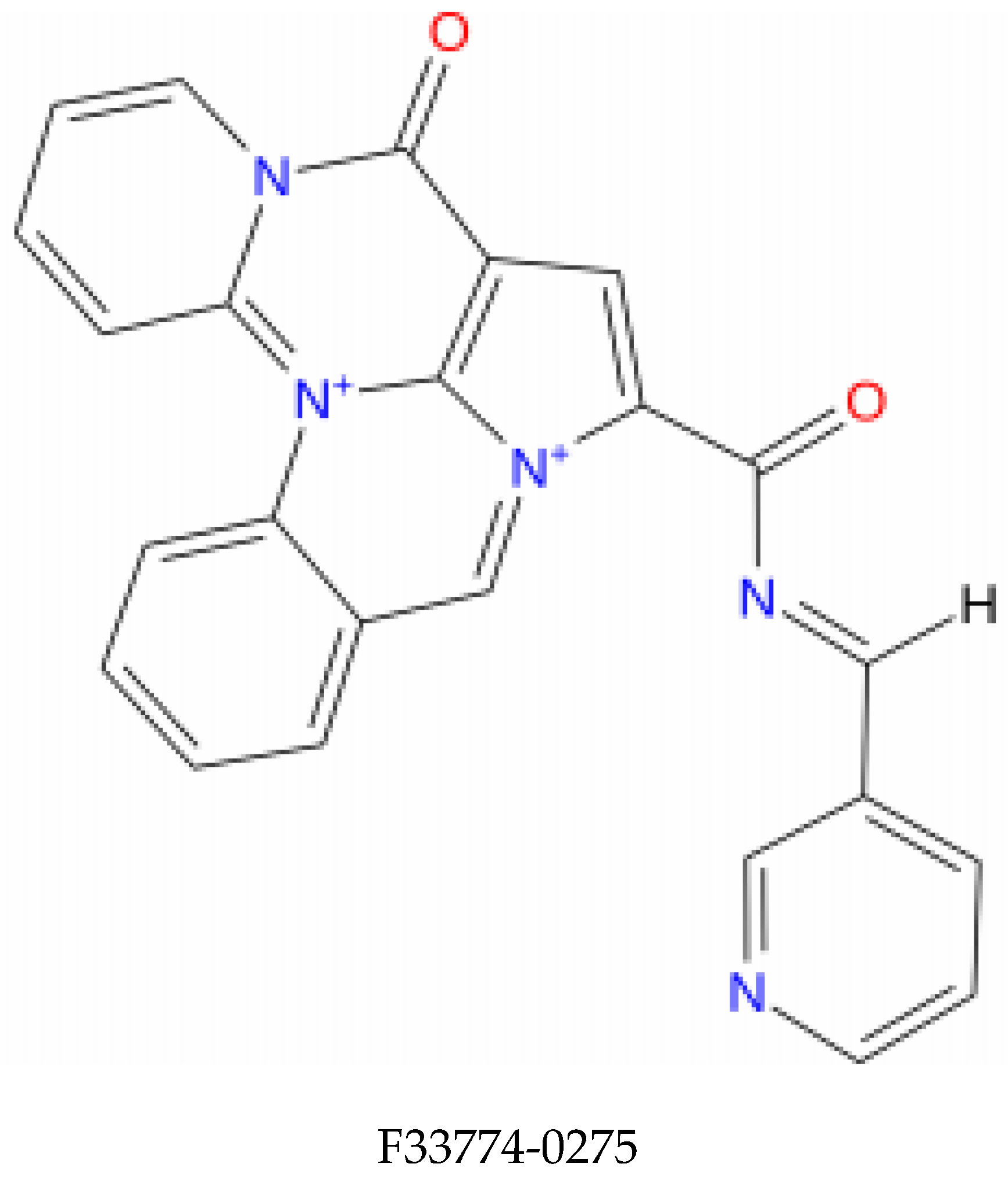
Molecular dynamics simulations are more in-depth studies that explore the dynamism and conformational changes of bimolecular complexes. Molecular dynamics simulations tend to calculate the motions of atoms as a function of time by integrating the classical Newtonian equation of motion. The binding state of the ligand in the physiological environment has been anticipated by using simulations. In our study, compounds with higher affinity for the E6 active site and favorable pharmacokinetic characteristics F0679-0355, F33774-0275, and F3345-0326 (Figure 1) were selected and investigated by performing 50 nanosecond molecular dynamics (MD) simulations by using Desmond, a Schrödinger LLC software (New York, NY, USA) [31]. Protein–ligand complexes were preprocessed using Maestro’s Protein Preparation Wizard, which includes optimization and minimization of complexes. The System Builder tool was used to prepare all systems. TIP3P (Transferable Intermolecular Interaction Potential 3 Points) was chosen as the solvent model with an orthorhombic box. The OPLS_2005 force field was used in the simulation [32]. Counter ions were added to the models to make them neutral. Salt (NaCl) at 0.15 M was added to mimic physiological conditions. For the entire simulation, the NPT ensemble with a temperature of 300 K and a pressure of 1 atm was used. The models were relaxed before the simulation. Every 100 ps, the trajectories were saved for analysis, and the stability of the simulation was determined by measuring the root mean square deviation (RMSD) of the protein and ligand over time. The trajectories of the Desmond simulation were analyzed. The root mean square fluctuation (RMSF) and protein–ligand contacts were also calculated from the MD trajectory analysis.
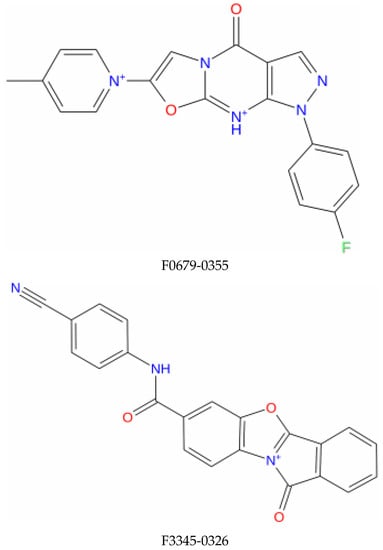
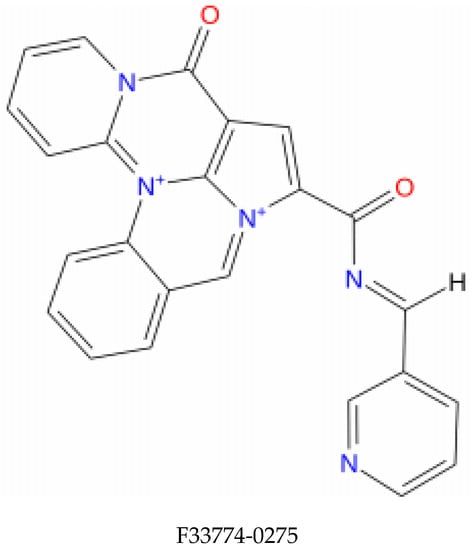
Figure 1.
Structure of the top identified lead compounds.
3. Results and Discussion
3.1. Virtual Screening
In the present study, the main objective is the identification of potential new drugs as E6 inhibitors. A total of 6340 molecules that already have anticancer activity are processed by setting up drug-like filters to evaluate the similarity of these molecules with drugs, a high-throughput screening was reread through PyRx 1.0 software [33]. The chemical database was subjected to a simple filter to remove non-drug molecules to generate targeted databases for HPV16 E6. Accordingly, using Lipinski based filter, we have removed 962 non-suitable molecules chemicals allowing to narrow the database to 5378 drug-like compounds.
The prepared chemical database was subjected to high-throughput screening to identify new candidate molecules potentially suitable for E6/E6AP/P53 inhibition, using AutoDoc vina involved in the PyRx tool which generated 9 distinct conformations for each ligand, ranked by binding affinity (kcal/mol).
3.2. Molecular Docking
Autodoc vina and MGL tools were used to dock the 28 candidate molecules for HPV16 E6 treatment. This resulted in a final list of three compounds; F0679-0355, F3345-0326, and F3374-0275 showed the highest affinities: −10.4 Kcal/mol, −10 Kcal/mol, and −9.9 Kcal/mol, respectively. Total energy, hydrogen bonds (HBond) and other interactions of the three selected compounds are reported in Table 1. From the molecular docking results of the three candidate compounds, we found that the majority of these compounds share interactions with the same amino acids responsible for the formation of the E6/E6AP/p53 complex, and are involved in Van Der Waals interaction and pi-alkyl and pi-sigma interactions with residues Val31, Tyr32, Leu50, Cys51, Val53, Val62, Leu67, Tyr70, Ile73, Leu96, Cys97, Asp98, Leu99 and Leu100 of E6 contribute to the LxxLL binding pocket [34]. Whereas, Gln6, Glu7, Arg8, Arg10, Gln14, Glu18, Arg40, Glu41, Val42, Tyr43, Asp44, Phe45, Ala46, Phe47, Asp49, Leu50, Leu100, Cys106, Gln107, Lys108, Pro109, Leu110, Cys111, Pro112, Glu113, Gln114, and Lys115 of E6 [35] contribute to the p53 binding pocket which is consistent with recent studies (Table 1).

Table 1.
Molecular Docking result analysis of the top three compounds.
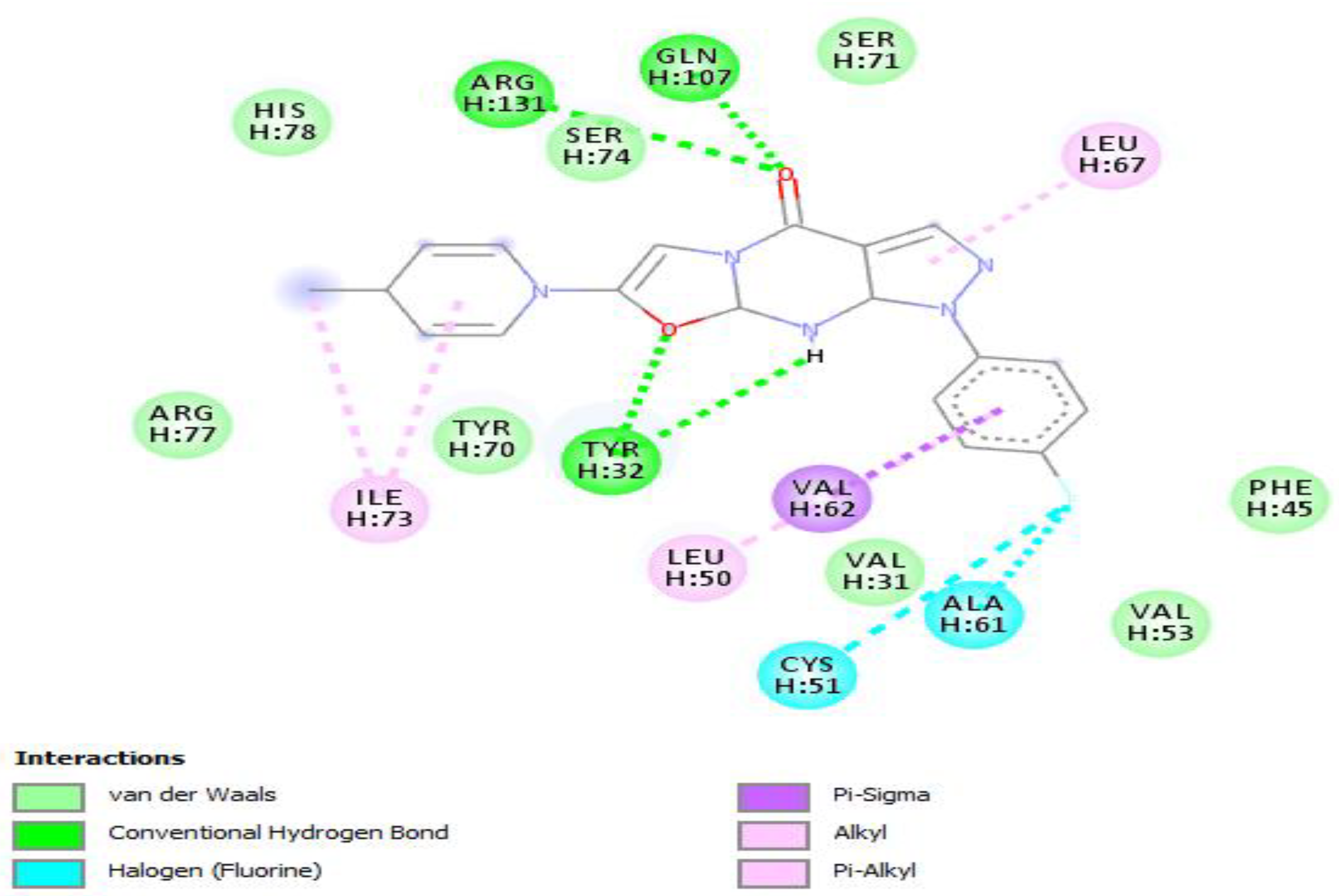
Molecular docking of the selected molecule F0679-0355 on HR E6 oncoprotein is reported in Figure 2 and showed that this ligand forms three strong hydrogen bonds with the amino acids Arg(131), Gln(107), and Tyr(32) of the HR E6 protein via 8 van der Waals bonds with the amino acids His(78), Ser(74), Ser(71), Arg(77), Tyr(70), Val(53), Val(31), and Phe(45). The docking study showed also the formation of 2 Halogen bonds between the ligand and the amino acids Cys(51) and Ala(61), and interacting via a π-Alkyl with the amino acids Ile(73) and Leu(50).
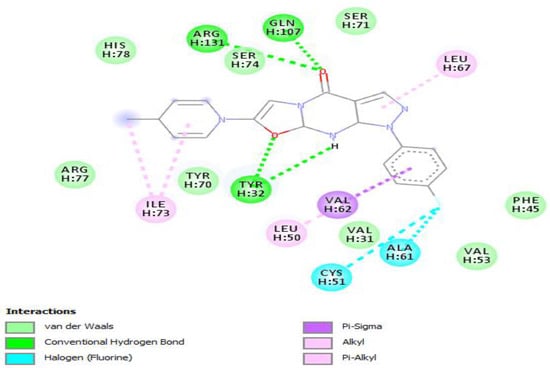
Figure 2.
Positioning and interactions of the molecule F0679-0355 inside the active site of receptor HPV16E6 (PDB ID 4xr8).
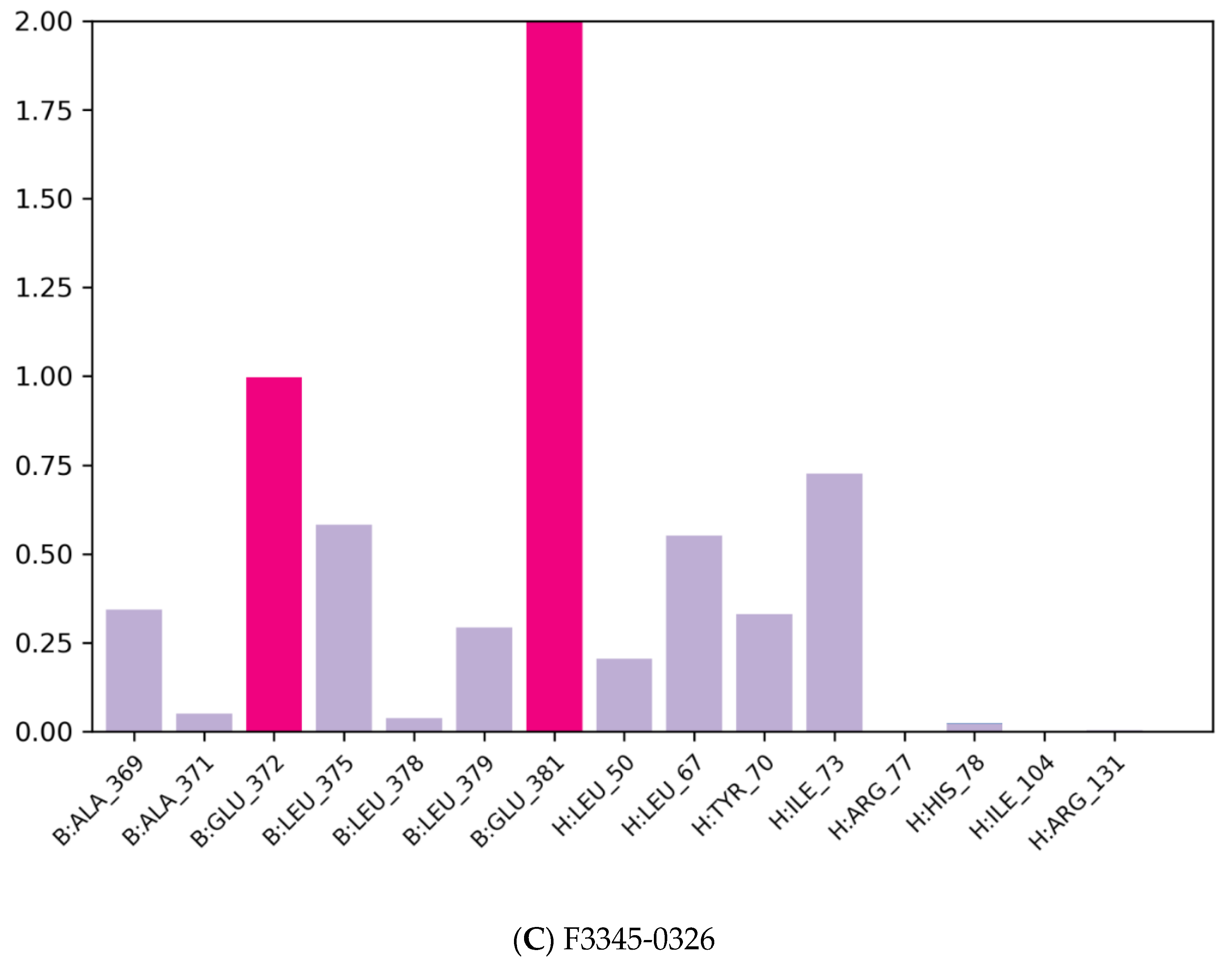
Results of molecular docking of the selected molecule F3345-0326 on HR E6 oncoprotein is reported in Figure 3 and clearly showed the formation of one strong hydrogen bond with the amino acids Ser(71) of the HR E6 protein and 9 van der Waals bonds with the amino acids Arg(129), His(78), Ser(74), Arg(131), Tyr(70), Leu(67),Val(53),Val(31),Gln(107), and 3 interacting via a π-Alkyl with the amino acids Val(62), Leu(50), and Ile(73). The docking study also showed the formation of 1 π-Sulfur bonds between the ligand and the amino acids Cys(51).
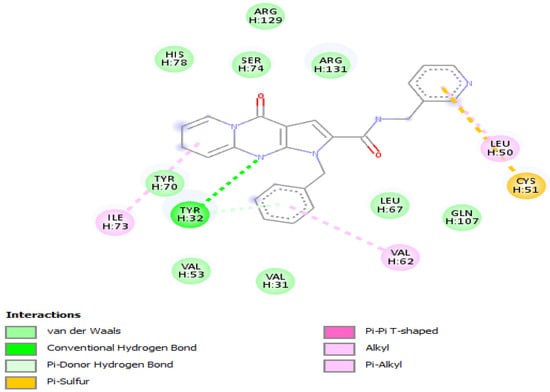
Figure 3.
Positioning and interactions of the molecule F3345-0326 inside the active site of receptor HPV16E6 (PDB ID 4xr8).
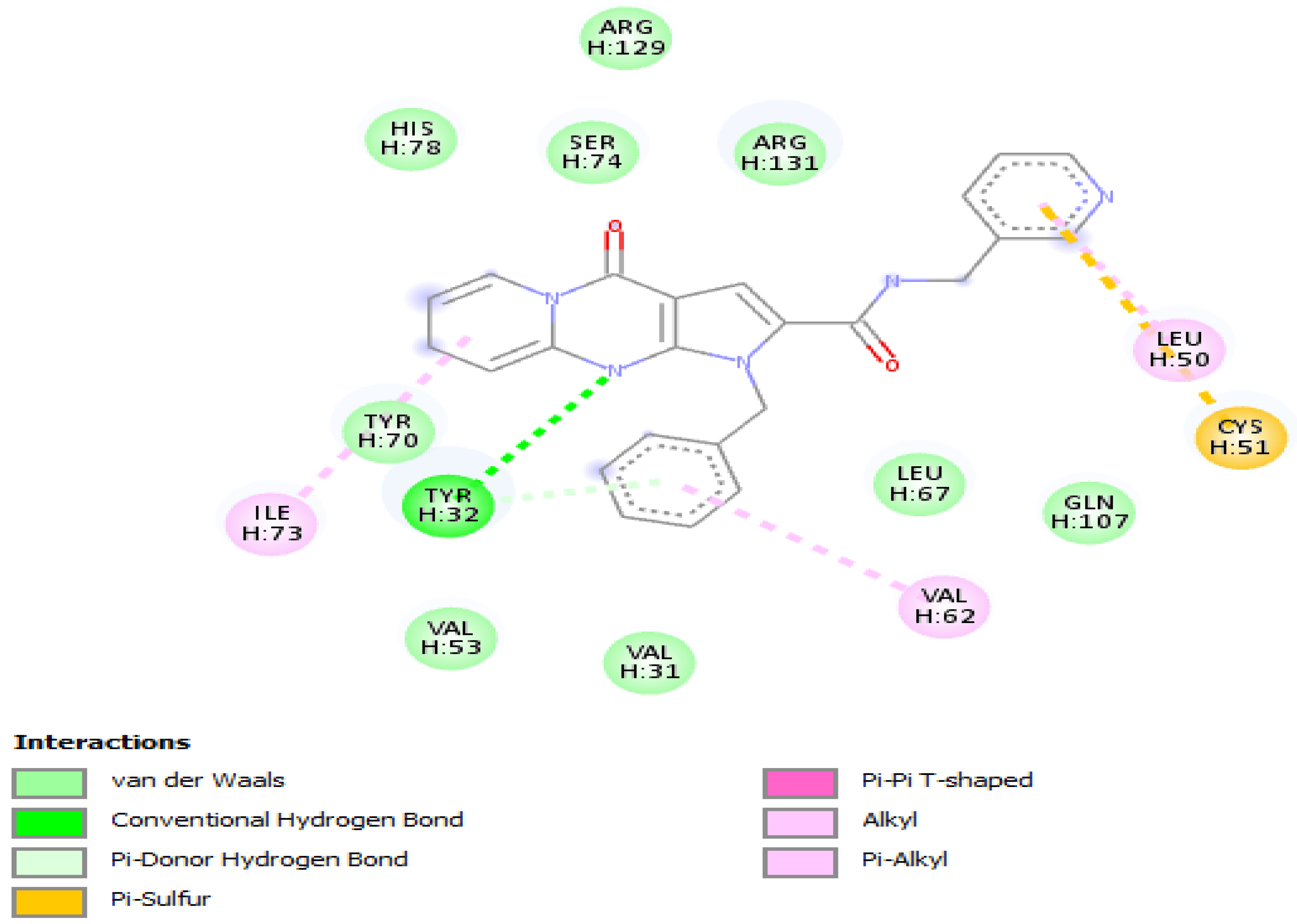
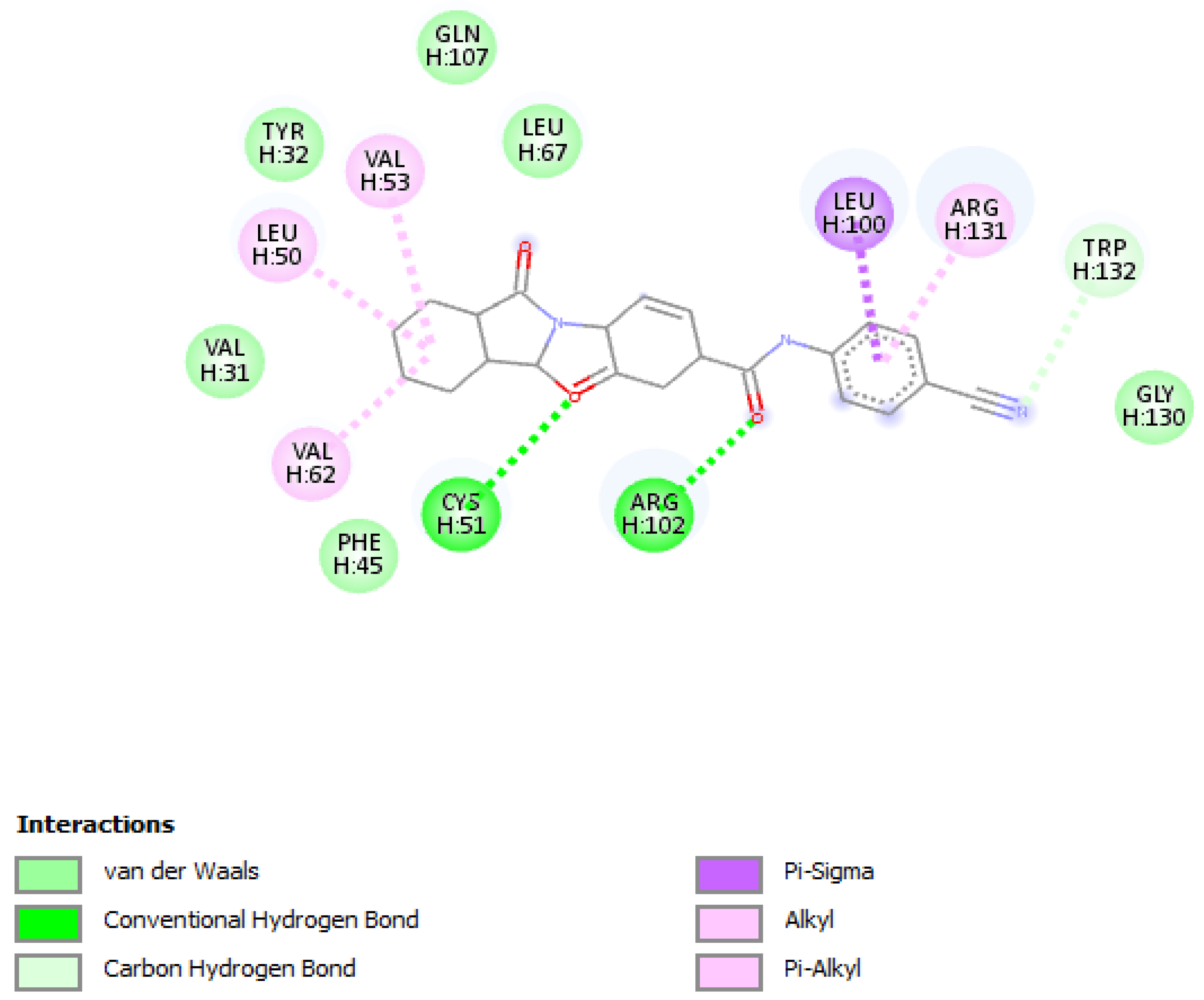
Molecular docking of the selected molecule F3374-0275 on HR E6 oncoprotein is reported in Figure 4 and showed the formation of two strong hydrogen bonds with the amino acids Cys(51) and Arg(102) of the HPV16 E6 protein and via 6 van der Waals bonds with the amino acids Gln(107), Leu(67), Tyr(32), Val(31), Phe(45), and Gly(130), and interacting via a π-Alkyl with the amino acids Val(53), Val(62), Leu(50), and Arg(131), and via π–sigma with Leu(100). The docking study showed also the formation of one carbon-hydrogen bond between the ligand and the amino acid Trp(132).
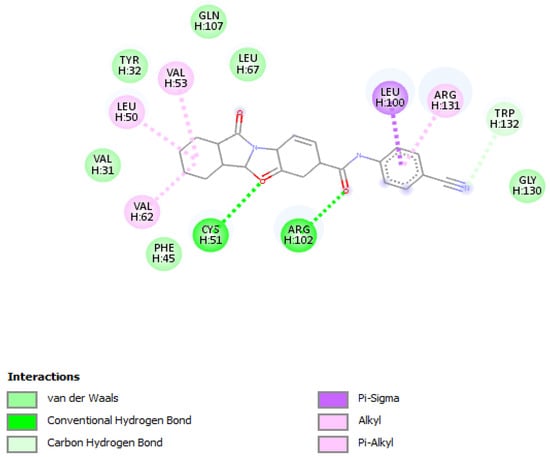
Figure 4.
Positioning and interactions of the molecule F3374-0275 inside the active site of receptor HPV16E6 (PDB ID 4xr8).
3.3. In Silico Pharmacokinetics (PK)/Pharmacodynamics (PD) Evaluation
The purpose of the ADMET preclinical study is to remove poorly performing molecules and focus on the best drug candidates. The present study investigated the suitability of the five proposed compounds for use as anti-cancer drugs based on properties, absorption, distribution, metabolism, excretion, and toxicity, which are critical elements in drug development.
These pharmacokinetic properties were obtained using the admet SAR approach evaluating blood-brain barrier (BBB) permeability, human intestinal absorption (HIA), Caco-2 cell permeability and the AMES assay that are the main elements used to determine drug capacities and results are reported in Table 2. Crossing the blood–brain barrier (BBB) is a crucial property widely used to determine the usefulness of the chemical as a drug, indicating whether drugs can cross the blood–brain barrier [36]. In this study, the three evaluated compounds have shown low BBB index and were, therefore, considered poorly distributed in the brain.

Table 2.
Analysis of the pharmacokinetic properties and toxicity of the three candidate molecules.
ADMET analysis showed also that the three ligands can be absorbed by the human gut, are not metabolized by most cytochrome P450 monooxygenases, and don’t exhibit any acute toxicity, mutagenic effect, or carcinogenic potential.
3.4. Molecular Dynamics
In this study, extensive pharmacokinetic analysis identified three candidate compounds for HPV16 E6 inhibition with the most advantageous binding interactions and the best pharmacokinetic profiles. These three molecules have already shown anticancer activity the first F0679-0355 in sienna cancer F3345-0326 kinase inhibition F3374-0275 for leucime [37].
Desmond simulation trajectories were analyzed, and the root mean square deviation (RMSD) Figure 5, root mean square fluctuation (RMSF) Figure 6 and protein–ligand contacts were calculated from the MD trajectory analysis.
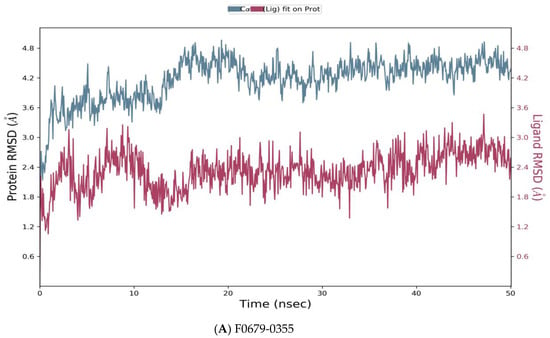
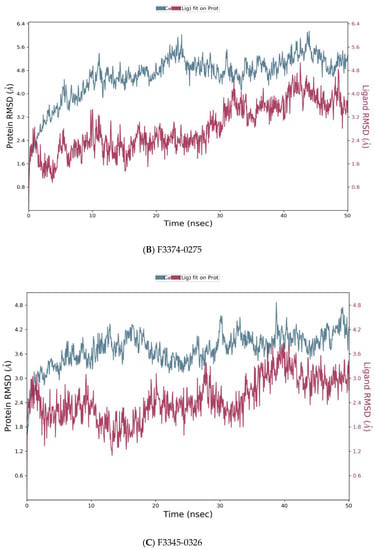
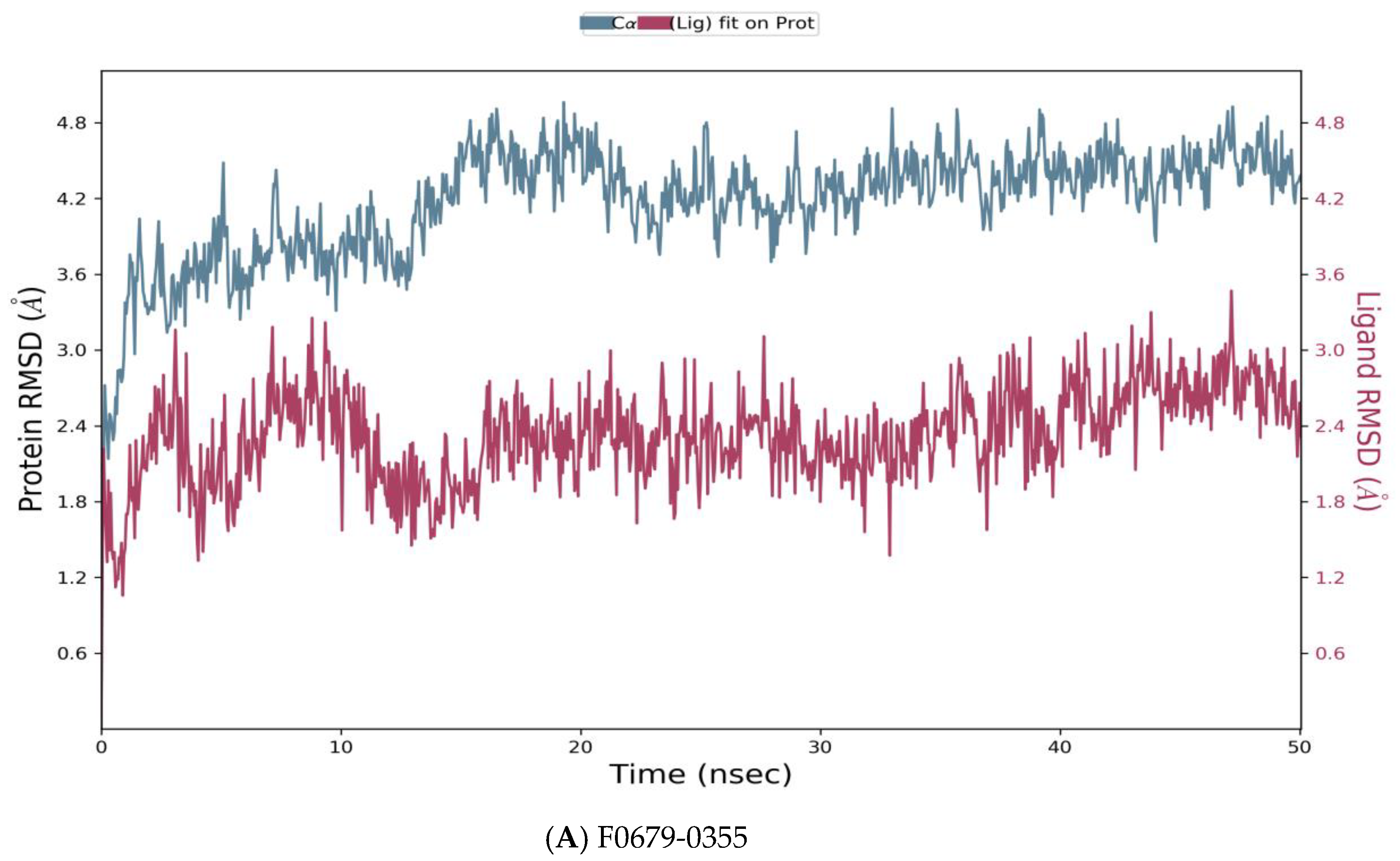
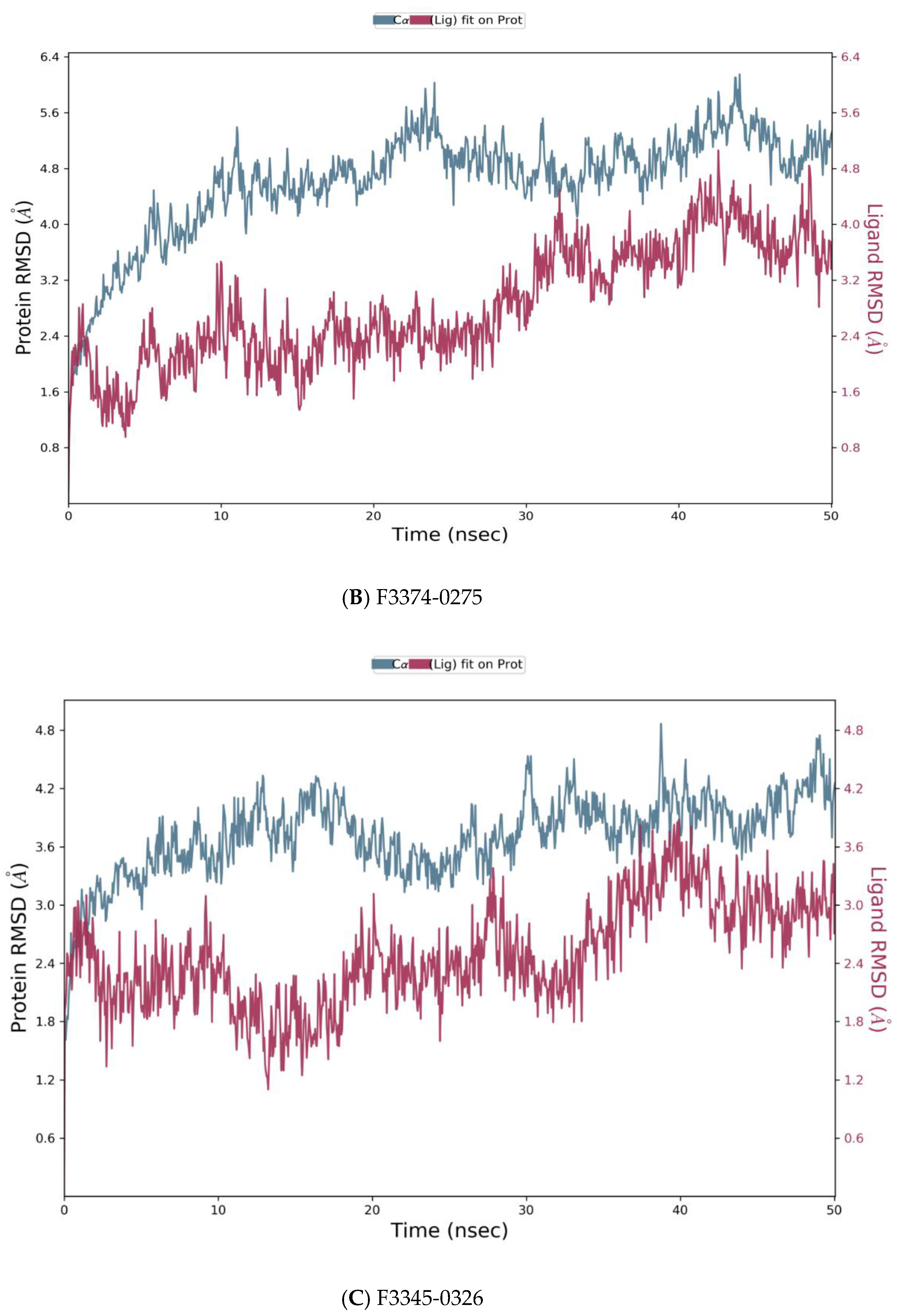
Figure 5.
Root means square deviation (RMSD) of the C-alpha atoms of protein and the ligand with time. The left Y-axis shows the variation of protein RMSD through time. The right Y-axis shows the variation of ligand RMSD through time. (A) RMSD of F0679-0355_4xr8 complex. (B) RMSD of F3374-0275_4xr8 complex. (C) RMSD of F3345-0326_4xr8 complex.
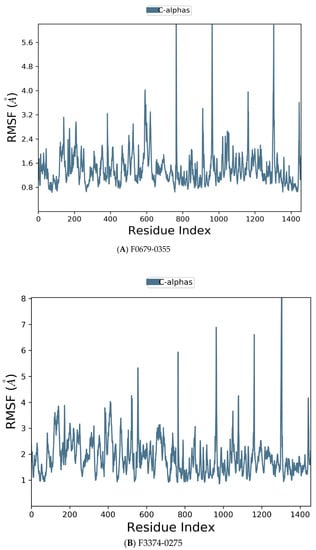
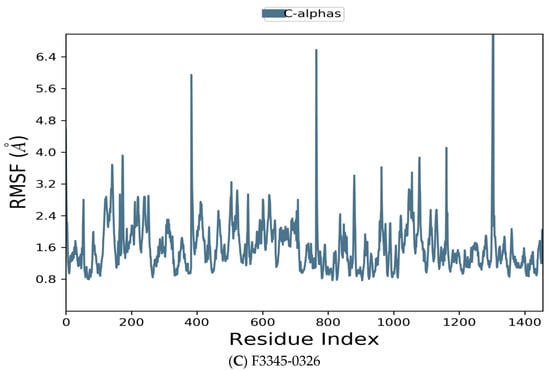
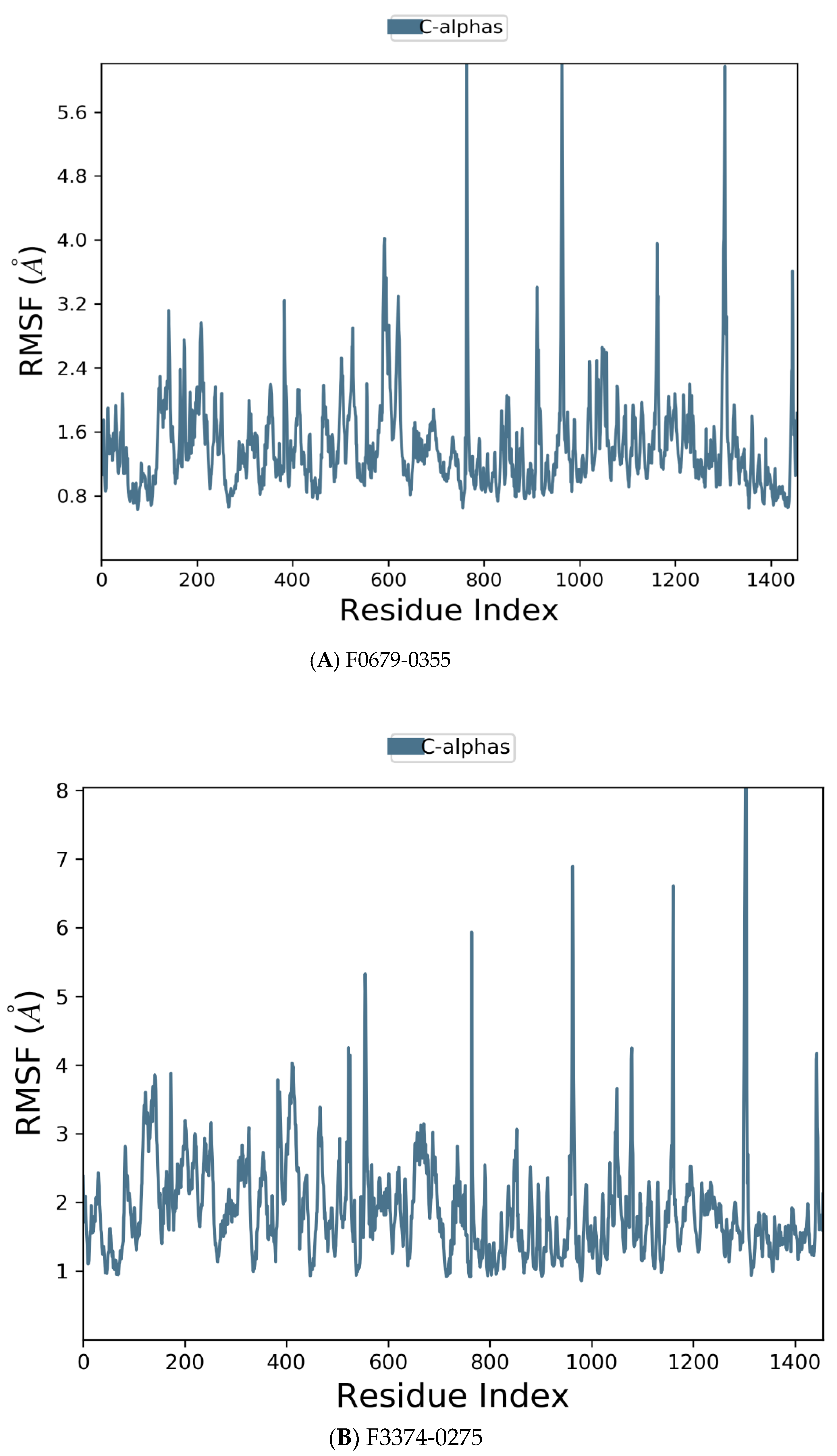
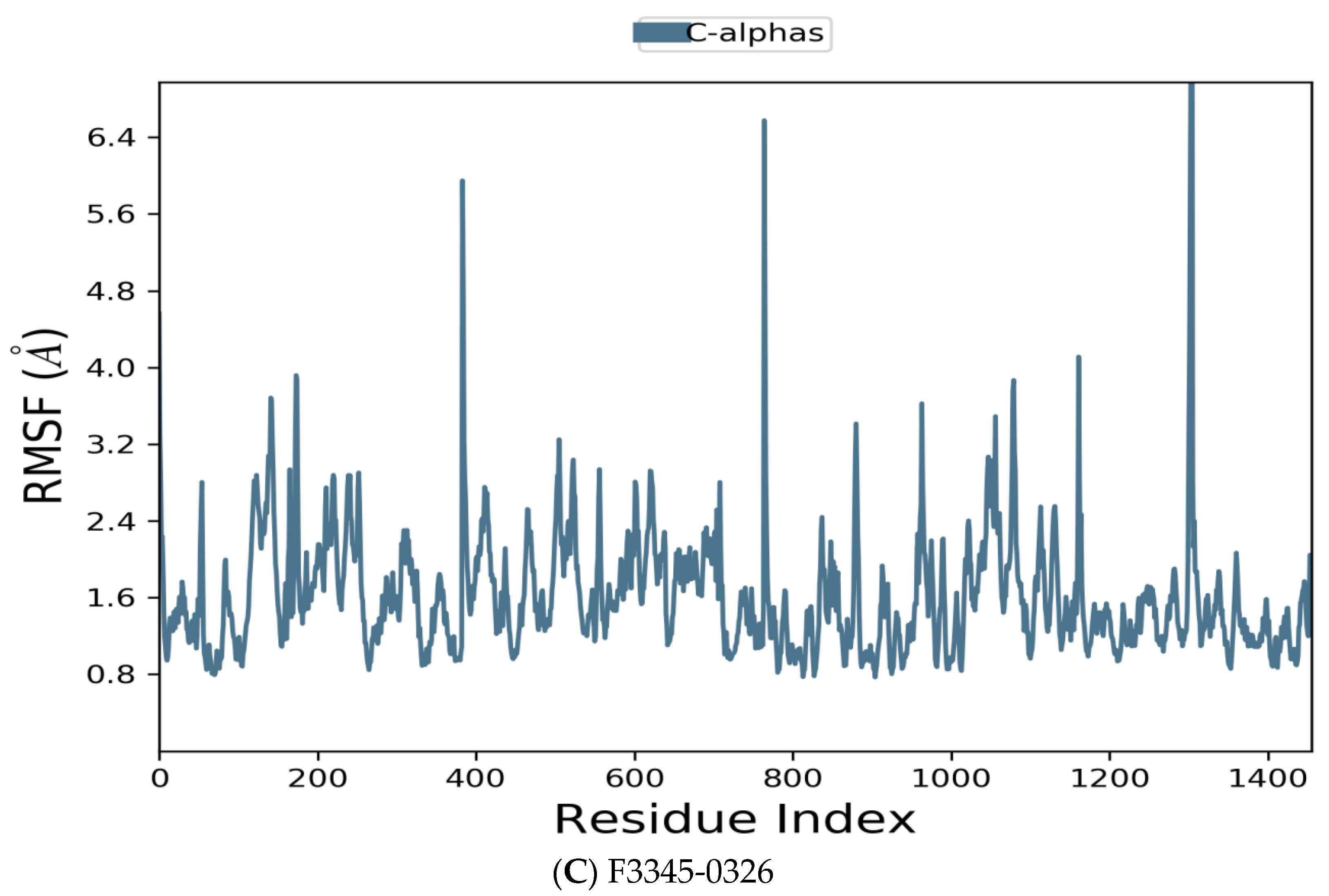
Figure 6.
Residue wise Root Mean Square Fluctuation (RMSF) of protein with F0679-0355 (A), F3374-0275 (B), and F3345-0326 (C) ligands.
As reported in Figure 5, the evolution of RMSD values with time for the C-alpha atoms of three complexes F0679-0355_4xr8, F3374-0275_4xr8, and F3345-0326_4xr8, indicates that the three complexes reach stability at 10 ns. From then, changes in RMSD values remain within 1.0 Å for the target (4xr8) during the simulation period, which is quite acceptable. Ligands fit to protein RMSD values fluctuate within 1.5 Å till 50 ns after being stable. These indicate that the ligands remained stably bound to the binding site of the receptor during the simulation period. Of particular interest, F0679-0355_4xr8 complex showed better results comparatively.
Figure 6 manifests the RMSF value per residue of the target bound to the ligands. Residues showing larger peaks belong to the loop regions, as determined from the MD trajectories (Figure 7), or to the N- and C-terminal regions. The target bound to F0679-0355 showed a comparatively better RMSF, as shown in Figure 6A. The low RMSF values of the binding site residues show the stability of the ligand binding to the protein.
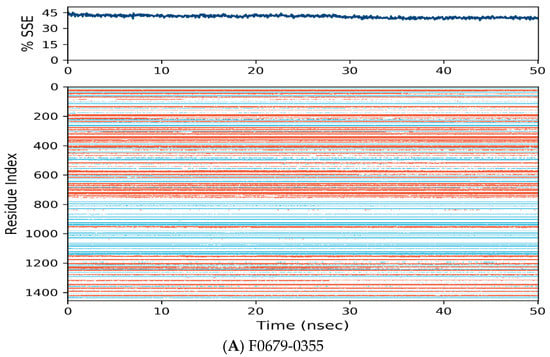
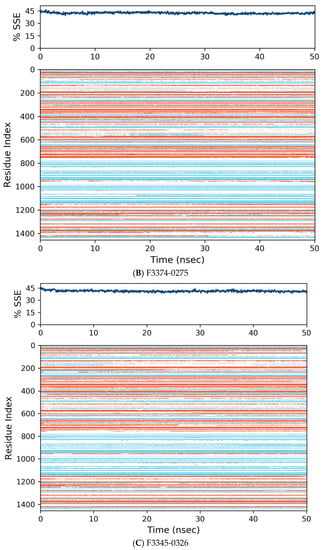
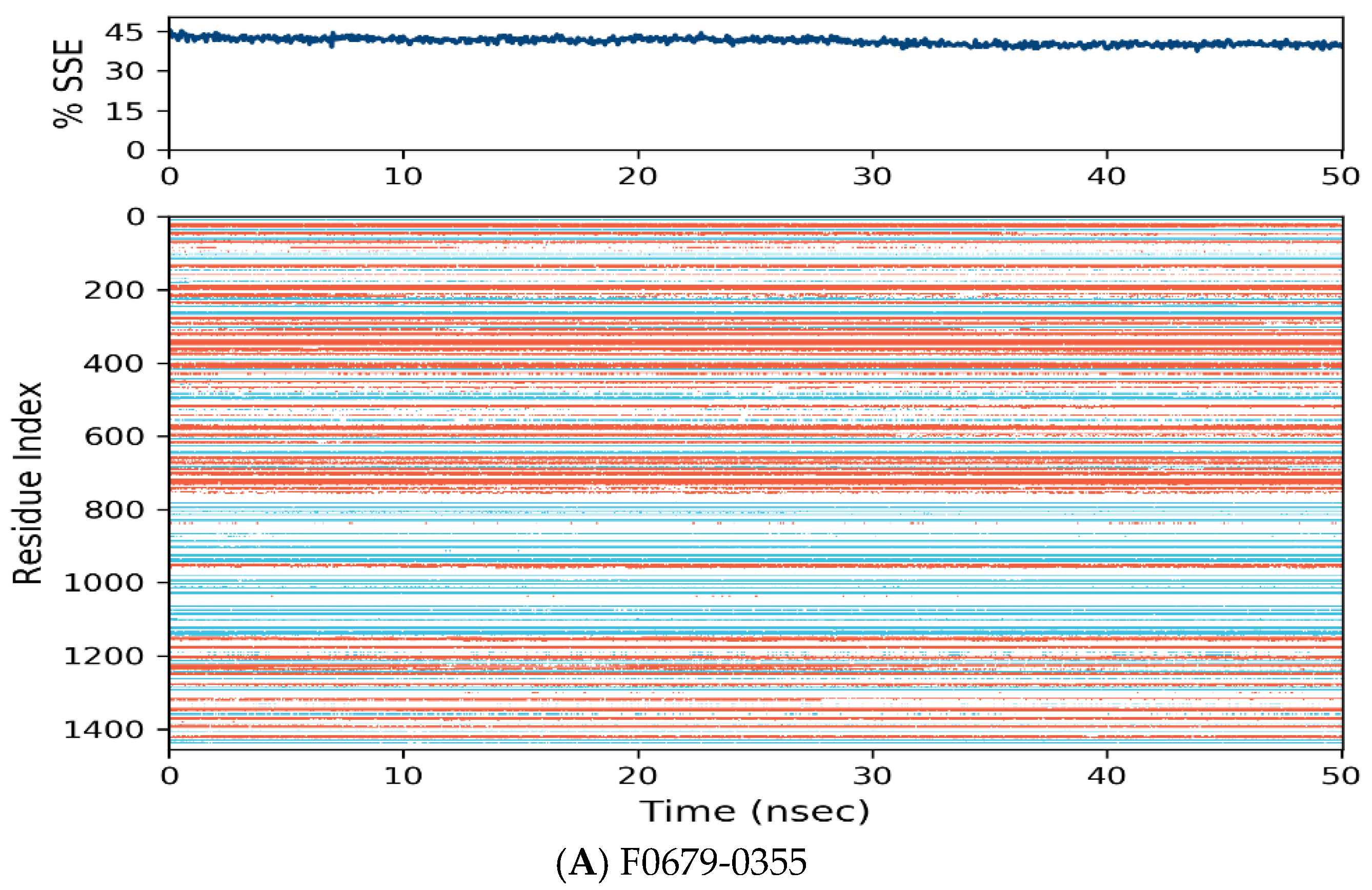
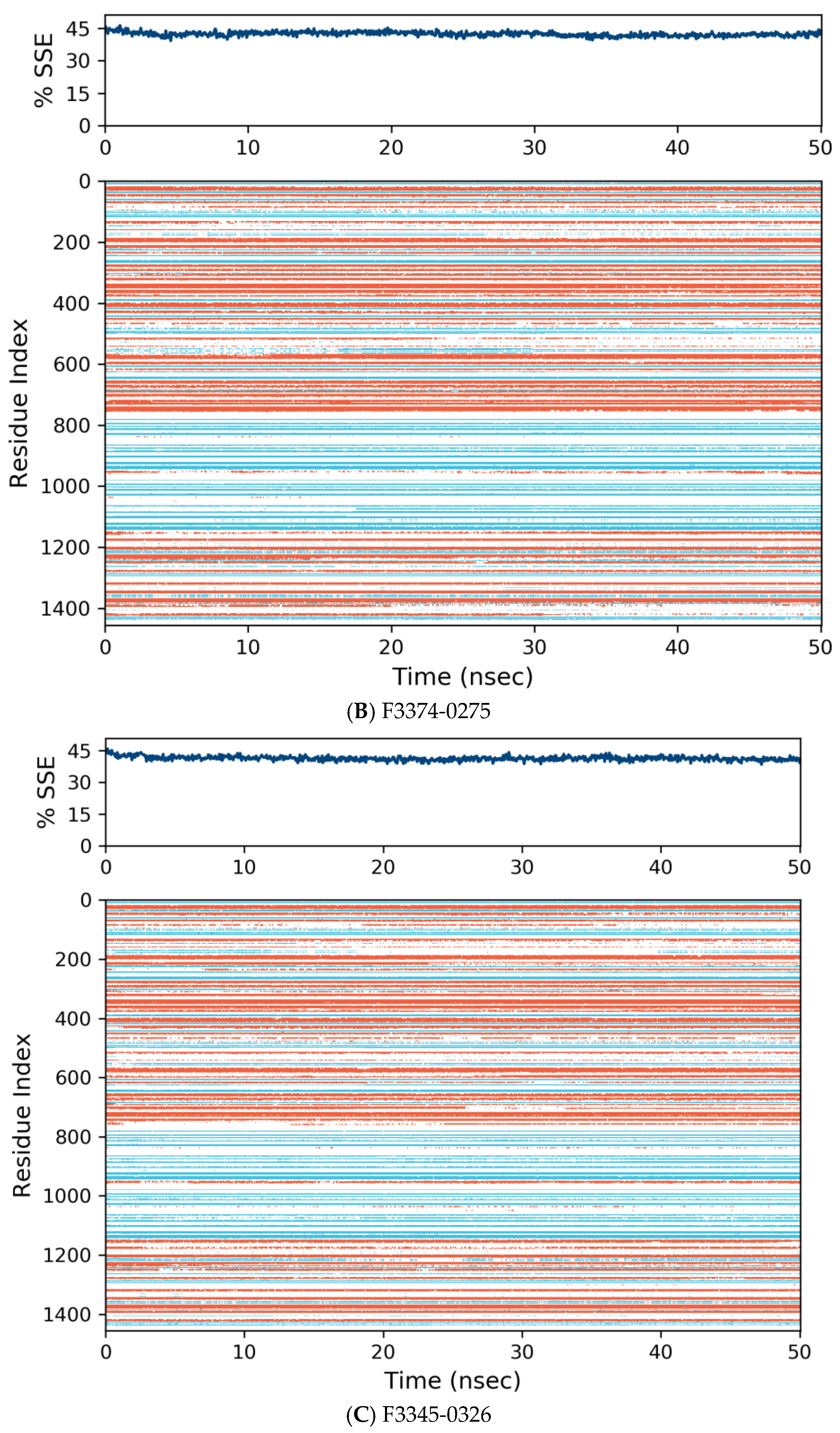
Figure 7.
Distribution of protein secondary structure components by residue index across the protein structure. Red denotes alpha helices, and blue denotes beta-strands.
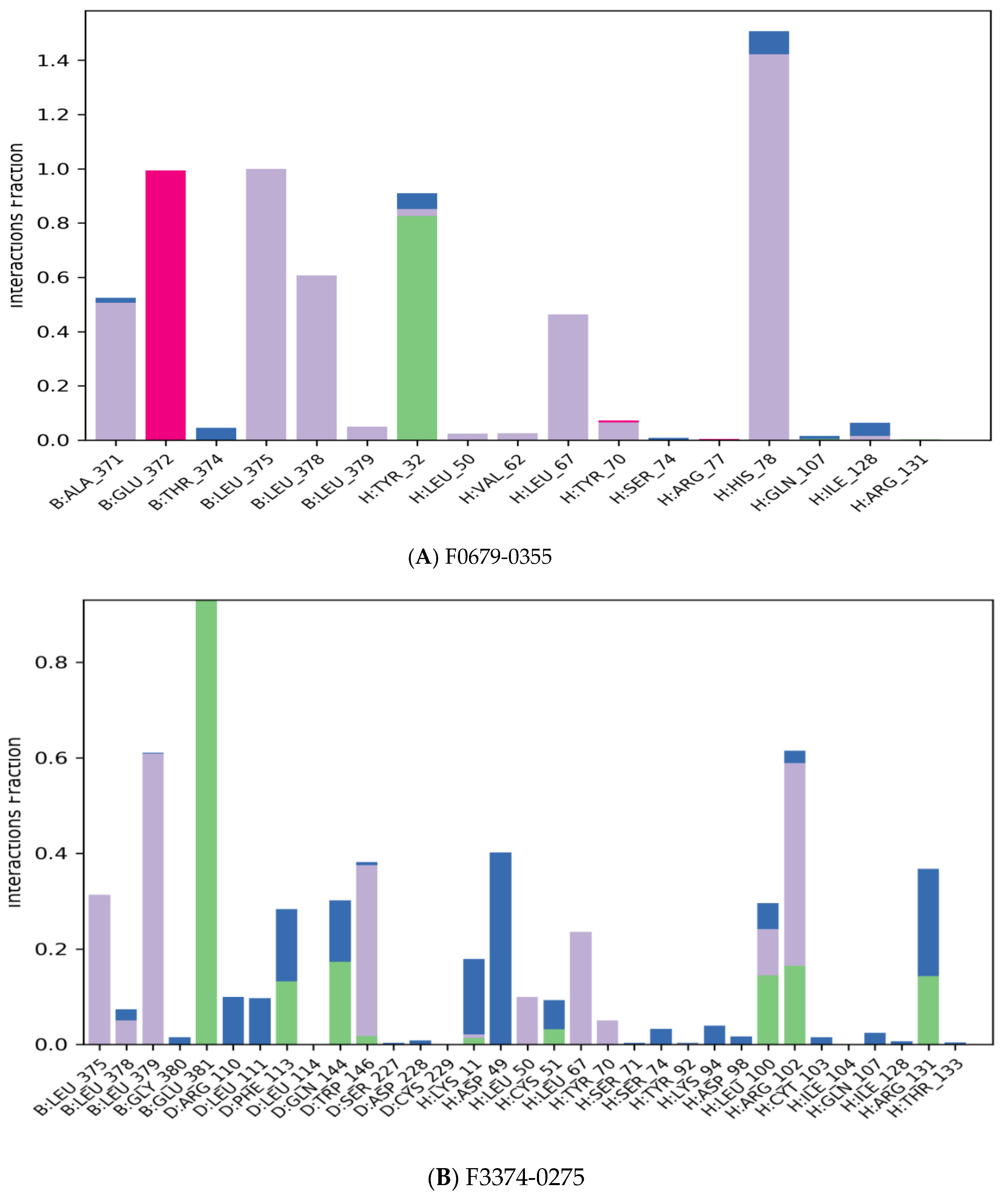
The majority of the essential ligand–protein interactions determined by MD are hydrogen bonds and hydrophobic interactions, as illustrated in Figure 5. The stacked bar graphs have been normalized over the pathway: therefore, a value of 1.0 indicates that for 100% of the simulation time the particular interaction was conserved Figure 8. Values above 1.0 are likely to be achieved, as some protein residues could have multiple contacts of the same subtype with the ligand.
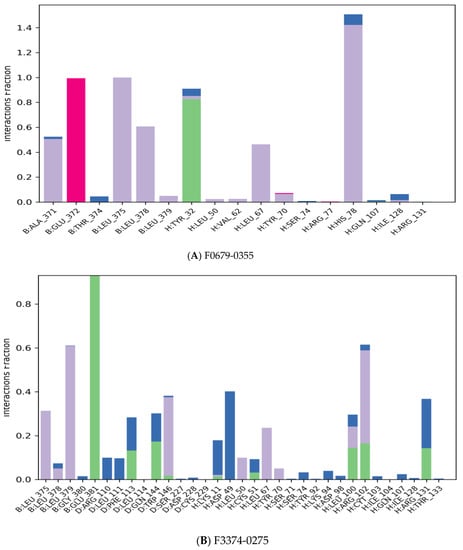
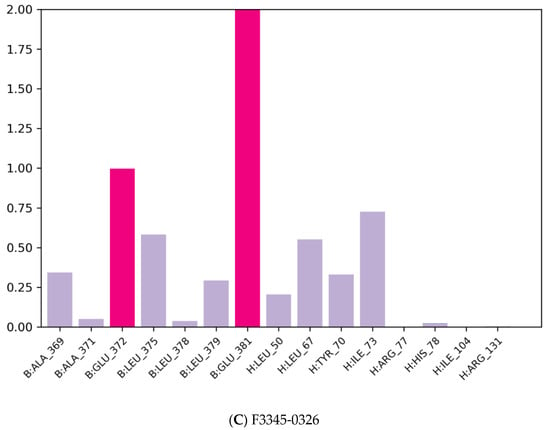
Figure 8.
Protein–ligand contact histogram.
4. Conclusions
Using the in silico protein–ligand interaction approach, we identified three promising candidates with the highest binding energy scores that could be potential inhibitors of HR E6 oncoproteins, likely without significant side effects. Furthermore, this study highlights the value of using the virtual screening approach as a time- and cost-saving strategy to identify chemicals with potential biological effects. However, further extensive in vitro and in vivo studies are needed to verify the antiviral activity of these three molecules.
In addition, to identify good molecules likely to bind to E6, but also to have medicinal properties, we have carried out an in silico study based on the identification of three molecules with anti-HPV activity.
Author Contributions
All authors confirmed they have contributed to the intellectual content of this paper and have met the following requirements: significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; drafting or revising the article for intellectual content; and final approval of the published article. Participation of authors by task: M.S. and H.H. conceived, designed and performed the study; M.S., H.H., M.E.M., F.Z.Y., and B.M. developed the methodology; M.S., B.M., and M.B. (Moualij Benaissa) wrote the original draft; M.S. and H.H. validated the data obtained; H.H., M.S., B.M., M.B. (Moualij Benaissa), and M.B. (Mohammed Bouachrine) analyzed and validated the data; all authors revised, edited and approved the submitted article; B.M., M.B. (Moualij Benaissa), and M.B. (Mohammed Bouachrine) supervised the study. All authors have read and agreed to the published version of the manuscript.
Funding
This research did not receive any external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
The authors would like to thank the anonymous reviewers for their valuable comments and suggestions to improve the paper’s quality.
Conflicts of Interest
We wish to confirm that there are no known conflict of interest associated with this publication, and there has been no significant financial support for this work that could have influenced its outcome.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Carper, M.B.; Goel, S.; Zhang, A.M.; Damrauer, J.S.; Cohen, S.; Zimmerman, M.P.; Gentile, G.M.; Parag-Sharma, K.; Murphy, R.M.; Sato, K.; et al. Activation of the CREB Coactivator CRTC2 by Aberrant Mitogen Signaling Promotes Oncogenic Functions in HPV16 Positive Head and Neck Cancer. Neoplasia 2022, 29, 100799. [Google Scholar] [CrossRef] [PubMed]
- Vonsky, M.; Shabaeva, M.; Runov, A.; Lebedeva, N.; Chowdhury, S.; Palefsky, J.M.; Isaguliants, M. Carcinogenesis Associated with Human Papillomavirus Infection. Mechanisms and Potential for Immunotherapy. Biochem. Moscow 2019, 84, 782–799. [Google Scholar] [CrossRef] [PubMed]
- Tognon, M.; Tagliapietra, A.; Magagnoli, F.; Mazziotta, C.; Oton-Gonzalez, L.; Lanzillotti, C.; Vesce, F.; Contini, C.; Rotondo, J.C.; Martini, F. Investigation on Spontaneous Abortion and Human Papillomavirus Infection. Vaccines 2020, 8, 473. [Google Scholar] [CrossRef]
- Condrat, C.E.; Filip, L.; Gherghe, M.; Cretoiu, D.; Suciu, N. Maternal HPV Infection: Effects on Pregnancy Outcome. Viruses 2021, 13, 2455. [Google Scholar] [CrossRef]
- Thomas, M.; Narayan, N.; Pim, D.; Tomaić, V.; Massimi, P.; Nagasaka, K.; Kranjec, C.; Gammoh, N.; Banks, L. Human papillomaviruses, cervical cancer and cell polarity. Oncogene 2008, 27, 7018–7030. [Google Scholar] [CrossRef]
- Burk, R.D.; Chen, Z.; Van Doorslaer, K. Human Papillomaviruses: Genetic Basis of Carcinogenicity. Public Health Genom. 2009, 12, 281–290. [Google Scholar] [CrossRef]
- Oyervides-Muñoz, M.A.; Pérez-Maya, A.A.; Rodríguez-Gutiérrez, H.F.; Gómez-Macias, G.S.; Fajardo, O.R.; Trevino, V.; Barrera-Saldaña, H.A.; Garza-Rodríguez, M.L. Understanding the HPV Integration and Its Progression to Cervical Cancer. Infect. Genet. Evol. 2018, 61, 134–144. [Google Scholar] [CrossRef]
- Hu, Z.; Ma, D. The Precision Prevention and Therapy of HPV-Related Cervical Cancer: New Concepts and Clinical Implications. Cancer Med. 2018, 7, 5217–5236. [Google Scholar] [CrossRef]
- Almeida, A.M.; Queiroz, J.; Sousa, F.; Sousa, Â. Cervical Cancer and HPV Infection: Ongoing Therapeutic Research to Counteract the Action of E6 and E7 Oncoproteins. Drug Discov. Today 2019, 24, 2044–2057. [Google Scholar] [CrossRef]
- Yeo-Teh, N.S.L.; Ito, Y.; Jha, S. High-Risk Human Papillomaviral Oncogenes E6 and E7 Target Key Cellular Pathways to Achieve Oncogenesis. Int. J. Mol. Sci. 2018, 19, 1706. [Google Scholar] [CrossRef] [PubMed]
- Nominé, Y.; Masson, M.; Charbonnier, S.; Zanier, K.; Ristriani, T.; Deryckère, F.; Sibler, A.P.; Desplancq, D.; Andrew Atkinson, R.; Weiss, E.; et al. Structural and functional analysis of E6 oncoprotein: Insights in the molecular pathways of human papillomavirus-mediated pathogenesis. Mol. cell 2006, 21, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Zapien, D.; Ruiz, F.X.; Poirson, J.; Mitschler, A.; Ramirez, J.; Forster, A.; Cousido-Siah, A.; Masson, M.; Vande Pol, S.; Podjarny, A.; et al. Structure of the E6/E6AP/p53 Complex Required for HPV-Mediated Degradation of P53. Nature 2016, 529, 541–545. [Google Scholar] [CrossRef]
- Huibregtse, J.M.; Scheffner, M.; Howley, P.M. Localization of the E6-AP Regions That Direct Human Papillomavirus E6 Binding, Association with P53, And Ubiquitination of Associated Proteins. Mol. Cell. Biol. 1993, 13, 4918–4927. [Google Scholar] [CrossRef] [PubMed]
- Zanier, K.; Charbonnier, S.; Sidi, A.O.M.O.; McEwen, A.G.; Ferrario, M.G.; Poussin-Courmontagne, P.; Cura, V.; Brimer, N.; Babah, K.O.; Ansari, T.; et al. Structural Basis for Hijacking of Cellular LxxLL Motifs by Papillomavirus E6 Oncoproteins. Science 2013, 339, 694–698. [Google Scholar] [CrossRef]
- Sailer, C.; Offensperger, F.; Julier, A.; Kammer, K.-M.; Walker-Gray, R.; Gold, M.G.; Scheffner, M.; Stengel, F. Structural Dynamics of the E6AP/UBE3A-E6-P53 Enzyme-Substrate Complex. Nat. Commun. 2018, 9, 4441. [Google Scholar] [CrossRef]
- Huibregtse, J.M.; Scheffner, M.; Howley, P.M. A Cellular Protein Mediates Association of P53 with the E6 Oncoprotein of Human Papillomavirus Types 16 or 18. EMBO J. 1991, 10, 4129–4135. [Google Scholar] [CrossRef]
- Pim, D.; Storey, A.; Thomas, M.; Massimi, P.; Banks, L. Mutational analysis of HPV-18 E6 identifies domains required for p53 degradation in vitro, abolition of p53 transactivation in vivo and immortalisation of primary BMK cells. Oncogene 1994, 9, 1869–1876. [Google Scholar]
- Patel, D.; Huang, S.M.; Baglia, L.A.; McCance, D.J. The E6 Protein of Human Papillomavirus Type 16 Binds to And Inhibits Co-Activation by CBP and p300. EMBO J. 1999, 18, 5061–5072. [Google Scholar] [CrossRef]
- Monie, A.; Hung, C.F.; Roden, R.; Wu, T.C. Cervarix™: A Vaccine for the Prevention of HPV 16, 18-Associated Cervical Cancer. Biol. Targets Ther. 2008, 2, 107. [Google Scholar]
- Shi, L.; Sings, H.L.; Bryan, J.T.; Wang, B.; Wang, Y.; Mach, H.; Kosinski, M.; Washabaugh, M.W.; Sitrin, R.; Barr, E. GARDASIL®: Prophylactic Human Papillomavirus Vaccine Development–from Bench Top to Bed-side. Clin. Pharmacol. Ther. 2007, 81, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Petrosky, E.; Bocchini, J.A., Jr.; Hariri, S.; Chesson, H.; Curtis, C.R.; Saraiya, M.; Unger, E.R.; Markowitz, L.E. Use of 9-Valent Human Papillomavirus (HPV) Vaccine: Updated HPV Vaccination Recommendations of the Advisory Committee on Immunization Practices. MMWR. Morb. Mortal. Wkly. Rep. 2015, 64, 300–304. [Google Scholar] [PubMed]
- Hampson, L.; Martin-Hirsch, P.; Hampson, I.N. An Overview of Early Investigational Drugs for the Treatment of Human Papilloma Virus Infection and Associated Dysplasia. Expert Opin. Investig. Drugs 2015, 24, 1529–1537. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and Drug-Like Compounds: The Rule-of-Five Revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- R.P.D. Bank. RCSB PDB: Homepage. Available online: http://www.rcsb.org/ (accessed on 19 April 2020).
- BIOVIA Discovery Studio. System Requirements for Discovery Studio 2020. Available online: https://www.3dsbiovia.com/products/collaborative-science/biovia-discovery-studio/requirements/technical-requirements-2020.html (accessed on 29 April 2020).
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Téléchargements-MGLTools. Available online: http://mgltools.scripps.edu/downloads (accessed on 29 April 2020).
- PyMOL. pymol.org. Available online: https://pymol.org/2/ (accessed on 12 May 2020).
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sac-erdoti, F.D.; et al. Molecular dynamics—Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing-SC ’06, Tampa, FL, USA, 11–17 November 2006. [Google Scholar] [CrossRef]
- Shivakumar, D.; Williams, J.; Wu, Y.; Damm, W.; Shelley, J.; Sherman, W. Prediction of Absolute Solvation Free Energies using Molecular Dynamics Free Energy Perturbation and the OPLS Force Field. J. Chem. Theory Comput. 2010, 6, 1509–1519. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with Pyrx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar]
- zur Hausen, H. Papillomaviruses and Cancer: From Basic Studies to Clinical Application. Nat. Rev. Cancer 2002, 2, 342–350. [Google Scholar] [CrossRef]
- Travé, G.; Zanier, K. HPV-Mediated Inactivation of Tumor Suppressor P53. Cell Cycle 2016, 15, 2231–2232. [Google Scholar] [CrossRef]
- Upadhyay, R.K. Drug Delivery Systems, CNS Protection, and the Blood Brain Barrier. BioMed Res. Int. 2014, 2014, 869269. [Google Scholar] [CrossRef]
- Mier, P.; Hurk, J.J. Lysosomal Hydrolases of the Epidermis. 2. Ester Hydrolases. Br. J. Dermatol. 1975, 93, 391–398. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).