Auxotrophic Lactobacillus Expressing Porcine Rotavirus VP4 Constructed Using CRISPR-Cas9D10A System Induces Effective Immunity in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain, Virus, Cell, Plasmid
2.2. Construction of Nutritionally-Deficient L. casei by CRISPR-Cas9D10A
2.3. Morphological Observation
2.4. Physiological and Biochemical Test
2.5. Complementary Plasmid Generation
2.6. Western Blotting
2.7. Animal Immunization
2.8. ELISA Assay
2.9. Neutralization Assay
2.10. Statistical Analysis
3. Results
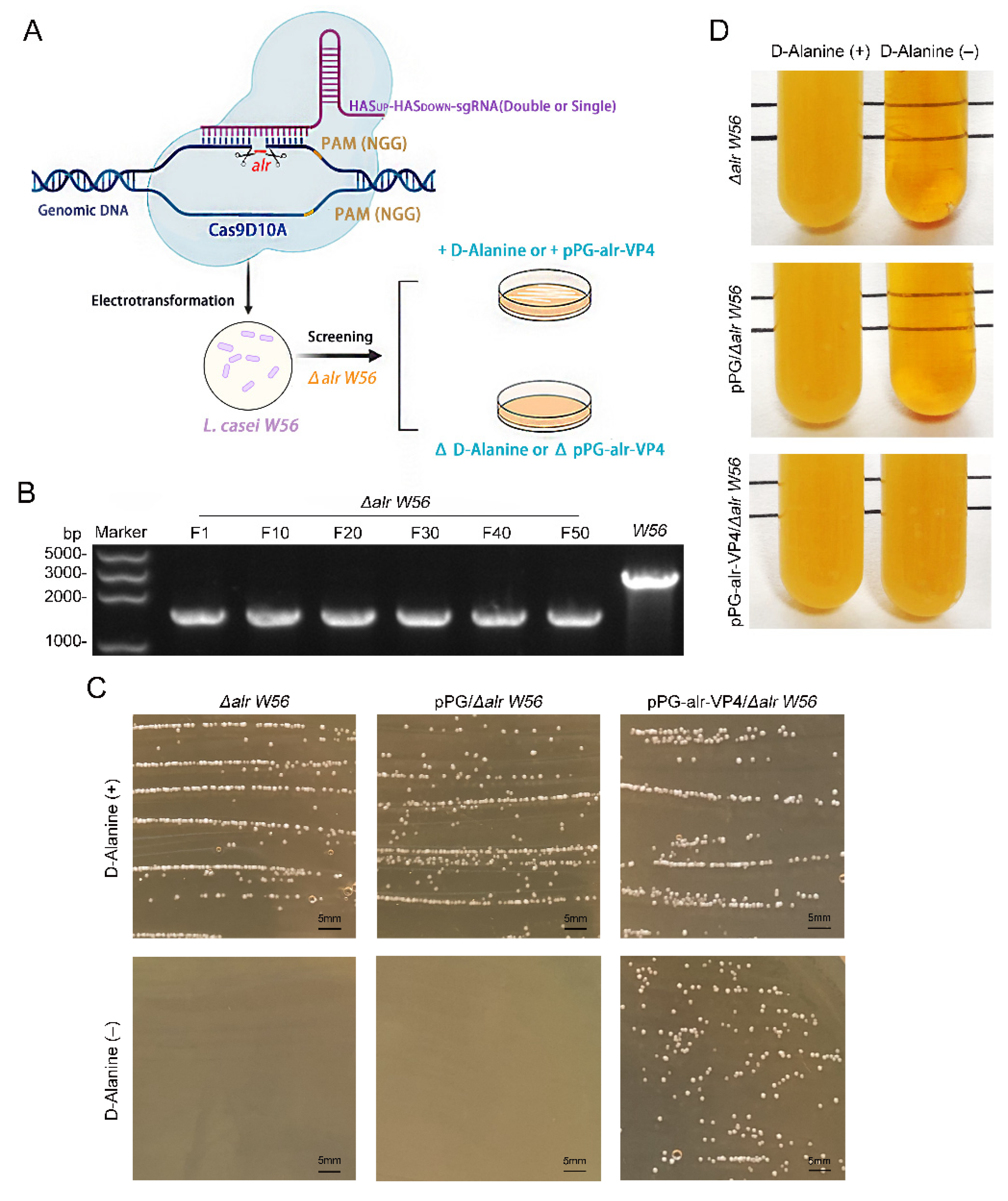
3.1. Construction of alr Nutrition-Deficient L. casei
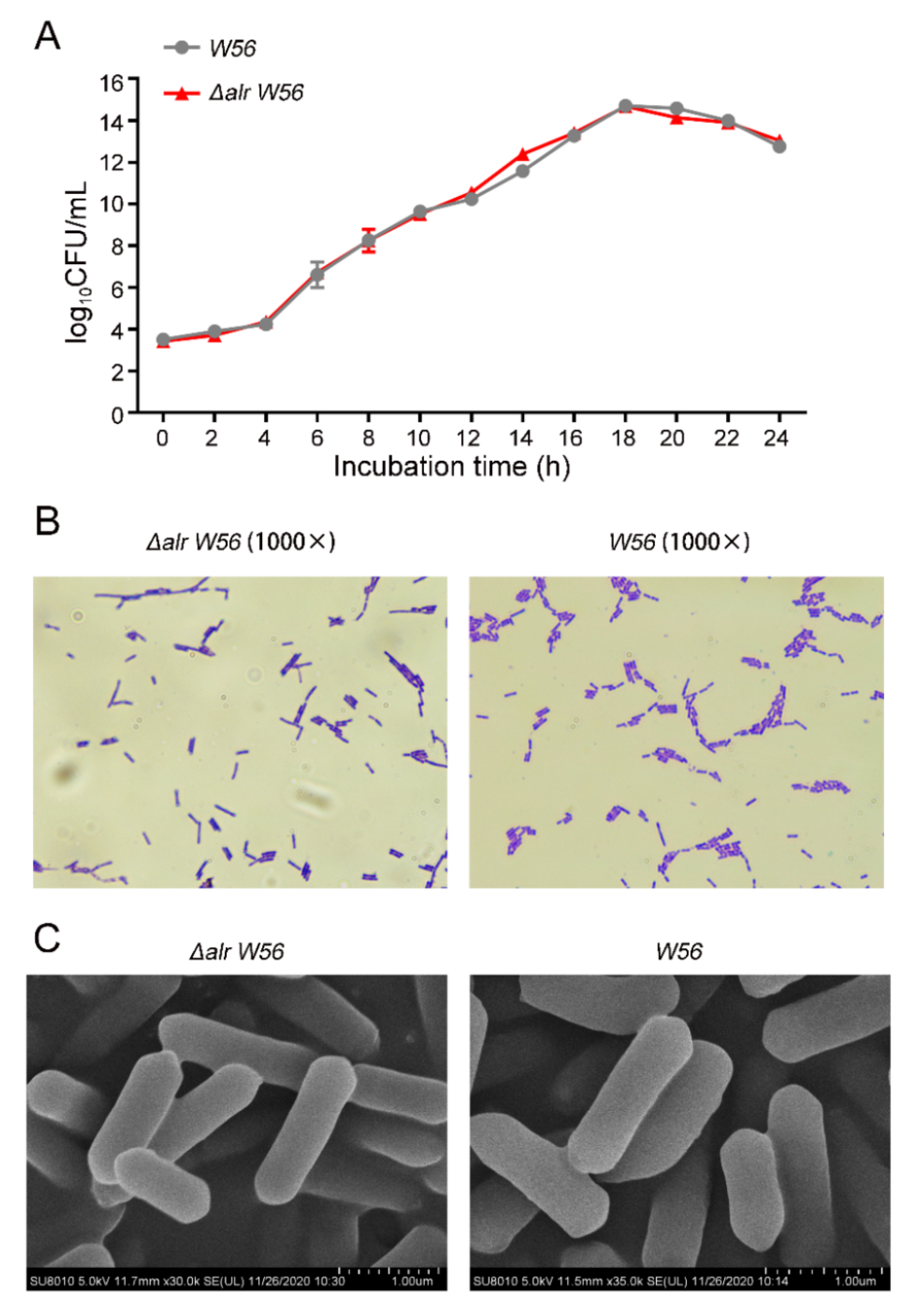
3.2. Biological Characterization of Δalr W56
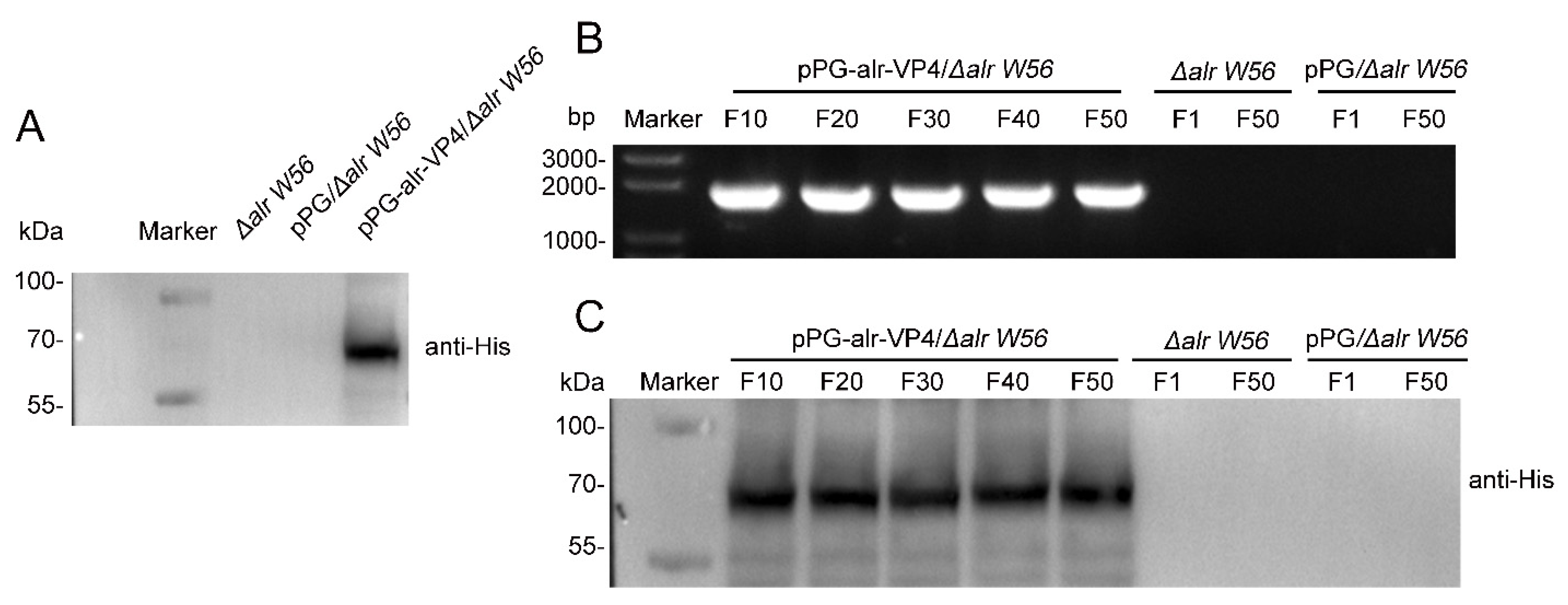
3.3. Inherited Stability of the Δalr W56 Expressing alr-VP4
3.4. Antibody Responses Post Oral Immunization
3.5. In Vitro PoRV-Neutralizing Activity of Antibodies
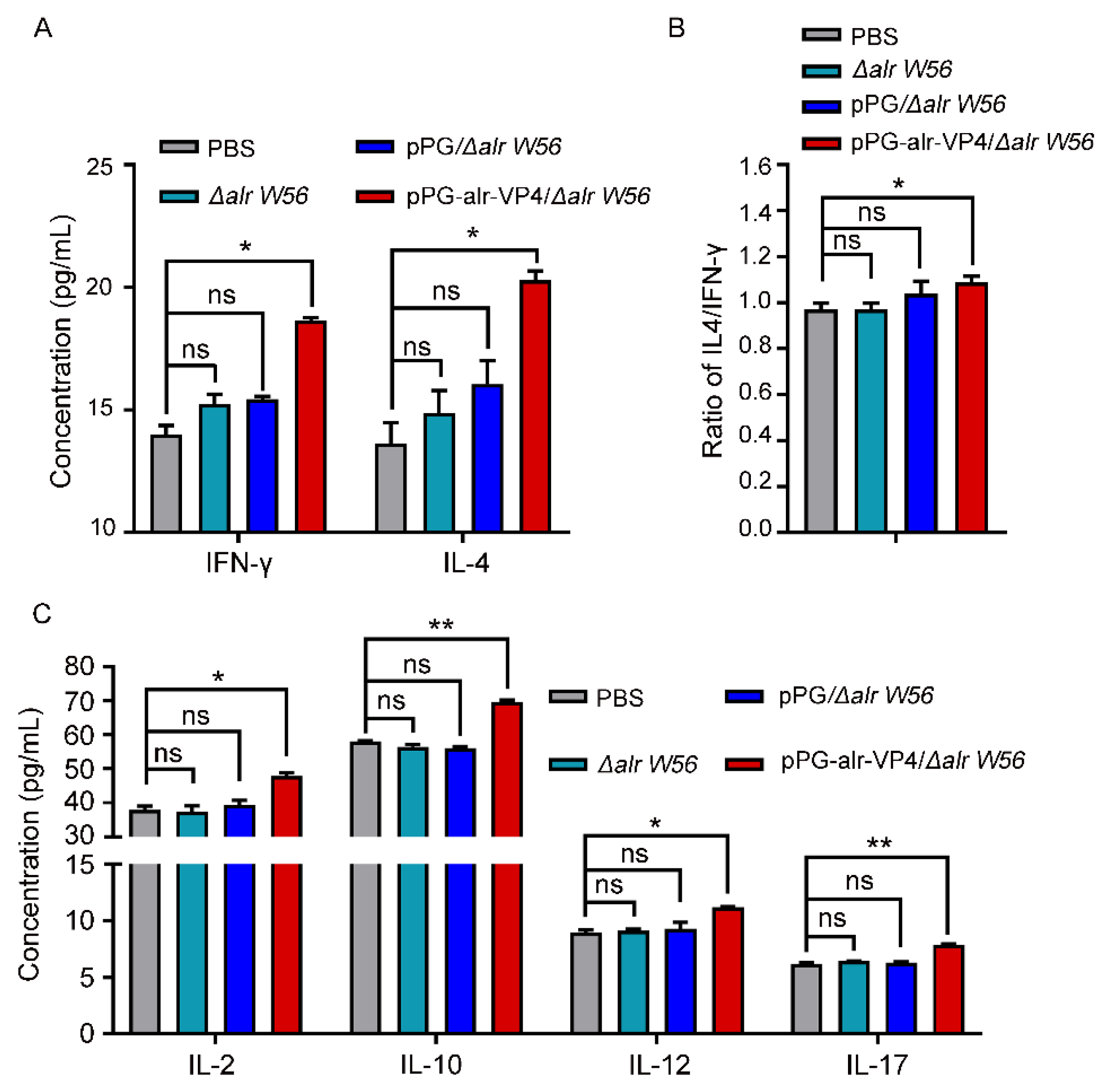
3.6. Cytokine Secretion Level in Immunized Mice
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vlasova, A.N.; Amimo, J.O.; Saif, L.J. Porcine Rotaviruses: Epidemiology, Immune Responses and Control Strategies. Viruses 2017, 9, 48. [Google Scholar] [CrossRef]
- Dhama, K.; Chauhan, R.S.; Mahendran, M.; Malik, S.V. Rotavirus diarrhea in bovines and other domestic animals. Vet. Res. Commun. 2009, 33, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Das, P.J.; Handique, P.J. Molecular characterization of porcine group A rotavirus to contain piglet diarrhea for productivity enhancement in North East India. Virusdisease 2021, 32, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Ma, G.P.; Li, G.W.; Qiao, X.Y.; Ge, J.W.; Tang, L.J.; Liu, M.; Liu, L.W. Oral vaccination with the porcine rotavirus VP4 outer capsid protein expressed by Lactococcus lactis induces specific antibody production. J. Biomed. Biotechnol. 2010, 2010, 708460. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.H.; Hogarty, M.P.; Harris, V.C.; Baldridge, M.T. The Complex Interactions Between Rotavirus and the Gut Microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 586751. [Google Scholar] [CrossRef]
- Suzuki, T.; Hasebe, A.; Miyazaki, A.; Tsunemitsu, H. Analysis of genetic divergence among strains of porcine rotavirus C, with focus on VP4 and VP7 genotypes in Japan. Virus Res. 2015, 197, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Park, J.G.; Alfajaro, M.M.; Cho, E.H.; Kim, J.Y.; Soliman, M.; Baek, Y.B.; Park, C.H.; Lee, J.H.; Son, K.Y.; Cho, K.O.; et al. Development of a live attenuated trivalent porcine rotavirus A vaccine against disease caused by recent strains most prevalent in South Korea. Vet. Res. 2019, 50, 2. [Google Scholar] [CrossRef]
- Carvalho, M.F.; Gill, D. Rotavirus vaccine efficacy: Current status and areas for improvement. Hum. Vaccines Immunother. 2019, 15, 1237–1250. [Google Scholar] [CrossRef] [PubMed]
- Boyaka, P.N. Inducing Mucosal IgA: A Challenge for Vaccine Adjuvants and Delivery Systems. J. Immunol. 2017, 199, 9–16. [Google Scholar] [CrossRef]
- Miquel-Clopes, A.; Bentley, E.G.; Stewart, J.P.; Carding, S.R. Mucosal vaccines and technology. Clin. Exp. Immunol. 2019, 196, 205–214. [Google Scholar] [CrossRef] [Green Version]
- Ding, C.; Ma, J.; Dong, Q.; Liu, Q. Live bacterial vaccine vector and delivery strategies of heterologous antigen: A review. Immunol. Lett. 2018, 197, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Francis, M.J. Recent Advances in Vaccine Technologies. Vet. Clin. N. Am. Small Anim. Pract. 2018, 48, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Vinod, N.; Noh, H.B.; Oh, S.; Ji, S.; Park, H.J.; Lee, K.S.; Kim, S.C.; Park, H.O.; Yang, J.S.; Choi, C.W. A Salmonella typhimurium ghost vaccine induces cytokine expression in vitro and immune responses in vivo and protects rats against homologous and heterologous challenges. PLoS ONE 2017, 12, e0185488. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Jiang, X.; Jiang, Y.; Tang, L.; Xu, Y.; Qiao, X.; Min, L.; Wen, C.; Ma, G.; Li, Y. Oral Immunization against PEDV with Recombinant Lactobacillus casei Expressing Dendritic Cell-Targeting Peptide Fusing COE Protein of PEDV in Piglets. Viruses 2018, 10, 106. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Hou, X.; Tang, L.; Jiang, Y.; Ma, G.; Li, Y. A phase trial of the oral Lactobacillus casei vaccine polarizes Th2 cell immunity against transmissible gastroenteritis coronavirus infection. Appl. Microbiol. Biotechnol. 2016, 100, 7457–7469. [Google Scholar] [CrossRef]
- Zhang, W.; Wen, K.; Azevedo, M.S.; Gonzalez, A.; Saif, L.J.; Li, G.; Yousef, A.E.; Yuan, L. Lactic acid bacterial colonization and human rotavirus infection influence distribution and frequencies of monocytes/macrophages and dendritic cells in neonatal gnotobiotic pigs. Vet. Immunol. Immunopathol. 2008, 121, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Blum, S.; Schiffrin, E.J. Intestinal microflora and homeostasis of the mucosal immune response: Implications for probiotic bacteria? Curr. Issues Intest. Microbiol. 2003, 4, 53–60. [Google Scholar]
- Breidt, F.; Fleming, H.P. Competitive Growth of Genetically Marked Malolactic-Deficient Lactobacillus plantarum in Cucumber Fermentations. Appl. Environ. Microbiol. 1992, 58, 3845–3849. [Google Scholar] [CrossRef]
- De Vrese, M.; Marteau, P.R. Probiotics and prebiotics: Effects on diarrhea. J. Nutr. 2007, 137, 803S–811S. [Google Scholar] [CrossRef]
- Bron, P.A.; Benchimol, M.G.; Lambert, J.; Palumbo, E.; Deghorain, M.; Delcour, J.; De Vos, W.M.; Kleerebezem, M.; Hols, P. Use of the alr gene as a food-grade selection marker in lactic acid bacteria. Appl. Environ. Microbiol. 2002, 68, 5663–5670. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Mathiesen, G.; Fredriksen, L.; Kittl, R.; Nguyen, T.H.; Eijsink, V.G.; Haltrich, D.; Peterbauer, C.K. A food-grade system for inducible gene expression in Lactobacillus plantarum using an alanine racemase-encoding selection marker. J. Agric. Food Chem. 2011, 59, 5617–5624. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Li, G.; Wang, X.; Li, X.; Liu, M.; Li, Y. Recombinant porcine rotavirus VP4 and VP4-LTB expressed in Lactobacillus casei induced mucosal and systemic antibody responses in mice. BMC Microbiol. 2009, 9, 249. [Google Scholar] [CrossRef]
- Wang, Y.; Feng, B.; Niu, C.; Jia, S.; Sun, C.; Wang, Z.; Jiang, Y.; Cui, W.; Wang, L.; Xu, Y. Dendritic Cell Targeting of Bovine Viral Diarrhea Virus E2 Protein Expressed by Lactobacillus casei Effectively Induces Antigen-Specific Immune Responses via Oral Vaccination. Viruses 2019, 11, 575. [Google Scholar] [CrossRef]
- Desselberger, U.; Huppertz, H.I. Immune responses to rotavirus infection and vaccination and associated correlates of protection. J. Infect. Dis. 2011, 203, 188–195. [Google Scholar] [CrossRef]
- Li, F.; Wang, X.; Ma, R.; Wu, W.; Teng, F.; Cheng, X.; Jiang, Y.; Zhou, H.; Wang, L.; Tang, L.; et al. Oral Immunization with Lactobacillus casei Expressing the Porcine Circovirus Type 2 Cap and LTB Induces Mucosal and Systemic Antibody Responses in Mice. Viruses 2021, 13, 1302. [Google Scholar] [CrossRef]
- Gorziglia, M.; Larralde, G.; Kapikian, A.Z.; Chanock, R.M. Antigenic relationships among human rotaviruses as determined by outer capsid protein VP4. Proc. Natl. Acad. Sci. USA 1990, 87, 7155–7159. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Candelero-Rueda, R.A.; Saif, L.J.; Vlasova, A.N. Infection of porcine small intestinal enteroids with human and pig rotavirus A strains reveals contrasting roles for histo-blood group antigens and terminal sialic acids. PLoS Pathog. 2021, 17, e1009237. [Google Scholar] [CrossRef]
- Bermudez-Humaran, L.G. Lactococcus lactis as a live vector for mucosal delivery of therapeutic proteins. Hum. Vaccines 2009, 5, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Matthijnssens, J.; Ciarlet, M.; McDonald, S.M.; Attoui, H.; Banyai, K.; Brister, J.R.; Buesa, J.; Esona, M.D.; Estes, M.K.; Gentsch, J.R.; et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch. Virol. 2011, 156, 1397–1413. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.W.; Myers, L.E.; Ray, L.; Song, S.C.; Nasr, T.R.; Berardinelli, A.J.; Kundu, K.; Murthy, N.; Hansen, J.M.; Neish, A.S. Lactobacillus rhamnosus blocks inflammatory signaling in vivo via reactive oxygen species generation. Free Radic. Biol. Med. 2009, 47, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.Y.; Guo, C.Q.; Wang, Z.; Yu, M.L.; Gao, S.; Bukhari, S.M.; Tang, L.J.; Xu, Y.G.; Li, Y.J. Directed chromosomal integration and expression of porcine rotavirus outer capsid protein VP4 in Lactobacillus casei ATCC393. Appl. Microbiol. Biotechnol. 2016, 100, 9593–9604. [Google Scholar] [CrossRef] [PubMed]
- Bleau, C.; Monges, A.; Rashidan, K.; Laverdure, J.P.; Lacroix, M.; Van Calsteren, M.R.; Millette, M.; Savard, R.; Lamontagne, L. Intermediate chains of exopolysaccharides from Lactobacillus rhamnosus RW-9595M increase IL-10 production by macrophages. J. Appl. Microbiol. 2010, 108, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Merenstein, D.; Murphy, M.; Fokar, A.; Hernandez, R.K.; Park, H.; Nsouli, H.; Sanders, M.E.; Davis, B.A.; Niborski, V.; Tondu, F.; et al. Use of a fermented dairy probiotic drink containing Lactobacillus casei (DN-114 001) to decrease the rate of illness in kids: The DRINK study. A patient-oriented, double-blind, cluster-randomized, placebo-controlled, clinical trial. Eur. J. Clin. Nutr. 2010, 64, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Mohamadzadeh, M.; Olson, S.; Kalina, W.V.; Ruthel, G.; Demmin, G.L.; Warfield, K.L.; Bavari, S.; Klaenhammer, T.R. Lactobacilli activate human dendritic cells that skew T cells toward T helper 1 polarization. Proc. Natl. Acad. Sci. USA 2005, 102, 2880–2885. [Google Scholar] [CrossRef]
- Wang, P.; Li, Y.; Xiao, H.; Shi, Y.; Le, G.W.; Sun, J. Isolation of lactobacillus reuteri from Peyer’s patches and their effects on sIgA production and gut microbiota diversity. Mol. Nutr. Food Res. 2016, 60, 2020–2030. [Google Scholar] [CrossRef]
- Yang, D.; Yu, X.; Wu, Y.; Chen, X.; Wei, H.; Shah, N.P.; Xu, F. Enhancing flora balance in the gastrointestinal tract of mice by lactic acid bacteria from Chinese sourdough and enzyme activities indicative of metabolism of protein, fat, and carbohydrate by the flora. J. Dairy Sci. 2016, 99, 7809–7820. [Google Scholar] [CrossRef]
- O’Sullivan, D.J.; Klaenhammer, T.R. High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene 1993, 137, 227–231. [Google Scholar] [CrossRef]
- Goh, Y.J.; Barrangou, R. Portable CRISPR-Cas9(N) System for Flexible Genome Engineering in Lactobacillus acidophilus, Lactobacillus gasseri, and Lactobacillus paracasei. Appl. Environ. Microbiol. 2021, 87, e02669-20. [Google Scholar] [CrossRef]
- Hidalgo-Cantabrana, C.; Goh, Y.J.; Pan, M.; Sanozky-Dawes, R.; Barrangou, R. Genome editing using the endogenous type I CRISPR-Cas system in Lactobacillus crispatus. Proc. Natl. Acad. Sci. USA 2019, 116, 15774–15783. [Google Scholar] [CrossRef]
- Schuster, J.A.; Vogel, R.F.; Ehrmann, M.A. Characterization and distribution of CRISPR-Cas systems in Lactobacillus sakei. Arch. Microbiol. 2019, 201, 337–347. [Google Scholar] [CrossRef]
- Song, X.; Huang, H.; Xiong, Z.; Ai, L.; Yang, S. CRISPR-Cas9(D10A) Nickase-Assisted Genome Editing in Lactobacillus casei. Appl. Environ. Microbiol. 2017, 83, e01259-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Li, W.; Ujiroghene, O.J.; Yang, Y.; Lu, J.; Zhang, S.; Pang, X.; Lv, J. Occurrence and Diversity of CRISPR Loci in Lactobacillus casei Group. Front. Microbiol. 2020, 11, 624. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.H.; van Pijkeren, J.P. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014, 42, e131. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Xin, Y.; Zhang, Y.; Gu, X.; Kong, J. A rapid and versatile tool for genomic engineering in Lactococcus lactis. Microb. Cell Factories 2019, 18, 22. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Luo, M.L.; Leenay, R.T.; Beisel, C.L. Current and future prospects for CRISPR-based tools in bacteria. Biotechnol. Bioeng. 2016, 113, 930–943. [Google Scholar] [CrossRef]
- Klasse, P.J.; Sattentau, Q.J. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J. Gen. Virol. 2002, 83, 2091–2108. [Google Scholar] [CrossRef]
- Brandtzaeg, P. Induction of secretory immunity and memory at mucosal surfaces. Vaccine 2007, 25, 5467–5484. [Google Scholar] [CrossRef]
- Hanson, L.A.; Ahlstedt, S.; Andersson, B.; Carlsson, B.; Cole, M.F.; Cruz, J.R.; Dahlgren, U.; Ericsson, T.H.; Jalil, F.; Khan, S.R.; et al. Mucosal immunity. Ann. N. Y. Acad. Sci. 1983, 409, 1–21. [Google Scholar] [CrossRef]
- Xu, F.; Newby, J.M.; Schiller, J.L.; Schroeder, H.A.; Wessler, T.; Chen, A.; Forest, M.G.; Lai, S.K. Modeling Barrier Properties of Intestinal Mucus Reinforced with IgG and Secretory IgA against Motile Bacteria. ACS Infect. Dis. 2019, 5, 1570–1580. [Google Scholar] [CrossRef]
- Bessay, M.; Le Vern, Y.; Kerboeuf, D.; Yvore, P.; Quere, P. Changes in intestinal intra-epithelial and systemic T-cell subpopulations after an Eimeria infection in chickens: Comparative study between E acervulina and E tenella. Vet. Res. 1996, 27, 503–514. [Google Scholar] [PubMed]
- Blumberg, R.S.; Lencer, W.I.; Zhu, X.; Kim, H.S.; Claypool, S.; Balk, S.P.; Saubermann, L.J.; Colgan, S.P. Antigen presentation by intestinal epithelial cells. Immunol. Lett. 1999, 69, 7–11. [Google Scholar] [CrossRef]
- Zinkernagel, R.M. Maternal antibodies, childhood infections, and autoimmune diseases. N. Engl. J. Med. 2001, 345, 1331–1335. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.H.; Cao, X.H.; Du, X.G.; Feng, H.B.; Di, W.; He, S.; Zeng, X.Y. Mucosal and systemic immunity in mice after intranasal immunization with recombinant Lactococcus lactis expressing ORF6 of PRRSV. Cell. Immunol. 2014, 287, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Nan, C.L.; Lei, Z.L.; Zhao, Z.J.; Shi, L.H.; Ouyang, Y.C.; Song, X.F.; Sun, Q.Y.; Chen, D.Y. Increased Th1/Th2 (IFN-gamma/IL-4) Cytokine mRNA ratio of rat embryos in the pregnant mouse uterus. J. Reprod. Dev. 2007, 53, 219–228. [Google Scholar] [CrossRef]
- Ivanov, I.I.; McKenzie, B.S.; Zhou, L.; Tadokoro, C.E.; Lepelley, A.; Lafaille, J.J.; Cua, D.J.; Littman, D.R. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 2006, 126, 1121–1133. [Google Scholar] [CrossRef] [Green Version]


| ID | sgRNAs Sequence (5′-3′) |
|---|---|
| Double-sgRNA-1 | GCAGCTTTTCGCTGTGGTCA |
| Double-sgRNA-2 | GGTTGGGCAAAGTGGCGATC |
| Single-sgRNA | GCAGCTTTTCGCTGTGGTCA |
| ID | Primer Sequence (5′-3′) | Target |
|---|---|---|
| alr-HASup-F/R | F: tctttttctaaactagggccc 1 GAATCGCGCCACTGGCCA R: aaagaaggtcctg 1 TGATGGTGGCTCGTATGCCC | alr-upstream homology arms |
| alr-HASdown-F/R | F: acaccatca 1 CAGGACCTTCTTTTTCTAAAATTACCT R: agtcggtgctttttttgag 1 CCAAACGAAATCGAATATTTGCA | alr-downstream homology arms |
| alr-F/R | F: GACCAGACCCACTGAAATCG R: AACCACCAACAGCAGAAGAA | alr |
| pPG-F/R | F: GACAGCCTTAAACAGAAAACC R: GCAGTTCCCTACTCTCGC | alr-VP4 |
| Groups | Dosage | Number of Mice |
|---|---|---|
| pPG-alr-VP4/Δalr W56 | 1010 CFU | 30 |
| pPG/Δalr W56 | 1010 CFU | 30 |
| L. casei Δalr W56 | 1010 CFU | 30 |
| PBS | 100 µL | 30 |
| Strains ID | Viability (%) | ||||
|---|---|---|---|---|---|
| 0.1% Bile Salt | 0.3% Bile Salt | pH2 | pH3 | pH4.5 | |
| L. casei Δalr W56 | 1.82 ± 0.51 | 0.42 ± 0.04 | 0.76 ± 0.12 | 5.05 ± 1.55 | 50.12 ± 1.4 |
| L. casei W56 | 1.88 ± 0.41 | 0.46 ± 0.19 | 0.83 ± 0.17 | 5.78 ± 1.18 | 53.52 ± 2.35 |
| Reaction Type | Nitrate Reduction | Catalase Test | H2S | Indole Test | Gelatin Liquefaction | Glucose Fermentation | Melezitose | Lactose | Maltose | Raffinose | Mannose | Salicin | Mannitol | Melibiose | Rhamnose | Ribose | Sorbose | Sucrose | Xylose | Mushroompolysaccharide | Esculoside | Fructose | Arabinose | Galactose | Cellobiose | Citrate | Malonate | Gluconate | Arginine Dihydrolase | Arginine | Amygdalin |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BIM1 | − | − | − | − | − | + | + | − | + | − | + | + | + | − | − | + | + | + | − | + | + | + | − | + | + | − | − | + | − | − | + |
| L. casei | − | − | − | − | − | + | − | − | + | − | + | + | + | − | − | + | + | + | − | + | + | + | − | + | + | − | − | + | − | − | + |
| Δalr W56 | − | − | − | − | − | + | − | − | + | − | + | + | − | − | − | + | + | + | − | + | + | + | − | + | + | − | − | + | − | − | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Zhao, H.; Zhao, Y.; Sui, L.; Li, F.; Zhang, H.; Li, J.; Jiang, Y.; Cui, W.; Ding, G.; et al. Auxotrophic Lactobacillus Expressing Porcine Rotavirus VP4 Constructed Using CRISPR-Cas9D10A System Induces Effective Immunity in Mice. Vaccines 2022, 10, 1510. https://doi.org/10.3390/vaccines10091510
Zhang H, Zhao H, Zhao Y, Sui L, Li F, Zhang H, Li J, Jiang Y, Cui W, Ding G, et al. Auxotrophic Lactobacillus Expressing Porcine Rotavirus VP4 Constructed Using CRISPR-Cas9D10A System Induces Effective Immunity in Mice. Vaccines. 2022; 10(9):1510. https://doi.org/10.3390/vaccines10091510
Chicago/Turabian StyleZhang, Hailin, Haiyuan Zhao, Yuliang Zhao, Ling Sui, Fengsai Li, Huijun Zhang, Jiaxuan Li, Yanping Jiang, Wen Cui, Guojie Ding, and et al. 2022. "Auxotrophic Lactobacillus Expressing Porcine Rotavirus VP4 Constructed Using CRISPR-Cas9D10A System Induces Effective Immunity in Mice" Vaccines 10, no. 9: 1510. https://doi.org/10.3390/vaccines10091510
APA StyleZhang, H., Zhao, H., Zhao, Y., Sui, L., Li, F., Zhang, H., Li, J., Jiang, Y., Cui, W., Ding, G., Zhou, H., Wang, L., Qiao, X., Tang, L., Wang, X., & Li, Y. (2022). Auxotrophic Lactobacillus Expressing Porcine Rotavirus VP4 Constructed Using CRISPR-Cas9D10A System Induces Effective Immunity in Mice. Vaccines, 10(9), 1510. https://doi.org/10.3390/vaccines10091510





