NOD1, NOD2, and NLRC5 Receptors in Antiviral and Antimycobacterial Immunity
Abstract
:1. Introduction
2. The NLR Family
2.1. NLRP
2.2. NLRC
2.3. NLRA and NLRB
3. Structure and Expression of NOD1 and NOD2
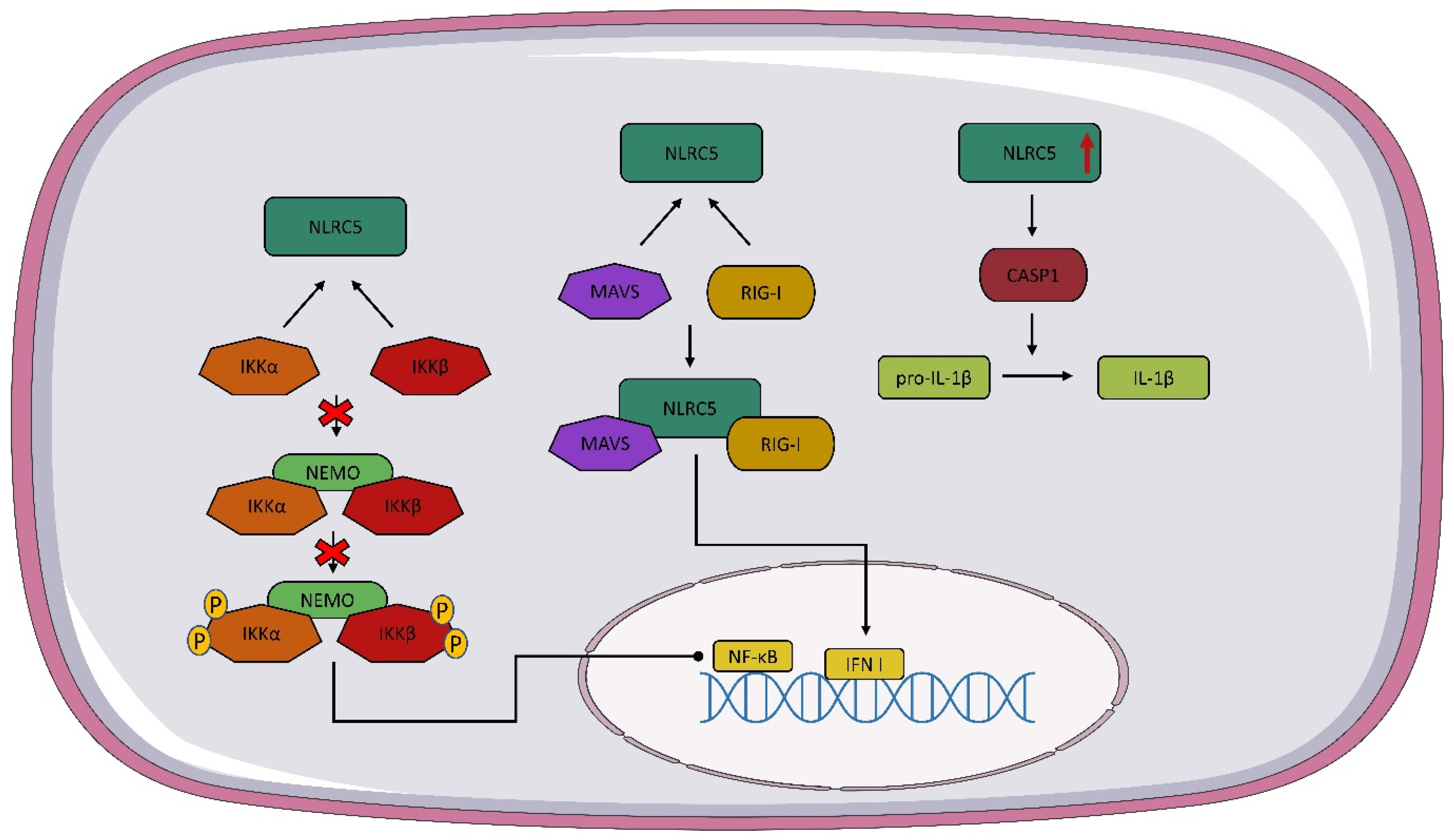
4. Structure and Expression NLRC5
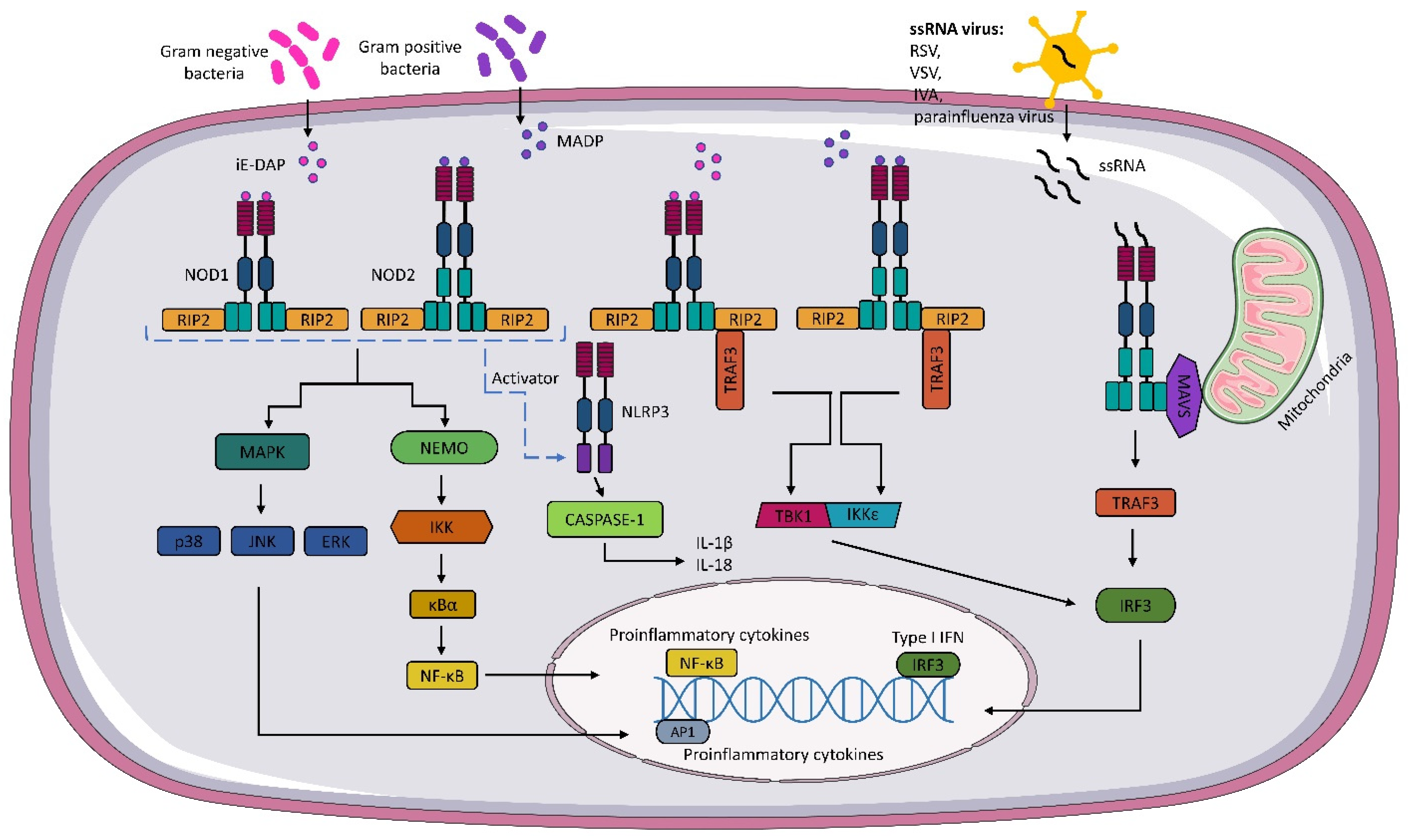
5. Interaction of NOD1, NOD2, and NLRC5 with Viruses
6. Interaction of NOD1, NOD2, and NLRC5 with Mycobacteria
7. NOD1, NOD2, and NLRC5 Gene Polymorphisms and Disease Predisposition
8. Potential Use of NOD1 and NOD2 Signaling Modulation in Therapy
9. Future Directions and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Fritz, J.H.; Ferrero, R.L.; Philpott, D.J.; Girardin, S.E. Nod-like Proteins in Immunity, Inflammation and Disease. Nat. Immunol. 2006, 7, 1250–1257. [Google Scholar]
- Zhong, Y.; Kinio, A.; Saleh, M. Functions of NOD-Like Receptors in Human Diseases. Front. Immunol. 2013, 4, 333. [Google Scholar] [PubMed]
- Zeromski, J.; Kierepa, A.; Boruczkowski, M.; Kowala-Piaskowska, A.; Mozer-Lisewska, I. The Role Of Nod-Like Receptors In Immunobiology And Medicine. Adv. Cell Biol. 2017, 44, 201–212. [Google Scholar]
- Macdonald, J.A.; Wijekoon, C.P.; Liao, K.C.; Muruve, D.A. Biochemical and Structural Aspects of the ATP-Binding Domain in Inflammasome-Forming Human NLRP Proteins. IUBMB Life 2013, 65, 851–862. [Google Scholar]
- Finger, J.N.; Lich, J.D.; Dare, L.C.; Cook, M.N.; Brown, K.K.; Duraiswamis, C.; Bertin, J.J.; Gough, P.J. Autolytic Proteolysis within the Function to Find Domain (FIIND) Is Required for NLRP1 Inflammasome Activity. J. Biol. Chem. 2012, 287, 25030–25037. [Google Scholar] [PubMed]
- Boucher, D.; Monteleone, M.; Coll, R.C.; Chen, K.W.; Ross, C.M.; Teo, J.L.; Gomez, G.A.; Holley, C.L.; Bierschenk, D.; Stacey, K.J.; et al. Caspase-1 self-cleavage is an intrinsic mechanism to terminate inflammasome activity. J. Exp. Med. 2018, 3, 827–840. [Google Scholar]
- Bronner, D.N.; Abuaita, B.H.; Chen, X.; Fitzgerald, K.A.; Nuñez, G.; He, Y.; Yin, X.M.; O’Riordan, M.X. Endoplasmic Reticulum Stress Activates the Inflammasome via NLRP3- and Caspase-2-Driven Mitochondrial Damage. Immunity 2015, 43, 451–462. [Google Scholar] [PubMed]
- Platnich, J.M.; Muruve, D.A. NOD-like Receptors and Inflammasomes: A Review of Their Canonical and Non-Canonical Signaling Pathways. Arch. Biochem. Biophys. 2019, 670, 4–14. [Google Scholar] [PubMed]
- Motta, V.; Soares, F.; Sun, T.; Philpott, D.J. Nod-like Receptors: Versatile Cytosolic Sentinels. Physiol. Rev. 2015, 95, 149–178. [Google Scholar]
- Proell, M.; Riedl, S.J.; Fritz, J.H.; Rojas, A.M.; Schwarzenbacher, R. The Nod-Like Receptor (NLR) Family: A Tale of Similarities and Differences. PLoS ONE 2008, 3, e2119. [Google Scholar]
- Bertsche, U.; Mayer, C.; Götz, F.; Gust, A.A. Peptidoglycan Perception—Sensing Bacteria by Their Common Envelope Structure. Int. J. Med. Microbiol. 2015, 305, 217–223. [Google Scholar]
- Girardin, S.E.; Boneca, I.G.; Viala, J.; Chamaillard, M.; Labigne, A.; Thomas, G.; Philpott, D.J.; Sansonetti, P.J. Nod2 Is a General Sensor of Peptidoglycan through Muramyl Dipeptide (MDP) Detection. J. Biol. Chem. 2003, 278, 8869–8872. [Google Scholar] [PubMed]
- Chamaillard, M.; Hashimoto, M.; Horie, Y.; Masumoto, J.; Qiu, S.; Saab, L.; Ogura, Y.; Kawasaki, A.; Fukase, K.; Kusumoto, S.; et al. An Essential Role for NOD1 in Host Recognition of Bacterial Peptidoglycan Containing Diaminopimelic Acid. Nat. Immunol. 2003, 4, 702–707. [Google Scholar]
- Sabbah, A.; Chang, T.H.; Harnack, R.; Frohlich, V.; Tominaga, K.; Dube, P.H.; Xiang, Y.; Bose, S. Activation of Innate Immune Antiviral Responses by Nod2. Nat. Immunol. 2009, 10, 1073–1080. [Google Scholar] [PubMed]
- Kim, J.; Yang, Y.L.; Jang, Y.S. Human β-Defensin 2 Is Involved in CCR2-Mediated Nod2 Signal Transduction, Leading to Activation of the Innate Immune Response in Macrophages. Immunobiology 2019, 224, 502–510. [Google Scholar]
- Chen, L.; Cao, S.Q.; Lin, Z.M.; He, S.J.; Zuo, J.P. NOD-like Receptors in Autoimmune Diseases. Acta Pharmacol. Sin. 2021, 42, 1742–1756. [Google Scholar]
- Bauernfeind, F.; Ablasser, A.; Bartok, E.; Kim, S.; Schmid-Burgk, J.; Cavlar, T.; Hornung, V. Inflammasomes: Current Understanding and Open Questions. Cell. Mol. Life Sci. 2010, 68, 765–783. [Google Scholar]
- Leibund Gut-Landmann, S.; Waldburger, J.M.; Krawczyk, M.; Otten, L.A.; Suter, T.; Fontana, A.; Acha-Orbea, H.; Reith, W. Mini-Review: Specificity and Expression of CIITA, the Master Regulator of MHC Class II Genes. Eur. J. Immunol. 2004, 34, 1513–1525. [Google Scholar]
- Accolla, R.S.; Ramia, E.; Tedeschi, A.; Forlani, G. CIITA-Driven MHC Class II Expressing Tumor Cells as Antigen Presenting Cell Performers: Toward the Construction of an Optimal Anti-Tumor Vaccine. Front. Immunol. 2019, 10, 1806. [Google Scholar] [PubMed]
- Velloso, F.J.; Trombetta-Lima, M.; Anschau, V.; Sogayar, M.C.; Correa, R.G. NOD-like Receptors: Major Players (and Targets) in the Interface between Innate Immunity and Cancer. Biosci. Rep. 2019, 39, 1–21. [Google Scholar]
- Inohara, N.; Koseki, T.; del Peso, L.; Hu, Y.; Yee, C.; Chen, S.; Carrio, R.; Merino, J.; Liu, D.; Ni, J.; et al. Nod1, an Apaf-1-like Activator of Caspase-9 and Nuclear Factor-KappaB. J. Biol. Chem. 1999, 274, 14560–14567. [Google Scholar] [CrossRef] [PubMed]
- Ogura, Y.; Inohara, N.; Benito, A.; Chen, F.F.; Yamaoka, S.; Nunez, G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J. Biol. Chem. 2001, 276, 4812–4818. [Google Scholar] [CrossRef] [PubMed]
- Sidiq, T.; Yoshihama, S.; Downs, I.; Kobayashi, K.S. Nod2: A Critical Regulator of Ileal Microbiota and Crohn’s Disease. Front. Immunol. 2016, 7, 367. [Google Scholar] [CrossRef] [Green Version]
- Moreira, L.O.; Zamboni, D.S. NOD1 and NOD2 Signaling in Infection and Inflammation. Front. Immunol. 2012, 3, 328. [Google Scholar] [CrossRef]
- Hasegawa, M.; Yang, K.; Hashimoto, M.; Park, J.H.; Kim, Y.G.; Fujimoto, Y.; Nuñez, G.; Fukase, K.; Inohara, N. Differential Release and Distribution of Nod1 and Nod2 Immunostimulatory Molecules among Bacterial Species and Environments. J. Biol. Chem. 2006, 281, 29054–29063. [Google Scholar] [CrossRef] [PubMed]
- Girardin, S.E.; Boneca, I.G.; Carneiro, L.A.M.; Antignac, A.; Jéhanno, M.; Viala, J.; Tedin, K.; Taha, M.K.; Labigne, A.; Zähringer, U.; et al. Nod1 Detects a Unique Muropeptide from Gram-Negative Bacterial Peptidoglycan. Science 2003, 300, 1584–1587. [Google Scholar] [CrossRef] [PubMed]
- Inohara, N.; Ogura, Y.; Fontalba, A.; Gutierrez, O.; Pons, F.; Crespo, J.; Fukase, K.; Inamura, S.; Kusumoto, S.; Hashimoto, M.; et al. Host Recognition of Bacterial Muramyl Dipeptide Mediated through NOD2: Implications for Crohn′s disease. J. Biol. Chem. 2003, 278, 5509–5512. [Google Scholar] [CrossRef]
- Grimes, C.L.; Ariyananda, L.D.Z.; Melnyk, J.E.; O’Shea, E.K. The Innate Immune Protein Nod2 Binds Directly to MDP, a Bacterial Cell Wall Fragment. J. Am. Chem. Soc. 2012, 134, 13535–13537. [Google Scholar] [CrossRef]
- Coulombe, F.; Divangahi, M.; Veyrier, F.; de Léséleuc, L.; Gleason, J.L.; Yang, Y.; Kelliher, M.A.; Pandey, A.K.; Sassetti, C.M.; Reed, M.B.; et al. Increased NOD2-Mediated Recognition of N-Glycolyl Muramyl Dipeptide. J. Exp. Med. 2009, 206, 1709. [Google Scholar] [CrossRef] [PubMed]
- Keestra-Gounder, A.M.; Byndloss, M.X.; Seyffert, N.; Young, B.M.; Chávez-Arroyo, A.; Tsai, A.Y.; Cevallos, S.A.; Winter, M.G.; Pham, O.H.; Tiffany, C.R.; et al. NOD1/NOD2 Signaling Links ER Stress with Inflammation. Nature 2016, 532, 394. [Google Scholar] [CrossRef] [PubMed]
- Marijke Keestra, A.; Winter, M.G.; Klein-Douwel, D.; Xavier, M.N.; Winter, S.E.; Kim, A.; Tsolis, R.M.; Bäumler, A.J. A Salmonella Virulence Factor Activates the NOD1/NOD2 Signaling Pathway. mBio 2011, 2, e00266-11. [Google Scholar] [CrossRef]
- Keestra, A.M.; Winter, M.G.; Auburger, J.J.; Fräßle, S.P.; Xavier, M.N.; Winter, S.E.; Kim, A.; Poon, V.; Ravesloot, M.M.; Waldenmaier, J.F.T.; et al. Manipulation of Small Rho GTPases Is a Pathogen-Induced Process Detected by Nod1. Nature 2013, 496, 233. [Google Scholar] [CrossRef] [PubMed]
- Fukazawa, A.; Alonso, C.; Kurachi, K.; Gupta, S.; Lesser, C.F.; McCormick, B.A.; Reinecker, H.C. GEF-H1 Mediated Control of NOD1 Dependent NF-ΚB Activation by Shigella Effectors. PLoS Pathog. 2008, 4, 1000228. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.S.; Chamaillard, M.; Ogura, Y.; Henegariu, O.; Inohara, N.; Nuñez, G.; Flavell, R.A. Nod2-Dependent Regulation of Innate and Adaptive Immunity in the Intestinal Tract. Science 2005, 307, 731–734. [Google Scholar] [CrossRef]
- Masumoto, J.; Yang, K.; Varambally, S.; Hasegawa, M.; Tomlins, S.A.; Qiu, S.; Fujimoto, Y.; Kawasaki, A.; Foster, S.J.; Horie, Y.; et al. Nod1 Acts as an Intracellular Receptor to Stimulate Chemokine Production and Neutrophil Recruitment in Vivo. J. Exp. Med. 2006, 203, 203–213. [Google Scholar] [CrossRef]
- Nakamura, N.; Lill, J.R.; Phung, Q.; Jiang, Z.; Bakalarski, C.; de Mazière, A.; Klumperman, J.; Schlatter, M.; Delamarre, L.; Mellman, I. Endosomes Are Specialized Platforms for Bacterial Sensing and NOD2 Signalling. Nature 2014, 509, 240–244. [Google Scholar] [CrossRef]
- Irving, A.T.; Mimuro, H.; Kufer, T.A.; Lo, C.; Wheeler, R.; Turner, L.J.; Thomas, B.J.; Malosse, C.; Gantier, M.P.; Casillas, L.N.; et al. The Immune Receptor NOD1 and Kinase RIP2 Interact with Bacterial Peptidoglycan on Early Endosomes to Promote Autophagy and Inflammatory Signaling. Cell Host Microbe 2014, 15, 623–635. [Google Scholar] [CrossRef]
- Travassos, L.H.; Carneiro, L.A.M.; Ramjeet, M.; Hussey, S.; Kim, Y.G.; Magalhes, J.G.; Yuan, L.; Soares, F.; Chea, E.; le Bourhis, L.; et al. Nod1 and Nod2 Direct Autophagy by Recruiting ATG16L1 to the Plasma Membrane at the Site of Bacterial Entry. Nat. Immunol. 2010, 11, 55–62. [Google Scholar] [CrossRef]
- Kufer, T.A.; Kremmer, E.; Adam, A.C.; Philpott, D.J.; Sansonetti, P.J. The Pattern-Recognition Molecule Nod1 Is Localized at the Plasma Membrane at Sites of Bacterial Interaction. Cell Microbiol. 2008, 10, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Legrand-Poels, S.; Kustermans, G.; Bex, F.; Kremmer, E.; Kufer, T.A.; Piette, J. Modulation of Nod2-Dependent NF-KappaB Signaling by the Actin Cytoskeleton. J. Cell Sci. 2007, 120, 1299–1310. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Westcott, N.P.; Griffin, M.E.; Hang, H.C. Peptidoglycan Metabolite Photoaffinity Reporters Reveal Direct Binding to Intracellular Pattern Recognition Receptors and Arf GTPases. ACS Chem. Biol. 2019, 14, 405. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zheng, Y.; Coyaud, É.; Zhang, C.; Selvabaskaran, A.; Yu, Y.; Xu, Z.; Weng, X.; Chen, J.S.; Meng, Y.; et al. Palmitoylation of NOD1 and NOD2 Is Required for Bacterial Sensing. Science 2019, 366, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Inohara, N.; Chamaillard, M.; McDonald, C.; Nuñez, G. NOD-LRR proteins: Role in Host-Microbial Interactions and Inflammatory Disease. Annu. Rev. Biochem. 2005, 74, 355–383. [Google Scholar] [CrossRef] [PubMed]
- Nembrini, C.; Kisielow, J.; Shamshiev, A.T.; Tortola, L.; Coyle, A.J.; Kopf, M.; Marsland, B.J. The Kinase Activity of Rip2 Determines Its Stability and Consequently Nod1- and Nod2-Mediated Immune Responses. J. Biol. Chem. 2009, 284, 19183–19188. [Google Scholar] [CrossRef] [PubMed]
- Hrdinka, M.; Schlicher, L.; Dai, B.; Pinkas, D.M.; Bufton, J.C.; Picaud, S.; Ward, J.A.; Rogers, C.; Suebsuwong, C.; Nikhar, S.; et al. Small Molecule Inhibitors Reveal an Indispensable Scaffolding Role of RIPK2 in NOD2 Signaling. EMBO J. 2018, 37, e99372. [Google Scholar] [CrossRef]
- Rahighi, S.; Ikeda, F.; Kawasaki, M.; Akutsu, M.; Suzuki, N.; Kato, R.; Kensche, T.; Uejima, T.; Bloor, S.; Komander, D.; et al. Specific Recognition of Linear Ubiquitin Chains by NEMO Is Important for NF-KappaB Activation. Cell 2009, 136, 1098–1109. [Google Scholar] [CrossRef]
- Jiang, X.; Chen, Z.J. The Role of Ubiquitylation in Immune Defence and Pathogen Evasion. Nat. Rev. Immunol. 2011, 12, 35–48. [Google Scholar] [CrossRef]
- Abbott, D.W.; Wilkins, A.; Asara, J.M.; Cantley, L.C. The Crohn’s Disease Protein, NOD2, Requires RIP2 in Order to Induce Ubiquitinylation of a Novel Site on NEMO. Curr. Biol. 2004, 14, 2217–2227. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, E.; Desfosses, A.; Wallmann, A.; Schulze, W.M.; Rehbein, K.; Mas, P.; Signor, L.; Gaudon, S.; Zenkeviciute, G.; Hons, M.; et al. RIP2 Filament Formation Is Required for NOD2 Dependent NF-ΚB Signalling. Nat. Commun. 2018, 9, 1–19. [Google Scholar] [CrossRef]
- Girardin, S.E.; Tournebize, R.; Mavris, M.; Page, A.L.; Li, X.; Stark, G.R.; Bertin, J.; Distefano, P.S.; Yaniv, M.; Sansonetti, P.J.; et al. CARD4/Nod1 Mediates NF-KappaB and JNK Activation by Invasive Shigella Flexneri. EMBO Rep. 2001, 2, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-H.; Kim, Y.-G.; Shaw, M.; Kanneganti, T.-D.; Fujimoto, Y.; Fukase, K.; Inohara, N.; Núñez, G. Nod1/RICK and TLR Signaling Regulate Chemokine and Antimicrobial Innate Immune Responses in Mesothelial Cells. J. Immunol. 2007, 179, 514–521. [Google Scholar] [CrossRef]
- Stockinger, S.; Reutterer, B.; Schaljo, B.; Schellack, C.; Brunner, S.; Materna, T.; Yamamoto, M.; Akira, S.; Taniguchi, T.; Murray, P.J.; et al. IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J. Immunol. 2004, 173, 7416–7425. [Google Scholar] [CrossRef] [PubMed]
- Allison, C.C.; Ferrand, J.; McLeod, L.; Hassan, M.; Kaparakis-Liaskos, M.; Grubman, A.; Bhathal, P.S.; Dev, A.; Sievert, W.; Jenkins, B.J.; et al. Nucleotide Oligomerization Domain 1 Enhances IFN-γ Signaling in Gastric Epithelial Cells during Helicobacter pylori Infection and Exacerbates Disease Severity. J. Immunol. 2013, 190, 3706–3715. [Google Scholar] [CrossRef] [Green Version]
- Barnich, N.; Hisamatsu, T.; Aguirre, J.E.; Xavier, R.; Reinecker, H.C.; Podolsky, D.K. GRIM-19 Interacts with Nucleotide Oligomerization Domain 2 and Serves as Downstream Effector of Anti-Bacterial Function in Intestinal Epithelial Cells. J. Biol. Chem. 2005, 280, 19021–19026. [Google Scholar] [CrossRef]
- Stevens, C.; Henderson, P.; Nimmo, E.R.; Soares, D.C.; Dogan, B.; Simpson, K.W.; Barrett, J.C.; Wilson, D.C.; Satsangi, J. The Intermediate Filament Protein, Vimentin, Is a Regulator of NOD2 Activity. Gut 2013, 62, 695–707. [Google Scholar] [CrossRef]
- McDonald, C.; Chen, F.F.; Ollendorff, V.; Ogura, Y.; Marchetto, S.; Lécine, P.; Borg, J.P.; Nuñez, G. A Role for Erbin in the Regulation of Nod2-Dependent NF-ΚB Signaling. J. Biol. Chem. 2005, 280, 40301–40309. [Google Scholar] [CrossRef]
- Yamamoto-Furusho, J.K.; Barnich, N.; Xavier, R.; Hisamatsu, T.; Podolsky, D.K. Centaurin Β1 Down-Regulates Nucleotide-Binding Oligomerization Domains 1- and 2-Dependent NF-ΚB Activation. J. Biol. Chem. 2006, 281, 36060–36070. [Google Scholar] [CrossRef]
- Kuenzel, S.; Till, A.; Winkler, M.; Häsler, R.; Lipinski, S.; Jung, S.; Grötzinger, J.; Fickenscher, H.; Schreiber, S.; Rosenstiel, P. The Nucleotide-Binding Oligomerization Domain-Like Receptor NLRC5 Is Involved in IFN-Dependent Antiviral Immune Responses. J. Immunol. 2010, 184, 1990–2000. [Google Scholar] [CrossRef] [PubMed]
- Benko, S.; Magalhaes, J.G.; Philpott, D.J.; Girardin, S.E. NLRC5 Limits the Activation of Inflammatory Pathways. J. Immunol. 2010, 185, 1681–1691. [Google Scholar] [CrossRef]
- Neerincx, A.; Lautz, K.; Menning, M.; Kremmer, E.; Zigrino, P.; Hösel, M.; Büning, H.; Schwarzenbacher, R.; Kufer, T.A. A Role for the Human Nucleotide-Binding Domain, Leucine-Rich Repeat-Containing Family Member NLRC5 in Antiviral Responses. J. Biol. Chem. 2010, 285, 26223–26232. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, C.; Bian, E.; Lei, T.; Lv, X.; Li, J. NLRC5 Negatively Regulates Inflammatory Responses in LPS-Induced Acute Lung Injury through NF-ΚB and P38 MAPK Signal Pathways. Toxicol. Appl. Pharmacol. 2020, 403, 115150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jiao, C.; Liu, L.; Wang, A.; Tang, L.; Ren, Y.; Huang, P.; Xu, J.; Mao, D.; Liu, L. NLRC5: A Potential Target for Central Nervous System Disorders. Front. Immunol. 2021, 12, 2451. [Google Scholar] [CrossRef]
- Meissner, T.B.; Li, A.; Biswas, A.; Lee, K.H.; Liu, Y.J.; Bayir, E.; Iliopoulos, D.; van den Elsen, P.J.; Kobayashi, K.S. NLR Family Member NLRC5 Is a Transcriptional Regulator of MHC Class I Genes. Proc. Natl. Acad. Sci. USA 2010, 107, 13794–13799. [Google Scholar] [CrossRef] [Green Version]
- Neerincx, A.; Rodriguez, G.M.; Steimle, V.; Kufer, T.A. NLRC5 Controls Basal MHC Class I Gene Expression in an MHC Enhanceosome-Dependent Manner. J. Immunol. 2012, 188, 4940–4950. [Google Scholar] [CrossRef]
- Cui, J.; Zhu, L.; Xia, X.; Wang, H.Y.; Legras, X.; Hong, J.; Ji, J.; Shen, P.; Zheng, S.; Chen, Z.J.; et al. NLRC5 Negatively Regulates the NF-ΚB and Type I Interferon Signaling Pathways and Antiviral Immunity. Cell 2010, 141, 483. [Google Scholar] [CrossRef]
- Ranjan, P.; Singh, N.; Kumar, A.; Neerincx, A.; Kremmer, E.; Cao, W.; Davis, W.G.; Katz, J.M.; Gangappa, S.; Lin, R.; et al. NLRC5 Interacts with RIG-I to Induce a Robust Antiviral Response against Influenza Virus Infection. Eur. J. Immunol. 2015, 45, 758–772. [Google Scholar] [CrossRef]
- Kumar, H.; Pandey, S.; Zou, J.; Kumagai, Y.; Takahashi, K.; Akira, S.; Kawai, T. NLRC5 Deficiency Does Not Influence Cytokine Induction by Virus and Bacteria Infections. J. Immunol. 2011, 186, 994–1000. [Google Scholar] [CrossRef]
- Davis, B.K.; Roberts, R.A.; Huang, M.T.; Willingham, S.B.; Conti, B.J.; Brickey, W.J.; Barker, B.R.; Kwan, M.; Taxman, D.J.; Accavitti-Loper, M.-A.; et al. Cutting Edge: NLRC5-Dependent Activation of the Inflammasome. J. Immunol. 2011, 186, 1333–1337. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Cui, J.; Li, Q.; Zou, J.; Wang, H.Y.; Wang, R.F. Enhanced TLR-Induced NF-ΚB Signaling and Type I Interferon Responses in NLRC5 Deficient Mice. Cell Res. 2012, 22, 822–835. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, M.; Zhang, X.; Meng, X.; Huang, C.; Li, J. NLRC5 Promotes Cell Proliferation via Regulating the AKT/VEGF-A Signaling Pathway in Hepatocellular Carcinoma. Toxicology 2016, 359–360, 47–57. [Google Scholar] [CrossRef]
- Peng, Y.; He, Y.; Chen, C.; Xu, T.; Li, L.; Ni, M.; Meng, X.; Huang, C.; Li, J. NLRC5 Regulates Cell Proliferation, Migration and Invasion in Hepatocellular Carcinoma by Targeting the Wnt/β-Catenin Signaling Pathway. Cancer Lett. 2016, 376, 10–21. [Google Scholar] [CrossRef]
- Ma, H.L.; Zhao, X.F.; Chen, G.Z.; Fang, R.H.; Zhang, F.R. Silencing NLRC5 Inhibits Extracellular Matrix Expression in Keloid Fibroblasts via Inhibition of Transforming Growth Factor-Β1/Smad Signaling Pathway. Biomed. Pharmacother. 2016, 83, 1016–1021. [Google Scholar] [CrossRef] [PubMed]
- Pichlmair, A.; Schulz, O.; Tan, C.P.; Näslund, T.I.; Liljeström, P.; Weber, F.; Reis E Sousa, C. RIG-I-Mediated Antiviral Responses to Single-Stranded RNA Bearing 5′-Phosphates. Science 2006, 314, 997–1001. [Google Scholar] [CrossRef] [Green Version]
- Coutermarsh-Ott, S.; Eden, K.; Allen, I.C. Beyond the Inflammasome: Regulatory Nod-like Receptor Modulation of the Host Immune Response Following Virus Exposure. J. Gen. Virol. 2016, 97, 825–838. [Google Scholar] [CrossRef]
- Vissers, M.; Remijn, T.; Oosting, M.; de Jong, D.J.; Diavatopoulos, D.A.; Hermans, P.W.M.; Ferwerda, G. Respiratory Syncytial Virus Infection Augments NOD2 Signaling in an IFN-β-Dependent Manner in Human Primary Cells. Eur. J. Immunol. 2012, 42, 2727–2735. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Liu, T.; Shi, H.; Wang, J.; Ji, P.; Wang, H.; Hou, Y.; Tan, R.X.; Li, E. Respiratory Syncytial Virus Infection Upregulates NLRC5 and Major Histocompatibility Complex Class I Expression through RIG-I Induction in Airway Epithelial Cells. J. Virol. 2015, 89, 7636–7645. [Google Scholar] [CrossRef]
- Lupfer, C.; Thomas, P.G.; Kanneganti, T.-D. Nucleotide Oligomerization and Binding Domain 2-Dependent Dendritic Cell Activation Is Necessary for Innate Immunity and Optimal CD8+ T Cell Responses to Influenza A Virus Infection. J. Virol. 2014, 88, 8946–8955. [Google Scholar] [CrossRef]
- Lupfer, C.R.; Stokes, K.L.; Kuriakose, T.; Kanneganti, T.-D. Deficiency of the NOD-Like Receptor NLRC5 Results in Decreased CD8 + T Cell Function and Impaired Viral Clearance. J. Virol. 2017, 91, e00377-17. [Google Scholar] [CrossRef] [PubMed]
- Limonta, D.; Dyna-Dagman, L.; Branton, W.; Mancinelli, V.; Makio, T.; Wozniak, R.W.; Power, C.; Hobman, T.C. Nodosome Inhibition as a Novel Broad-Spectrum Antiviral Strategy against Arboviruses, Enteroviruses, and SARS-CoV-2. Antimicrob. Agents Chemother. 2021, 65, e0049121. [Google Scholar] [CrossRef]
- Wang, F.; Liu, R.; Yang, J.; Chen, B. New Insights into Genetic Characteristics between Multiple Myeloma and COVID-19: An Integrative Bioinformatics Analysis of Gene Expression Omnibus Microarray and the Cancer Genome Atlas Data. Int. J. Lab. Hematol. 2021, 43, 1325–1333. [Google Scholar] [CrossRef]
- Yin, X.; Riva, L.; Pu, Y.; Martin-Sancho, L.; Kanamune, J.; Yamamoto, Y.; Sakai, K.; Gotoh, S.; Miorin, L.; de Jesus, P.D.; et al. MDA5 Governs the Innate Immune Response to SARS-CoV-2 in Lung Epithelial Cells. Cell Rep. 2021, 34, 108628. [Google Scholar] [CrossRef]
- Lupfer, C.; Kanneganti, T.D. The Expanding Role of NLRs in Antiviral Immunity. Immunol. Rev. 2013, 255, 13. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.S.; Sasaki, M.; Cho, S.X.; Kasuga, Y.; Zhu, B.; Ouda, R.; Orba, Y.; de Figueiredo, P.; Sawa, H.; Kobayashi, K.S. SARS-CoV-2 Inhibits Induction of the MHC Class I Pathway by Targeting the STAT1-IRF1-NLRC5 Axis. Nat. Commun. 2021, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.L.; Jeong, H.G.; Kasper, C.A.; Arrieumerlou, C. IKKα Contributes to Canonical NF-ΚB Activation Downstream of Nod1-Mediated Peptidoglycan Recognition. PLoS ONE 2010, 5, e15371. [Google Scholar] [CrossRef]
- Kim, J.G.; Lee, S.J.; Kagnoff, M.F. Nod1 Is an Essential Signal Transducer in Intestinal Epithelial Cells Infected with Bacteria That Avoid Recognition by Toll-Like Receptors. Infect. Immun. 2004, 72, 1487. [Google Scholar] [CrossRef] [PubMed]
- Allison, C.C.; Kufer, T.A.; Kremmer, E.; Kaparakis, M.; Ferrero, R.L. Helicobacter Pylori Induces MAPK Phosphorylation and AP-1 Activation via a NOD1-Dependent Mechanism. J. Immunol. 2009, 183, 8099–8109. [Google Scholar] [CrossRef]
- Watanabe, T.; Asano, N.; Fichtner-Feigl, S.; Gorelick, P.L.; Tsuji, Y.; Matsumoto, Y.; Chiba, T.; Fuss, I.J.; Kitani, A.; Strober, W. NOD1 Contributes to Mouse Host Defense against Helicobacter pylori via Induction of Type I IFN and Activation of the ISGF3 Signaling Pathway. J. Clin. Investig. 2010, 120, 1645–1662. [Google Scholar] [CrossRef]
- Zilbauer, M.; Dorrell, N.; Elmi, A.; Lindley, K.J.; Schüller, S.; Jones, H.E.; Klein, N.J.; Núňez, G.; Wren, B.W.; Bajaj-elliott, M. A Major Role for Intestinal Epithelial Nucleotide Oligomerization Domain 1 (NOD1) in Eliciting Host Bactericidal Immune Responses to Campylobacter jejuni. Cell Microbiol. 2007, 9, 2404–2416. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Yamazaki, T.; Kamada, N.; Tawaratsumida, K.; Kim, Y.-G.; Núñez, G.; Inohara, N. Nucleotide-Binding Oligomerization Domain 1 Mediates Recognition of Clostridium difficile and Induces Neutrophil Recruitment and Protection against the Pathogen. J. Immunol. 2011, 186, 4872–4880. [Google Scholar] [CrossRef]
- Frutuoso, M.S.; Hori, J.I.; Pereira, M.S.F.; Junior, D.S.L.; Sônego, F.; Kobayashi, K.S.; Flavell, R.A.; Cunha, F.Q.; Zamboni, D.S. The Pattern Recognition Receptors Nod1 and Nod2 Account for Neutrophil Recruitment to the Lungs of Mice Infected with Legionella pneumophila. Microbes Infect. 2010, 12, 819–827. [Google Scholar] [CrossRef]
- Ferwerda, G.; Girardin, S.E.; Kullberg, B.J.; le Bourhis, L.; de Jong, D.J.; Langenberg, D.M.L.; van Crevel, R.; Adema, G.J.; Ottenhoff, T.H.M.; van der Meer, J.W.M.; et al. NOD2 and Toll-Like Receptors Are Nonredundant Recognition Systems of Mycobacterium tuberculosis. PLoS Pathog. 2005, 1, e34. [Google Scholar] [CrossRef]
- Kufer, T.A.; Kremmer, E.; Banks, D.J.; Philpott, D.J. Role for Erbin in Bacterial Activation of Nod2. Infect. Immun. 2006, 74, 3115. [Google Scholar] [CrossRef]
- Hisamatsu, T.; Suzuki, M.; Reinecker, H.C.; Nadeau, W.J.; McCormick, B.A.; Podolsky, D.K. CARD15/NOD2 Functions as an Antibacterial Factor in Human Intestinal Epithelial Cells. Gastroenterology 2003, 124, 993–1000. [Google Scholar] [CrossRef] [PubMed]
- Magalhaes, J.G.; Fritz, J.H.; Le Bourhis, L.; Sellge, G.; Travassos, L.H.; Selvanantham, T.; Girardin, S.E.; Gommerman, J.L.; Philpott, D.J. Nod2-Dependent Th2 Polarization of Antigen-Specific Immunity. J. Immunol. 2008, 181, 7925–7935. [Google Scholar] [CrossRef] [PubMed]
- Shimada, K.; Chen, S.; Dempsey, P.W.; Sorrentino, R.; Alsabeh, R.; Slepenkin, A.v.; Peterson, E.; Doherty, T.M.; David, U.; Crother, T.R.; et al. The NOD/RIP2 Pathway Is Essential for Host Defenses against Chlamydophila pneumoniae Lung Infection. PLoS Pathog. 2009, 5, e1000379. [Google Scholar] [CrossRef]
- Berrington, W.R.; Iyer, R.; Wells, R.D.; Smith, K.D.; Skerrett, S.J.; Hawn, T.R. NOD1 and NOD2 Regulation of Pulmonary Innate Immunity to Legionella pneumophila. Eur. J. Immunol. 2010, 40, 3519–3527. [Google Scholar] [CrossRef]
- Davis, K.M.; Nakamura, S.; Weiser, J.N. Nod2 Sensing of Lysozyme-Digested Peptidoglycan Promotes Macrophage Recruitment and Clearance of S. pneumoniae Colonization in Mice. J. Clin. Investig. 2011, 121, 3666. [Google Scholar] [CrossRef]
- Juárez, E.; Carranza, C.; Hernández-Sánchez, F.; León-Contreras, J.C.; Hernández-Pando, R.; Escobedo, D.; Torres, M.; Sada, E. NOD2 Enhances the Innate Response of Alveolar Macrophages to Mycobacterium tuberculosis in Humans. Eur. J. Immunol. 2012, 42, 880–889. [Google Scholar] [CrossRef]
- Gandotra, S.; Jang, S.; Murray, P.J.; Salgame, P.; Ehrt, S. Nucleotide-Binding Oligomerization Domain Protein 2-Deficient Mice Control Infection with Mycobacterium tuberculosis. Infect. Immun. 2007, 75, 5127–5134. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yin, C.; Pandey, A.; Abbott, D.; Sassetti, C.; Kelliher, M.A. NOD2 Pathway Activation by MDP or Mycobacterium tuberculosis Infection Involves the Stable Polyubiquitination of Rip2. J. Biol. Chem. 2007, 282, 36223–36229. [Google Scholar] [CrossRef]
- Leber, J.H.; Crimmins, G.T.; Raghavan, S.; Meyer-Morse, N.P.; Cox, J.S.; Portnoy, D.A. Distinct TLR- and NLR-Mediated Transcriptional Responses to an Intracellular Pathogen. PLoS Pathog. 2008, 4, e6. [Google Scholar] [CrossRef]
- Brooks, M.N.; Rajaram, M.V.S.; Azad, A.K.; Amer, A.O.; Valdivia-Arenas, M.A.; Park, J.H.; Núñez, G.; Schlesinger, L.S. NOD2 Controls the Nature of the Inflammatory Response and Subsequent Fate of Mycobacterium tuberculosis and M. bovis BCG in Human Macrophages. Cell. Microbiol. 2011, 13, 402. [Google Scholar] [CrossRef] [PubMed]
- Landes, M.B.; Rajaram, M.V.S.; Nguyen, H.; Schlesinger, L.S. Role for NOD2 in Mycobacterium tuberculosis-Induced INOS Expression and NO Production in Human Macrophages. J. Leukoc. Biol. 2015, 97, 1111–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Divangahi, M.; Mostowy, S.; Coulombe, F.; Kozak, R.; Guillot, L.; Veyrier, F.; Kobayashi, K.S.; Flavell, R.A.; Gros, P.; Behr, M.A. NOD2-Deficient Mice Have Impaired Resistance to Mycobacterium tuberculosis Infection through Defective Innate and Adaptive Immunity. J. Immunol. 2008, 181, 7157–7165. [Google Scholar] [CrossRef]
- Austin, C.M.; Ma, X.; Graviss, E.A. Common Nonsynonymous Polymorphisms in the NOD2 Gene Are Associated with Resistance or Susceptibility to Tuberculosis Disease in African Americans. J. Infect. Dis. 2008, 197, 1713–1716. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, S.; Till, A.; Sina, C.; Arlt, A.; Grasberger, H.; Schreiber, S.; Rosenstiel, P. DUOX2-Derived Reactive Oxygen Species Are Effectors of NOD2-Mediated Antibacterial Responses. J. Cell Sci. 2009, 122, 3522–3530. [Google Scholar] [CrossRef] [PubMed]
- Kleinnijenhuis, J.; Quintin, J.; Preijers, F.; Joosten, L.A.B.; Ifrim, D.C.; Saeed, S.; Jacobs, C.; van Loenhout, J.; de Jong, D.; Hendrik, S.; et al. Bacille Calmette-Guérin Induces NOD2-Dependent Nonspecific Protection from Reinfection via Epigenetic Reprogramming of Monocytes. Proc. Natl. Acad. Sci. USA 2012, 109, 17537–17542. [Google Scholar] [CrossRef]
- Arts, R.J.W.; Blok, B.A.; Aaby, P.; Joosten, L.A.B.; de Jong, D.; van der Meer, J.W.M.; Benn, C.S.; van Crevel, R.; Netea, M.G. Long-Term in Vitro and in Vivo Effects of γ-Irradiated BCG on Innate and Adaptive Immunity. J. Leukoc. Biol. 2015, 98, 995–1001. [Google Scholar] [CrossRef]
- Wannigama, D.L.; Jacquet, A. NOD2-Dependent BCG-Induced Trained Immunity: A Way to Regulate Innate Responses to SARS-CoV2? Int. J. Infect. Dis. 2020, 101, 52–55. [Google Scholar] [CrossRef] [PubMed]
- Israelow, B.; Song, E.; Mao, T.; Lu, P.; Meir, A.; Liu, F.; Alfajaro, M.M.; Wei, J.; Dong, H.; Homer, R.J.; et al. Mouse Model of SARS-CoV-2 Reveals Inflammatory Role of Type I Interferon Signaling. J. Exp. Med. 2020, 217, e20201241. [Google Scholar] [CrossRef]
- Koeken, V.A.C.M.; Verrall, A.J.; Netea, M.G.; Hill, P.C.; van Crevel, R. Trained Innate Immunity and Resistance to Mycobacterium tuberculosis Infection. Clin. Microbiol. Infect. 2019, 25, 1468–1472. [Google Scholar] [CrossRef]
- Hilligan, K.L.; Namasivayam, S.; Clancy, C.S.; O’Mard, D.; Oland, S.D.; Robertson, S.J.; Baker, P.J.; Castro, E.; Garza, N.L.; Lafont, B.A.P.; et al. Intravenous Administration of BCG Protects Mice against Lethal SARS-CoV-2 Challenge. J. Exp. Med. 2022, 219, e20211862. [Google Scholar] [CrossRef]
- Kang, T.J.; Chae, G.-T. The Role of Intracellular Receptor NODs for Cytokine Production by Macrophages Infected with Mycobacterium leprae. Immun. Netw. 2011, 11, 424–427. [Google Scholar] [CrossRef] [Green Version]
- Mahapatra, S.; Crick, D.C.; McNeil, M.R.; Brennan, P.J. Unique Structural Features of the Peptidoglycan of Mycobacterium leprae. J. Bacteriol. 2008, 190, 655–661. [Google Scholar] [CrossRef]
- Schenk, M.; Mahapatra, S.; Le, P.; Kim, H.J.; Choi, A.W.; Brennan, P.J.; Belisle, J.T.; Modlin, R.L. Human NOD2 Recognizes Structurally Unique Muramyl Dipeptides from Mycobacterium leprae. Infect. Immun. 2016, 84, 2429–2438. [Google Scholar] [CrossRef]
- Lee, J.Y.; Lee, M.S.; Kim, D.J.; Yang, S.J.; Lee, S.J.; Noh, E.J.; Shin, S.J.; Park, J.H. Nucleotide-Binding Oligomerization Domain 2 Contributes to Limiting Growth of Mycobacterium abscessus in the Lung of Mice by Regulating Cytokines and Nitric Oxide Production. Front. Immunol. 2017, 8, 1477. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.H.; Park, J.Y.; Kim, D.Y.; Lee, T.S.; Jung, D.H.; Kim, Y.J.; Lee, Y.J.; Lee, Y.J.; Seo, I.S.; Song, E.J.; et al. Type I Interferons Are Involved in the Intracellular Growth Control of Mycobacterium abscessus by Mediating NOD2-Induced Production of Nitric Oxide in Macrophages. Front. Immunol. 2021, 12, 738070. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, N.B.; Oliveira, F.S.; Marinho, F.A.V.; de Almeida, L.A.; Fahel, J.S.; Báfica, A.; Rothfuchs, A.G.; Zamboni, D.S.; Caliari, M.v.; Oliveira, S.C. Nucleotide-Binding Oligomerization Domain-2 (NOD2) Regulates Type-1 Cytokine Responses to Mycobacterium avium but Is Not Required for Host Control of Infection. Microbes Infect. 2015, 17, 337–344. [Google Scholar] [CrossRef]
- Ferwerda, G.; Kullberg, B.J.; de Jong, D.J.; Girardin, S.E.; Langenberg, D.M.L.; van Crevel, R.; Ottenhoff, T.H.M.; van der Meer, J.W.M.; Netea, M.G. Mycobacterium paratuberculosis Is Recognized by Toll-like Receptors and NOD2. J. Leukoc. Biol. 2007, 82, 1011–1018. [Google Scholar] [CrossRef]
- Biswas, A.; Meissner, T.B.; Kawai, T.; Kobayashi, K.S. Cutting Edge: Impaired MHC Class I Expression in Mice Deficient for Nlrc5/CITA. J. Immunol. 2012, 189, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wang, Y.; Chen, F.; Huang, Y.; Zhu, S.; Leng, Q.; Wang, H.; Shi, Y.; Qian, Y. NLRC5 Regulates MHC Class I Antigen Presentation in Host Defense against Intracellular Pathogens. Cell Res. 2012, 22, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Caruso, R.; Warner, N.; Inohara, N.; Núñez, G. NOD1 and NOD2: Signaling, Host Defense, and Inflammatory Disease. Immunity 2014, 41, 898–908. [Google Scholar] [CrossRef]
- Davis, B.K.; Wen, H.; Ting, J.P.Y. The Inflammasome NLRs in Immunity, Inflammation, and Associated Diseases. Annu. Rev. Immunol. 2011, 29, 707. [Google Scholar] [CrossRef] [Green Version]
- Ogura, Y.; Bonen, D.K.; Inohara, N.; Nicolae, D.L.; Chen, F.F.; Ramos, R.; Britton, H.; Moran, T.; Karaliuskas, R.; Duerr, R.H.; et al. A Frameshift Mutation in NOD2 Associated with Susceptibility to Crohn’s Disease. Nature 2001, 411, 603–606. [Google Scholar] [CrossRef]
- Dugan, J.; Griffiths, E.; Snow, P.; Rosenzweig, H.; Lee, E.; Brown, B.; Carr, D.W.; Rose, C.; Rosenbaum, J.; Davey, M.P. A Blau Syndrome-Associated Nod2 Mutation Alters Expression of Full Length NOD2 and Limits Responses to Muramyl Dipeptide in Knock-in Mice. J. Immunol. 2015, 194, 349. [Google Scholar] [CrossRef]
- Kanazawa, N.; Okafuji, I.; Kambe, N.; Nishikomori, R.; Nakata-Hizume, M.; Nagai, S.; Fuji, A.; Yuasa, T.; Manki, A.; Sakurai, Y.; et al. Early-Onset Sarcoidosis and CARD15 Mutations with Constitutive Nuclear Factor-ΚB Activation: Common Genetic Etiology with Blau Syndrome. Blood 2005, 105, 1195–1197. [Google Scholar] [CrossRef]
- Chamaillard, M.; Philpott, D.; Girardin, S.E.; Zouali, H.; Lesage, S.; Chareyre, F.; Bui, T.H.; Giovannini, M.; Zaehringer, U.; Penard-Lacronique, V.; et al. Gene–Environment Interaction Modulated by Allelic Heterogeneity in Inflammatory Diseases. Proc. Natl. Acad. Sci. USA 2003, 100, 3455. [Google Scholar] [CrossRef]
- Hugot, J.P.; Zaccaria, I.; Cavanaugh, J.; Yang, H.; Vermeire, S.; Lappalainen, M.; Schreiber, S.; Annese, V.; Jewell, D.P.; Fowler, E.V.; et al. Prevalence of CARD15/NOD2 mutations in Caucasian healthy people. Am. J. Gastroenterol. 2007, 6, 1259–1267. [Google Scholar] [CrossRef]
- Li, J.; Moran, T.; Swanson, E.; Julian, C.; Harris, J.; Bonen, D.K.; Hedl, M.; Nicolae, D.L.; Abraham, C.; Cho, J.H. Regulation of IL-8 and IL-1beta Expression in Crohn’s Disease Associated NOD2/CARD15 Mutations. Hum. Mol. Genet. 2004, 13, 1715–1725. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Ferwerda, G.; de Jong, D.J.; Jansen, T.; Jacobs, L.; Kramer, M.; Naber, T.H.J.; Drenth, J.P.H.; Girardin, S.E.; Kullberg, B.J.; et al. Nucleotide-Binding Oligomerization Domain-2 Modulates Specific TLR Pathways for the Induction of Cytokine Release. J. Immunol. 2005, 174, 6518–6523. [Google Scholar] [CrossRef] [PubMed]
- Bonen, D.K.; Ogura, Y.; Nicolae, D.L.; Inohara, N.; Saab, L.; Tanabe, T.; Chen, F.F.; Foster, S.J.; Duerr, R.H.; Brant, S.R.; et al. Crohn’s Disease-Associated NOD2 Variants Share a Signaling Defect in Response to Lipopolysaccharide and Peptidoglycan. Gastroenterology 2003, 124, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Caso, F.; Galozzi, P.; Costa, L.; Sfriso, P.; Cantarini, L.; Punzi, L. Autoinflammatory Granulomatous Diseases: From Blau Syndrome and Early-Onset Sarcoidosis to NOD2-Mediated Disease and Crohn’s Disease. RMD Open 2015, 1, e000097. [Google Scholar] [CrossRef]
- Hall, N.B.; Igo, R.P.; Malone, L.L.; Truitt, B.; Schnell, A.; Tao, L.; Okware, B.; Nsereko, M.; Chervenak, K.; Lancioni, C.; et al. Polymorphisms in TICAM2 and IL1B Are Associated with TB. Genes Immun. 2015, 16, 127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, M.; Jiang, F.; Zhang, W.; Li, F.; Wei, L.; Liu, J.; Xue, Y.; Deng, X.; Wu, F.; Zhang, L.; et al. A Novel Single Nucleotide Polymorphism within the NOD2 Gene Is Associated with Pulmonary Tuberculosis in the Chinese Han, Uygur and Kazak Populations. BMC Infect. Dis. 2012, 12, 91. [Google Scholar] [CrossRef]
- Zhang, F.-R.; Huang, W.; Chen, S.-M.; Sun, L.-D.; Liu, H.; Li, Y.; Cui, Y.; Yan, X.-X.; Yang, H.-T.; Yang, R.-D.; et al. Genomewide Association Study of Leprosy. NEJM 2009, 361, 2609–2618. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Dai, Y.; Tang, S.; Wang, J. Polymorphisms of NOD2 and the Risk of Tuberculosis: A Validation Study in the Chinese Population. Int. J. Immunogenet. 2012, 39, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Grant, A.V.; Alter, A.; Huong, N.T.; Orlova, M.; van Thuc, N.; Ba, N.N.; Thai, V.H.; Abel, L.; Schurr, E.; Alcais, A. Crohn’s Disease Susceptibility Genes Are Associated With Leprosy in the Vietnamese Population. J. Infect. Dis. 2012, 206, 1763–1767. [Google Scholar] [CrossRef]
- Leturiondo, A.L.; Noronha, A.B.; Mendonca, C.Y.R.; de Oliveira Ferreira, C.; Alvarado-Arnez, L.E.; de Neves Manta, F.S.; de Lima Bezerra, O.C.; de Carvalho, E.F.; Moraes, M.O.; da Costa Rodrigues, F.; et al. Association of NOD2 and IFNG Single Nucleotide Polymorphisms with Leprosy in the Amazon Ethnic Admixed Population. PLoS Negl. Trop. Dis. 2020, 14, e0008247. [Google Scholar] [CrossRef] [PubMed]
- Hysi, P.; Kabesch, M.; Moffatt, M.F.; Schedel, M.; Carr, D.; Zhang, Y.; Boardman, B.; von Mutius, E.; Weiland, S.K.; Leupold, W.; et al. NOD1 Variation, Immunoglobulin E and Asthma. Hum. Mol. Genet. 2005, 14, 935–941. [Google Scholar] [CrossRef] [PubMed]
- McGovern, D.P.B.; Hysi, P.; Ahmad, T.; van Heel, D.A.; Moffatt, M.F.; Carey, A.; Cookson, W.O.C.; Jewell, D.P. Association between a Complex Insertion/Deletion Polymorphism in NOD1 (CARD4) and Susceptibility to Inflammatory Bowel Disease. Hum Mol. Genet. 2005, 14, 1245–1250. [Google Scholar] [CrossRef] [PubMed]
- Zupin, L.; Navarra, C.O.; Robino, A.; Bevilacqua, L.; di Lenarda, R.; Gasparini, P.; Crovella, S. NLRC5 Polymorphism Is Associated with Susceptibility to Chronic Periodontitis. Immunobiology 2017, 222, 704–708. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Liu, L.; Lu, Y.; Gu, Y.; Zhao, J.; Chen, B.; Zhou, W.; Su, X. NLRP3, NLRC4 and NLRC5 Gene Polymorphisms Associate with Susceptibility of Pulmonary Aspergillosis in Non-Neutropenic Patients. J. Clin. Med. 2022, 11, 1870. [Google Scholar] [CrossRef]
- Huang, S.; Zhao, L.; Kim, K.; Dong, S.L.; Hwang, D.H. Inhibition of Nod2 Signaling and Target Gene Expression by Curcumin. Mol. Pharmacol. 2008, 74, 274. [Google Scholar] [CrossRef] [Green Version]
- Rüngeler, P.; Castro, V.; Mora, G.; Gören, N.; Vichnewski, W.; Pahl, H.L.; Merfort, I.; Schmidt, T.J. Inhibition of Transcription Factor NF-ΚB by Sesquiterpene Lactones: A Proposed Molecular Mechanism of Action. Bioorg. Med. Chem. 1999, 7, 2343–2352. [Google Scholar] [CrossRef]
- Ly, G.; Knorre, A.; Schmidt, T.J.; Pahl, H.L.; Merfort, I. The Anti-Inflammatory Sesquiterpene Lactone Helenalin Inhibits the Transcription Factor NF-KappaB by Directly Targeting P65. J. Biol. Chem. 1998, 273, 33508–33516. [Google Scholar]
- Zhao, L.; Kwon, M.J.; Huang, S.; Lee, J.Y.; Fukase, K.; Inohara, N.; Hwang, D.H. Differential Modulation of Nods Signaling Pathways by Fatty Acids in Human Colonic Epithelial HCT116 Cells. J. Biol. Chem. 2007, 282, 11618–11628. [Google Scholar] [CrossRef] [PubMed]
- Magnuson, G.; Khan, P.; Yuan, H.; Brown, B.; Divlianska, D.B.; Stonich, D.; Peddibhotla, M.; Su, Y.; Dad, S.; Sergienko, E.; et al. High Throughput Screening Assays for NOD1. In Probe Reports from the NIH Molecular Libraries Program; National Center for Biotechnology Information: Bethesda, MD, USA, 2010. Available online: https://www.ncbi.nlm.nih.gov/books/NBK50683/ (accessed on 5 September 2022).
- Magnuson, G.; Khan, P.; Yuan, H.; Brown, B.; Divlianska, D.B.; Stonich, D.; Peddibhotla, M.; Su, Y.; Dad, S.; Sergienko, E.; et al. High Throughput Screening Assays for NOD1 Inhibitors—Probe 2. In Probe Reports from the NIH Molecular Libraries Program; National Center for Biotechnology Information: Bethesda, MD, USA, 2010. Available online: https://www.ncbi.nlm.nih.gov/books/NBK50701/ (accessed on 5 September 2022).
- Rickard, D.J.; Sehon, C.A.; Kasparcova, V.; Kallal, L.A.; Zeng, X.; Montoute, M.N.; Chordia, T.; Poore, D.D.; Li, H.; Wu, Z.; et al. Identification of Benzimidazole Diamides as Selective Inhibitors of the Nucleotide-Binding Oligomerization Domain 2 (NOD2) Signaling Pathway. PLoS ONE 2013, 8, e69619. [Google Scholar] [CrossRef]
- Jakopin, Ž. Nucleotide-Binding Oligomerization Domain (NOD) Inhibitors: A Rational Approach toward Inhibition of NOD Signaling Pathway. J. Med. Chem. 2014, 57, 6897–6918. [Google Scholar] [CrossRef]
| NLR Family | Member | Stucture | Reference |
|---|---|---|---|
| NLRA | CIITA | 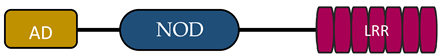 | [16,17] |
| NLRB | NAIP |  | [16,17] |
| NLRC | NOD1 |  | [9,10] |
| NOD2 |  | [9,10] | |
| NLRC4 | 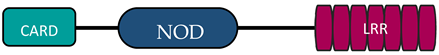 | [9,10] | |
| NLRC3/C5/X1 |  | [9,10] | |
| NLRP | NLRP1 | 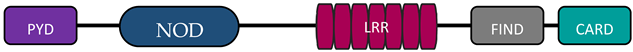 | [4] |
| NLRP2-9, 11-14 | 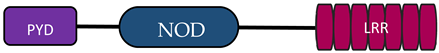 | [4] | |
| NLRP10 |  | [4] |
| Viruses | Receptor | Potiential Signaling Pathways | Refs |
|---|---|---|---|
| Respiratory Syncytial Virus | NOD2 | MAVS-IRF3-IFN pathway | [14] |
| Parainfluenza Virus 3 | NOD2 | MAVS-IRF3-IFN pathway | [14] |
| Influenza A Virus | NOD2 | MAVS-IRF3-IFN pathway | [14] |
| NLRC5 | NLRC5-RFX-CREB1-ATF1-NF-Y-X1/2-Y-MHCI pathway | [77] | |
| Vesicular Stomatitis Virus | NOD2 | MAVS-IRF3-IFN pathway | [14] |
| Middle East Respiratory Syndrome | NOD2 | MAVS-IRF3-IFN pathway | [15] |
| Zika Virus | NOD2 | Unknown | [79] |
| SARS-CoV-2 | NOD1 | LGP2+MDA5-MAVS-IRF3/5-IFN pathway | [81] |
| NLRC5 | STAT1-IRF1-NLRC5 pathway | [83] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Godkowicz, M.; Druszczyńska, M. NOD1, NOD2, and NLRC5 Receptors in Antiviral and Antimycobacterial Immunity. Vaccines 2022, 10, 1487. https://doi.org/10.3390/vaccines10091487
Godkowicz M, Druszczyńska M. NOD1, NOD2, and NLRC5 Receptors in Antiviral and Antimycobacterial Immunity. Vaccines. 2022; 10(9):1487. https://doi.org/10.3390/vaccines10091487
Chicago/Turabian StyleGodkowicz, Magdalena, and Magdalena Druszczyńska. 2022. "NOD1, NOD2, and NLRC5 Receptors in Antiviral and Antimycobacterial Immunity" Vaccines 10, no. 9: 1487. https://doi.org/10.3390/vaccines10091487





