Updated Insights into the T Cell-Mediated Immune Response against SARS-CoV-2: A Step towards Efficient and Reliable Vaccines
Abstract
1. Introduction
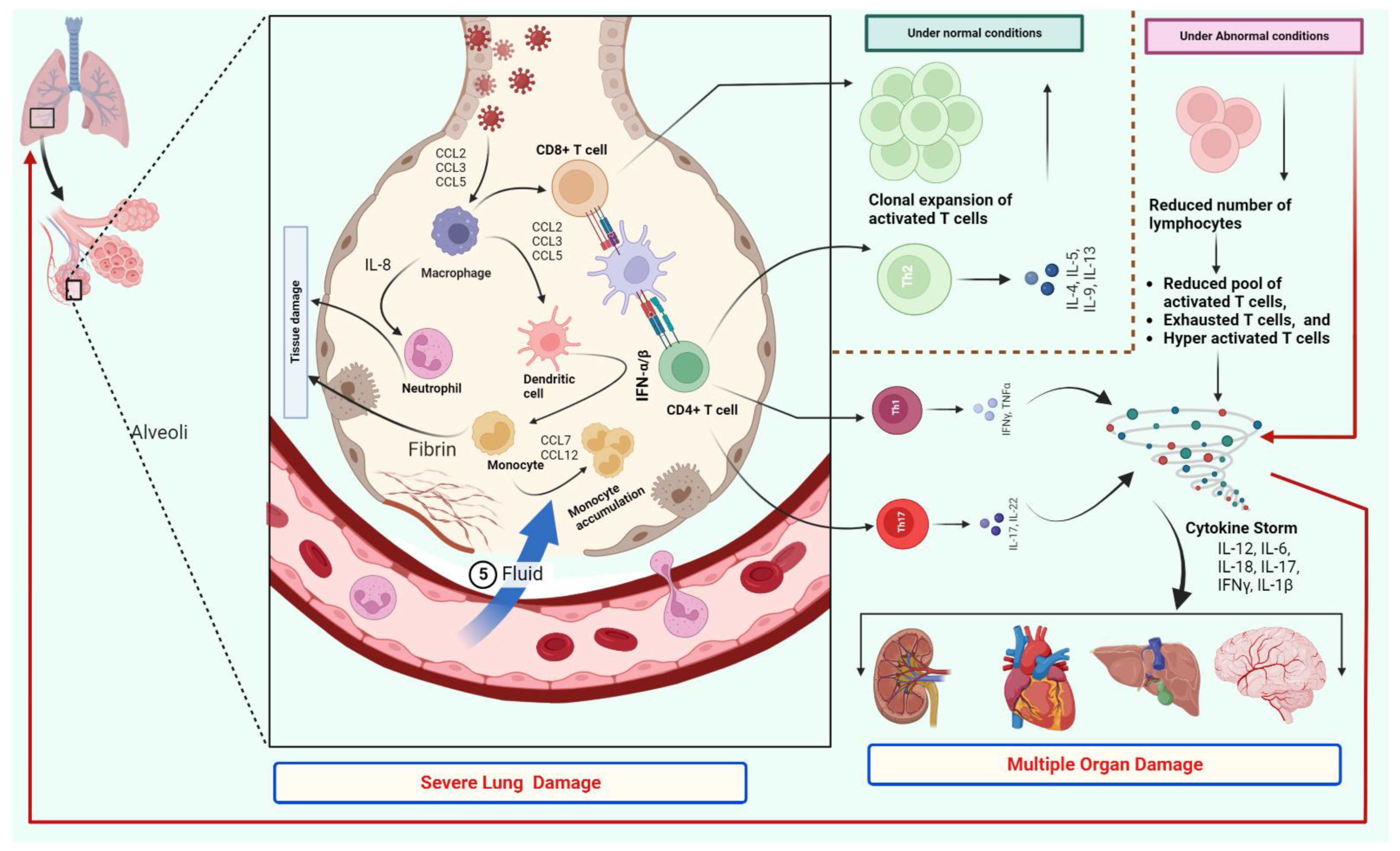
2. T Cells Response and Cytokine Storm
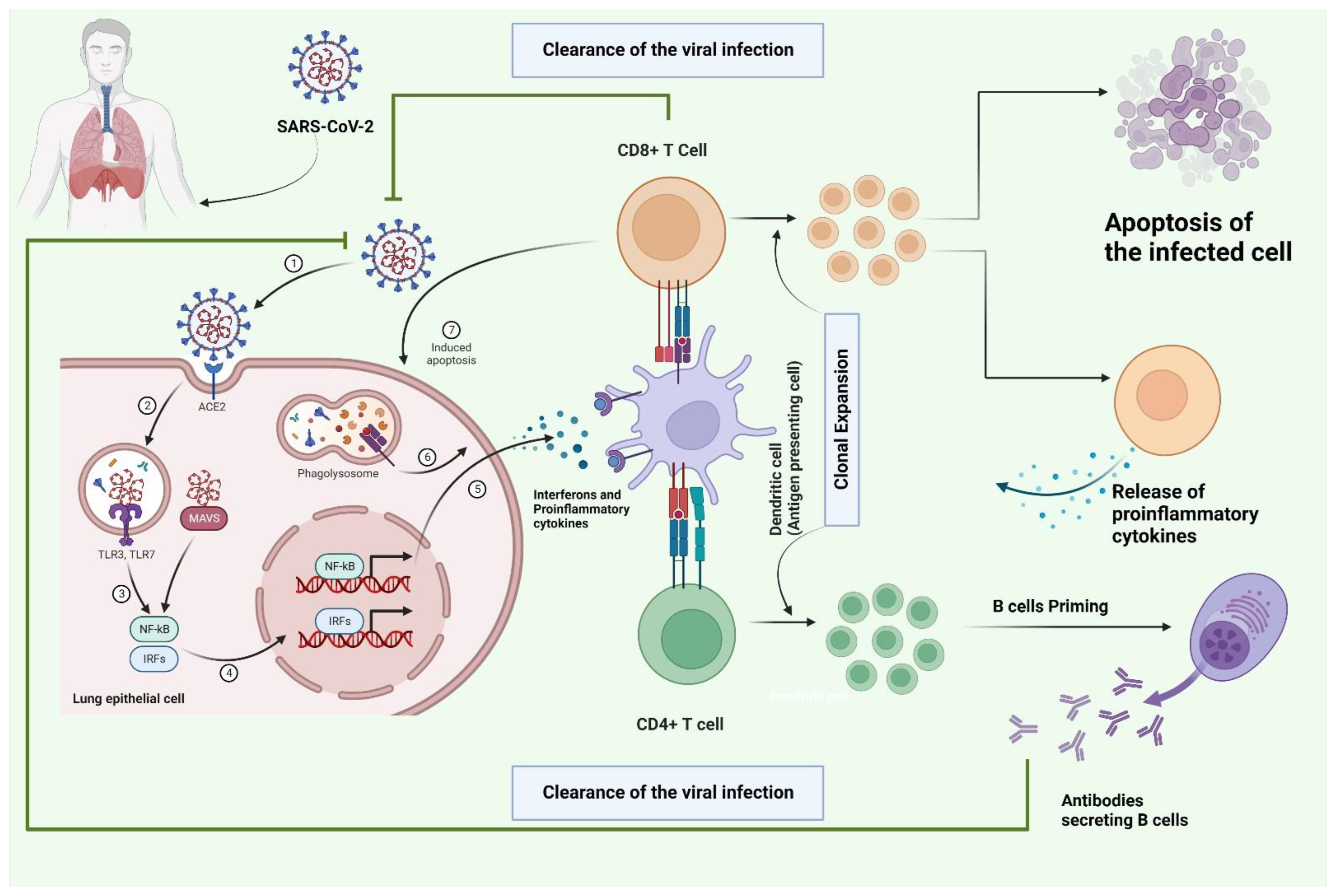
3. The Role of T Cells in the Infection Mechanism
4. T Cells and Disease Severity
5. T Cell Responses to Vaccines
6. SARS-CoV-2 Variants and Vaccine-Induced T Cell Immune Responses
7. T-Cell Exhaustion and Disease Severity
8. Immunomodulatory Approaches to Overcome T-Cell Exhaustion
9. Other Strategies to Improve T-Cells Mediated Immune Response
10. Methods for Large-Scale Screening of T-Cell Responses
11. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACE2 | angiotensin-converting enzyme 2 |
| APCs | antigen-presenting cells |
| BCR | B cell receptor |
| BALF | bronchoalveolar lavage fluid |
| COVID-19 | coronavirus disease 19 |
| MHC | major histocompatibility complex |
| mAbs | monoclonal antibodies |
| nAbs | neutralising antibodies |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| Tregs | T regulatory cells |
| TLRs | toll-like receptors |
| TCR | T cell receptor |
| VOCs | variants of concern |
References
- Bertoletti, A.; Le Bert, N.; Tan, A.T. SARS-CoV-2-Specific T Cells in the Changing Landscape of the COVID-19 Pandemic. Immunity 2022, 55, 1764–1778. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, M.; Priyanka; Choudhary, O.P. Emergence of Omicron Sub-Variant BA.2: Is It a Matter of Concern amid the COVID-19 Pandemic? Int. J. Surg. 2022, 99, 106581. [Google Scholar] [CrossRef] [PubMed]
- Zinatizadeh, M.R.; Zarandi, P.K.; Zinatizadeh, M.; Yousefi, M.H.; Amani, J.; Rezaei, N. Efficacy of mRNA, Adenoviral Vector, and Perfusion Protein COVID-19 Vaccines. Biomed. Pharmacother. 2022, 146, 112527. [Google Scholar] [CrossRef]
- Dhawan, M.; Saied, A.A.; Mitra, S.; Alhumaydhi, F.A.; Emran, T.B.; Wilairatana, P. Omicron Variant (B.1.1.529) and Its Sublineages: What Do We Know so Far amid the Emergence of Recombinant Variants of SARS-CoV-2? Biomed. Pharmacother. 2022, 154, 113522. [Google Scholar] [CrossRef] [PubMed]
- Islam, F.; Dhawan, M.; Nafady, M.H.; Emran, T.B.; Mitra, S.; Choudhary, O.P.; Akter, A. Understanding the Omicron Variant (B.1.1.529) of SARS-CoV-2: Mutational Impacts, Concerns, and the Possible Solutions. Ann. Med. Surg. 2022, 78, 103737. [Google Scholar] [CrossRef]
- Kudlay, D.; Kofiadi, I.; Khaitov, M. Peculiarities of the T Cell Immune Response in COVID-19. Vaccines 2022, 10, 242. [Google Scholar] [CrossRef]
- Moss, P. The T Cell Immune Response against SARS-CoV-2. Nat. Immunol. 2022, 23, 186–193. [Google Scholar] [CrossRef]
- Kedzierska, K.; Thomas, P.G. Count on Us: T Cells in SARS-CoV-2 Infection and Vaccination. Cell Rep. Med. 2022, 3, 100562. [Google Scholar] [CrossRef]
- Bertoletti, A.; Le Bert, N.; Tan, A.T. Act Early and at the Right Location: SARS-CoV-2 T Cell Kinetics and Tissue Localization. Int. J. Mol. Sci. 2022, 23, 10679. [Google Scholar] [CrossRef]
- Sette, A.; Crotty, S. Adaptive Immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880. [Google Scholar] [CrossRef]
- Zhao, J.; Zhao, J.; Mangalam, A.K.; Channappanavar, R.; Fett, C.; Meyerholz, D.K.; Agnihothram, S.; Baric, R.S.; David, C.S.; Perlman, S. Airway Memory CD4 + T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity 2016, 44, 1379–1391. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chandrashekar, A.; Sellers, D.; Barrett, J.; Jacob-Dolan, C.; Lifton, M.; McMahan, K.; Sciacca, M.; VanWyk, H.; Wu, C.; et al. Vaccines Elicit Highly Conserved Cellular Immunity to SARS-CoV-2 Omicron. Nature 2022, 603, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Rydyznski Moderbacher, C.; Ramirez, S.I.; Dan, J.M.; Grifoni, A.; Hastie, K.M.; Weiskopf, D.; Belanger, S.; Abbott, R.K.; Kim, C.; Choi, J.; et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 2020, 183, 996–1012.e19. [Google Scholar] [CrossRef]
- Madden, E.A.; Diamond, M.S. Host Cell-Intrinsic Innate Immune Recognition of SARS-CoV-2. Curr. Opin. Virol. 2022, 52, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Notarbartolo, S.; Ranzani, V.; Bandera, A.; Gruarin, P.; Bevilacqua, V.; Putignano, A.R.; Gobbini, A.; Galeota, E.; Manara, C.; Bombaci, M.; et al. Integrated Longitudinal Immunophenotypic, Transcriptional, and Repertoire Analyses Delineate Immune Responses in Patients with COVID-19. Sci. Immunol. 2021, 6, eabg5021. [Google Scholar] [CrossRef] [PubMed]
- Bergamaschi, L.; Mescia, F.; Turner, L.; Hanson, A.L.; Kotagiri, P.; Dunmore, B.J.; Ruffieux, H.; De Sa, A.; Huhn, O.; Morgan, M.D.; et al. Longitudinal Analysis Reveals That Delayed Bystander CD8+ T Cell Activation and Early Immune Pathology Distinguish Severe COVID-19 from Mild Disease. Immunity 2021, 54, 1257–1275.e8. [Google Scholar] [CrossRef]
- Lucas, C.; Klein, J.; Sundaram, M.E.; Liu, F.; Wong, P.; Silva, J.; Mao, T.; Oh, J.E.; Mohanty, S.; Huang, J.; et al. Delayed Production of Neutralizing Antibodies Correlates with Fatal COVID-19. Nat. Med. 2021, 27, 1178–1186. [Google Scholar] [CrossRef]
- Graham, M.B.; Braciale, V.L.; Braciale, T.J. Influenza Virus-Specific CD4+ T Helper Type 2 T Lymphocytes Do Not Promote Recovery from Experimental Virus Infection. J. Exp. Med. 1994, 180, 1273–1282. [Google Scholar] [CrossRef]
- Maurice, N.J.; Taber, A.K.; Prlic, M. The Ugly Duckling Turned to Swan: A Change in Perception of Bystander-Activated Memory CD8 T Cells. J. Immunol. 2021, 206, 455–462. [Google Scholar] [CrossRef]
- Kim, T.-S.; Shin, E.-C. The Activation of Bystander CD8+ T Cells and Their Roles in Viral Infection. Exp. Mol. Med. 2019, 51, 1–9. [Google Scholar] [CrossRef]
- Tye, E.X.C.; Jinks, E.; Haigh, T.A.; Kaul, B.; Patel, P.; Parry, H.M.; Newby, M.L.; Crispin, M.; Kaur, N.; Moss, P.; et al. Mutations in SARS-CoV-2 Spike Protein Impair Epitope-Specific CD4+ T Cell Recognition. Nat. Immunol. 2022, 23, 1726–1734. [Google Scholar] [CrossRef] [PubMed]
- Kent, S.J.; Khoury, D.S.; Reynaldi, A.; Juno, J.A.; Wheatley, A.K.; Stadler, E.; John Wherry, E.; Triccas, J.; Sasson, S.C.; Cromer, D.; et al. Disentangling the Relative Importance of T Cell Responses in COVID-19: Leading Actors or Supporting Cast? Nat. Rev. Immunol. 2022, 22, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Khatamzas, E.; Antwerpen, M.H.; Rehn, A.; Graf, A.; Hellmuth, J.C.; Hollaus, A.; Mohr, A.-W.; Gaitzsch, E.; Weiglein, T.; Georgi, E.; et al. Accumulation of Mutations in Antibody and CD8 T Cell Epitopes in a B Cell Depleted Lymphoma Patient with Chronic SARS-CoV-2 Infection. Nat. Commun. 2022, 13, 5586. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Chen, D.; Yuan, D.; Lausted, C.; Choi, J.; Dai, C.L.; Voillet, V.; Duvvuri, V.R.; Scherler, K.; Troisch, P.; et al. Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell 2020, 183, 1479–1495.e20. [Google Scholar] [CrossRef]
- Mathew, D.; Giles, J.R.; Baxter, A.E.; Oldridge, D.A.; Greenplate, A.R.; Wu, J.E.; Alanio, C.; Kuri-Cervantes, L.; Pampena, M.B.; D’Andrea, K.; et al. Deep Immune Profiling of COVID-19 Patients Reveals Distinct Immunotypes with Therapeutic Implications. Science 2020, 369, eabc8511. [Google Scholar] [CrossRef]
- Le Bert, N.; Clapham, H.E.; Tan, A.T.; Chia, W.N.; Tham, C.Y.L.; Lim, J.M.; Kunasegaran, K.; Tan, L.W.L.; Dutertre, C.-A.; Shankar, N.; et al. Highly Functional Virus-Specific Cellular Immune Response in Asymptomatic SARS-CoV-2 Infection. J. Exp. Med. 2021, 218, e20202617. [Google Scholar] [CrossRef]
- Grau-Expósito, J.; Sánchez-Gaona, N.; Massana, N.; Suppi, M.; Astorga-Gamaza, A.; Perea, D.; Rosado, J.; Falcó, A.; Kirkegaard, C.; Torrella, A.; et al. Peripheral and Lung Resident Memory T Cell Responses against SARS-CoV-2. Nat. Commun. 2021, 12, 3010. [Google Scholar] [CrossRef]
- Goswami, T.K.; Singh, M.; Dhawan, M.; Mitra, S.; Emran, T.B.; Rabaan, A.A.; Mutair, A.A.; Alawi, Z.A.; Alhumaid, S.; Dhama, K. Regulatory T Cells (Tregs) and Their Therapeutic Potential against Autoimmune Disorders – Advances and Challenges. Hum. Vaccines Immunother. 2022, 18, 2035117. [Google Scholar] [CrossRef]
- Gao, M.; Liu, Y.; Guo, M.; Wang, Q.; Wang, Y.; Fan, J.; Shen, Y.; Hou, J.; Wan, Y.; Zhu, Z. Regulatory CD4 + and CD8 + T Cells Are Negatively Correlated with CD4 + /CD8 + T Cell Ratios in Patients Acutely Infected with SARS-CoV-2. J. Leukoc. Biol. 2020, 109, 91–97. [Google Scholar] [CrossRef]
- Hillaire, M.; Rimmelzwaan, G.; Kreijtz, J. Clearance of Influenza Virus Infections by T Cells: Risk of Collateral Damage? Curr. Opin. Virol. 2013, 3, 430–437. [Google Scholar] [CrossRef]
- GeurtsvanKessel, C.H.; Geers, D.; Schmitz, K.S.; Mykytyn, A.Z.; Lamers, M.M.; Bogers, S.; Scherbeijn, S.; Gommers, L.; Sablerolles, R.S.G.; Nieuwkoop, N.N.; et al. Divergent SARS-CoV-2 Omicron–Reactive T and B Cell Responses in COVID-19 Vaccine Recipients. Sci. Immunol. 2022, 7, eabo2202. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. SARS-CoV-2 T Cell Responses Elicited by COVID-19 Vaccines or Infection Are Expected to Remain Robust against Omicron. Viruses 2022, 14, 79. [Google Scholar] [CrossRef]
- Thevarajan, I.; Nguyen, T.H.O.; Koutsakos, M.; Druce, J.; Caly, L.; van de Sandt, C.E.; Jia, X.; Nicholson, S.; Catton, M.; Cowie, B.; et al. Breadth of Concomitant Immune Responses Prior to Patient Recovery: A Case Report of Non-Severe COVID-19. Nat. Med. 2020, 26, 453–455. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and Immunological Features of Severe and Moderate Coronavirus Disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Li, S.; Jiang, L.; Li, X.; Lin, F.; Wang, Y.; Li, B.; Jiang, T.; An, W.; Liu, S.; Liu, H.; et al. Clinical and Pathological Investigation of Patients with Severe COVID-19. JCI Insight 2020, 5. [Google Scholar] [CrossRef]
- Liu, J.; Li, S.; Liu, J.; Liang, B.; Wang, X.; Wang, H.; Li, W.; Tong, Q.; Yi, J.; Zhao, L.; et al. Longitudinal Characteristics of Lymphocyte Responses and Cytokine Profiles in the Peripheral Blood of SARS-CoV-2 Infected Patients. EBioMedicine 2020, 55, 102763. [Google Scholar] [CrossRef] [PubMed]
- Urra, J.M.; Cabrera, C.M.; Porras, L.; Ródenas, I. Selective CD8 Cell Reduction by SARS-CoV-2 Is Associated with a Worse Prognosis and Systemic Inflammation in COVID-19 Patients. Clin. Immunol. 2020, 217, 108486. [Google Scholar] [CrossRef] [PubMed]
- Giamarellos-Bourboulis, E.J.; Netea, M.G.; Rovina, N.; Akinosoglou, K.; Antoniadou, A.; Antonakos, N.; Damoraki, G.; Gkavogianni, T.; Adami, M.-E.; Katsaounou, P.; et al. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe 2020, 27, 992–1000.e3. [Google Scholar] [CrossRef]
- Bordoni, V.; Sacchi, A.; Cimini, E.; Notari, S.; Grassi, G.; Tartaglia, E.; Casetti, R.; Giancola, M.L.; Bevilacqua, N.; Maeurer, M.; et al. An Inflammatory Profile Correlates With Decreased Frequency of Cytotoxic Cells in Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 2272–2275. [Google Scholar] [CrossRef] [PubMed]
- Rabaan, A.A.; Mutair, A.A.; Hajissa, K.; Alfaraj, A.H.; Al-Jishi, J.M.; Alhajri, M.; Alwarthan, S.; Alsuliman, S.A.; Al-Najjar, A.H.; Al Zaydani, I.A.; et al. A Comprehensive Review on the Current Vaccines and Their Efficacies to Combat SARS-CoV-2 Variants. Vaccines 2022, 10, 1655. [Google Scholar] [CrossRef]
- De Candia, P.; Prattichizzo, F.; Garavelli, S.; Matarese, G. T Cells: Warriors of SARS-CoV-2 Infection. Trends Immunol. 2021, 42, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Rabaan, A.A.; Al-Ahmed, S.H.; Garout, M.A.; Al-Qaaneh, A.M.; Sule, A.A.; Tirupathi, R.; Mutair, A.A.; Alhumaid, S.; Hasan, A.; Dhawan, M.; et al. Diverse Immunological Factors Influencing Pathogenesis in Patients with COVID-19: A Review on Viral Dissemination, Immunotherapeutic Options to Counter Cytokine Storm and Inflammatory Responses. Pathogens 2021, 10, 565. [Google Scholar] [CrossRef]
- Kaneko, N.; Kuo, H.-H.; Boucau, J.; Farmer, J.R.; Allard-Chamard, H.; Mahajan, V.S.; Piechocka-Trocha, A.; Lefteri, K.; Osborn, M.; Bals, J.; et al. Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19. Cell 2020, 183, 143–157.e13. [Google Scholar] [CrossRef] [PubMed]
- Yuki, K.; Fujiogi, M.; Koutsogiannaki, S. COVID-19 Pathophysiology: A Review. Clin. Immunol. 2020, 215, 108427. [Google Scholar] [CrossRef]
- Pacha, O.; Sallman, M.A.; Evans, S.E. COVID-19: A Case for Inhibiting IL-17? Nat. Rev. Immunol. 2020, 20, 345–346. [Google Scholar] [CrossRef]
- Wong, J.J.M.; Leong, J.Y.; Lee, J.H.; Albani, S.; Yeo, J.G. Insights into the Immuno-Pathogenesis of Acute Respiratory Distress Syndrome. Ann. Transl. Med. 2019, 7, 504. [Google Scholar] [CrossRef]
- Sekine, T.; Perez-Potti, A.; Rivera-Ballesteros, O.; Strålin, K.; Gorin, J.-B.; Olsson, A.; Llewellyn-Lacey, S.; Kamal, H.; Bogdanovic, G.; Muschiol, S.; et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell 2020, 183, 158–168.e14. [Google Scholar] [CrossRef]
- Nelde, A.; Bilich, T.; Heitmann, J.S.; Maringer, Y.; Salih, H.R.; Roerden, M.; Lübke, M.; Bauer, J.; Rieth, J.; Wacker, M.; et al. SARS-CoV-2-Derived Peptides Define Heterologous and COVID-19-Induced T Cell Recognition. Nat. Immunol. 2020, 22, 74–85. [Google Scholar] [CrossRef]
- Da Silva Antunes, R.; Pallikkuth, S.; Williams, E.; Dawen, Y.E.; Mateus, J.; Quiambao, L.; Wang, E.; Rawlings, S.A.; Stadlbauer, D.; Jiang, K.; et al. Differential T-Cell Reactivity to Endemic Coronaviruses and SARS-CoV-2 in Community and Health Care Workers. J. Infect. Dis. 2021, 224, 70–80. [Google Scholar] [CrossRef]
- Rowland-Jones, S. Immune Responses in HIV-Exposed Seronegatives: Have They Repelled the Virus? Curr. Opin. Immunol. 1995, 7, 448–455. [Google Scholar] [CrossRef]
- Vidya Vijayan, K.K.; Karthigeyan, K.P.; Tripathi, S.P.; Hanna, L.E. Pathophysiology of CD4+ T-Cell Depletion in HIV-1 and HIV-2 Infections. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Kuri-Cervantes, L.; Pampena, M.B.; Meng, W.; Rosenfeld, A.M.; Ittner, C.A.G.; Weisman, A.R.; Agyekum, R.S.; Mathew, D.; Baxter, A.E.; Vella, L.A.; et al. Comprehensive Mapping of Immune Perturbations Associated with Severe COVID-19. Sci. Immunol. 2020, 5, eabd7114. [Google Scholar] [CrossRef]
- Laing, A.G.; Lorenc, A.; del Molino del Barrio, I.; Das, A.; Fish, M.; Monin, L.; Muñoz-Ruiz, M.; McKenzie, D.R.; Hayday, T.S.; Francos-Quijorna, I.; et al. A Dynamic COVID-19 Immune Signature Includes Associations with Poor Prognosis. Nat. Med. 2020, 26, 1623–1635. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal Analyses Reveal Immunological Misfiring in Severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Schulien, I.; Kemming, J.; Oberhardt, V.; Wild, K.; Seidel, L.M.; Killmer, S.; Sagar; Daul, F.; Salvat Lago, M.; Decker, A.; et al. Characterization of Pre-Existing and Induced SARS-CoV-2-Specific CD8+ T Cells. Nat. Med. 2020, 27, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Kreutmair, S.; Unger, S.; Núñez, N.G.; Ingelfinger, F.; Alberti, C.; De Feo, D.; Krishnarajah, S.; Kauffmann, M.; Friebel, E.; Babaei, S.; et al. Distinct Immunological Signatures Discriminate Severe COVID-19 from Non-SARS-CoV-2-Driven Critical Pneumonia. Immunity 2021, 54, 1578–1593.e5. [Google Scholar] [CrossRef]
- Schwabenland, M.; Salié, H.; Tanevski, J.; Killmer, S.; Lago, M.S.; Schlaak, A.E.; Mayer, L.; Matschke, J.; Püschel, K.; Fitzek, A.; et al. Deep Spatial Profiling of Human COVID-19 Brains Reveals Neuroinflammation with Distinct Microanatomical Microglia-T-Cell Interactions. Immunity 2021, 54, 1594–1610.e11. [Google Scholar] [CrossRef]
- Szabo, P.A.; Dogra, P.; Gray, J.I.; Wells, S.B.; Connors, T.J.; Weisberg, S.P.; Krupska, I.; Matsumoto, R.; Poon, M.M.L.; Idzikowski, E.; et al. Longitudinal Profiling of Respiratory and Systemic Immune Responses Reveals Myeloid Cell-Driven Lung Inflammation in Severe COVID-19. Immunity 2021, 54, 797–814.e6. [Google Scholar] [CrossRef]
- Vijayakumar, B.; Boustani, K.; Ogger, P.P.; Papadaki, A.; Tonkin, J.; Orton, C.M.; Ghai, P.; Suveizdyte, K.; Hewitt, R.J.; Desai, S.R.; et al. Immuno-Proteomic Profiling Reveals Aberrant Immune Cell Regulation in the Airways of Individuals with Ongoing Post-COVID-19 Respiratory Disease. Immunity 2022, 55, 542–556.e5. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Jeong, S.; Shin, E.-C. Significance of Bystander T Cell Activation in Microbial Infection. Nat. Immunol. 2021, 23, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, I. Interfering with SARS-CoV-2: Are Interferons Friends or Foes in COVID-19? Curr. Opin. Virol. 2021, 50, 119–127. [Google Scholar] [CrossRef]
- Domizio, J.D.; Gulen, M.F.; Saidoune, F.; Thacker, V.V.; Yatim, A.; Sharma, K.; Nass, T.; Guenova, E.; Schaller, M.; Conrad, C.; et al. The cGAS–STING Pathway Drives Type I IFN Immunopathology in COVID-19. Nature 2022, 603, 145–151. [Google Scholar] [CrossRef]
- Junqueira, C.; Crespo, Â.; Ranjbar, S.; de Lacerda, L.B.; Lewandrowski, M.; Ingber, J.; Parry, B.; Ravid, S.; Clark, S.; Schrimpf, M.R.; et al. FcγR-Mediated SARS-CoV-2 Infection of Monocytes Activates Inflammation. Nature 2022, 606, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Cardone, J.; Le Friec, G.; Vantourout, P.; Roberts, A.; Fuchs, A.; Jackson, I.; Suddason, T.; Lord, G.; Atkinson, J.P.; Cope, A.; et al. Complement Regulator CD46 Temporally Regulates Cytokine Production by Conventional and Unconventional T Cells. Nat. Immunol. 2010, 11, 862–871. [Google Scholar] [CrossRef] [PubMed]
- Chauss, D.; Freiwald, T.; McGregor, R.; Yan, B.; Wang, L.; Nova-Lamperti, E.; Kumar, D.; Zhang, Z.; Teague, H.; West, E.E.; et al. Autocrine Vitamin D Signaling Switches off Pro-Inflammatory Programs of TH1 Cells. Nat. Immunol. 2021, 23, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Madan, R.; Karp, C.L.; Braciale, T.J. Effector T Cells Control Lung Inflammation during Acute Influenza Virus Infection by Producing IL-10. Nat. Med. 2009, 15, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Alshukairi, A.N.; Baharoon, S.A.; Ahmed, W.A.; Bokhari, A.A.; Nehdi, A.M.; Layqah, L.A.; Alghamdi, M.G.; Al Gethamy, M.M.; Dada, A.M.; et al. Recovery from the Middle East Respiratory Syndrome Is Associated with Antibody and T Cell Responses. Sci. Immunol. 2017, 2, eaan5393. [Google Scholar] [CrossRef]
- Yan, B.; Freiwald, T.; Chauss, D.; Wang, L.; West, E.; Mirabelli, C.; Zhang, C.J.; Nichols, E.-M.; Malik, N.; Gregory, R.; et al. SARS-CoV-2 Drives JAK1/2-Dependent Local Complement Hyperactivation. Sci. Immunol. 2021, 6, eabg0833. [Google Scholar] [CrossRef]
- Dhawan, M.; Priyanka; Choudhary, O.P. Immunomodulatory and Therapeutic Implications of Vitamin D in the Management of COVID-19. Hum. Vaccines Immunother. 2022, 18, 2025734. [Google Scholar] [CrossRef] [PubMed]
- Akbar, M.R.; Wibowo, A.; Pranata, R.; Setiabudiawan, B. Low Serum 25-Hydroxyvitamin D (Vitamin D) Level Is Associated With Susceptibility to COVID-19, Severity, and Mortality: A Systematic Review and Meta-Analysis. Front. Nutr. 2021, 8, 660420. [Google Scholar] [CrossRef] [PubMed]
- Witkowski, M.; Tizian, C.; Ferreira-Gomes, M.; Niemeyer, D.; Jones, T.C.; Heinrich, F.; Frischbutter, S.; Angermair, S.; Hohnstein, T.; Mattiola, I.; et al. Untimely TGFβ Responses in COVID-19 Limit Antiviral Functions of NK Cells. Nature 2021, 600, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Vick, S.C.; Frutoso, M.; Mair, F.; Konecny, A.J.; Greene, E.; Wolf, C.R.; Logue, J.K.; Franko, N.M.; Boonyaratanakornkit, J.; Gottardo, R.; et al. A Regulatory T Cell Signature Distinguishes the Immune Landscape of COVID-19 Patients from Those with Other Respiratory Infections. Sci. Adv. 2021, 7, eabj0274. [Google Scholar] [CrossRef]
- Shrotri, M.; van Schalkwyk, M.C.I.; Post, N.; Eddy, D.; Huntley, C.; Leeman, D.; Rigby, S.; Williams, S.V.; Bermingham, W.H.; Kellam, P.; et al. T Cell Response to SARS-CoV-2 Infection in Humans: A Systematic Review. PLoS ONE 2021, 16, e0245532. [Google Scholar] [CrossRef]
- Tan, A.T.; Linster, M.; Tan, C.W.; Le Bert, N.; Chia, W.N.; Kunasegaran, K.; Zhuang, Y.; Tham, C.Y.L.; Chia, A.; Smith, G.J.D.; et al. Early Induction of Functional SARS-CoV-2-Specific T Cells Associates with Rapid Viral Clearance and Mild Disease in COVID-19 Patients. Cell Rep. 2021, 34, 108728. [Google Scholar] [CrossRef]
- Braun, J.; Loyal, L.; Frentsch, M.; Wendisch, D.; Georg, P.; Kurth, F.; Hippenstiel, S.; Dingeldey, M.; Kruse, B.; Fauchere, F.; et al. SARS-CoV-2-Reactive T Cells in Healthy Donors and Patients with COVID-19. Nature 2020, 587, 270–274. [Google Scholar] [CrossRef]
- Zheng, M.; Gao, Y.; Wang, G.; Song, G.; Liu, S.; Sun, D.; Xu, Y.; Tian, Z. Functional Exhaustion of Antiviral Lymphocytes in COVID-19 Patients. Cell. Mol. Immunol. 2020, 17, 533–535. [Google Scholar] [CrossRef]
- Bertoletti, A.; Tan, A.T.; Le Bert, N. The T-Cell Response to SARS-CoV-2: Kinetic and Quantitative Aspects and the Case for Their Protective Role. Oxf. Open Immunol. 2021, 2, iqab006. [Google Scholar] [CrossRef]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-Cell Landscape of Bronchoalveolar Immune Cells in Patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Wimmers, F.; Mok, C.K.P.; Perera, R.A.P.M.; Scott, M.; Hagan, T.; Sigal, N.; Feng, Y.; Bristow, L.; Tak-Yin Tsang, O.; et al. Systems Biological Assessment of Immunity to Mild versus Severe COVID-19 Infection in Humans. Science 2020, 369, 1210–1220. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.; Ni, Z.; Hu, Y.; Liang, W.; Ou, C.; He, J.; Liu, L.; Shan, H.; Lei, C.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tan, Y.; Ling, Y.; Lu, G.; Liu, F.; Yi, Z.; Jia, X.; Wu, M.; Shi, B.; Xu, S.; et al. Viral and Host Factors Related to the Clinical Outcome of COVID-19. Nature 2020, 583, 437–440. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef]
- Saini, S.K.; Hersby, D.S.; Tamhane, T.; Povlsen, H.R.; Hernandez, S.P.A.; Nielsen, M.; Gang, A.O.; Hadrup, S.R. SARS-CoV-2 Genome-Wide T Cell Epitope Mapping Reveals Immunodominance and Substantial CD8 + T Cell Activation in COVID-19 Patients. Sci. Immunol. 2021, 6, eabf7550. [Google Scholar] [CrossRef]
- Kaneko, N.; Boucau, J.; Kuo, H.-H.; Perugino, C.; Mahajan, V.S.; Farmer, J.R.; Liu, H.; Diefenbach, T.J.; Piechocka-Trocha, A.; Lefteri, K.; et al. Temporal Changes in T Cell Subsets and Expansion of Cytotoxic CD4+ T Cells in the Lungs in Severe COVID-19. Clin. Immunol. 2022, 237, 108991. [Google Scholar] [CrossRef]
- Meckiff, B.J.; Ramírez-Suástegui, C.; Fajardo, V.; Chee, S.J.; Kusnadi, A.; Simon, H.; Eschweiler, S.; Grifoni, A.; Pelosi, E.; Weiskopf, D.; et al. Imbalance of Regulatory and Cytotoxic SARS-CoV-2-Reactive CD4+ T Cells in COVID-19. Cell 2020, 183, 1340–1353.e16. [Google Scholar] [CrossRef]
- Stephenson, E.; Reynolds, G.; Botting, R.A.; Calero-Nieto, F.J.; Morgan, M.D.; Tuong, Z.K.; Bach, K.; Sungnak, W.; Worlock, K.B.; Yoshida, M.; et al. Single-Cell Multi-Omics Analysis of the Immune Response in COVID-19. Nat. Med. 2021, 27, 904–916. [Google Scholar] [CrossRef]
- DiPiazza, A.T.; Graham, B.S.; Ruckwardt, T.J. T Cell Immunity to SARS-CoV-2 Following Natural Infection and Vaccination. Biochem. Biophys. Res. Commun. 2021, 538, 211–217. [Google Scholar] [CrossRef]
- Boettler, T.; Csernalabics, B.; Salié, H.; Luxenburger, H.; Wischer, L.; Salimi Alizei, E.; Zoldan, K.; Krimmel, L.; Bronsert, P.; Schwabenland, M.; et al. SARS-CoV-2 Vaccination Can Elicit a CD8 T-Cell Dominant Hepatitis. J. Hepatol. 2022, 77, 653–659. [Google Scholar] [CrossRef]
- Goel, R.R.; Apostolidis, S.A.; Painter, M.M.; Mathew, D.; Pattekar, A.; Kuthuru, O.; Gouma, S.; Hicks, P.; Meng, W.; Rosenfeld, A.M.; et al. Distinct Antibody and Memory B Cell Responses in SARS-CoV-2 Naïve and Recovered Individuals after mRNA Vaccination. Sci. Immunol. 2021, 6, eabi6950. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yin, S.; Tong, X.; Tao, Y.; Ni, J.; Pan, J.; Li, M.; Wan, Y.; Mao, M.; Xiong, Y.; et al. Dynamic SARS-CoV-2-Specific B-Cell and T-Cell Responses Following Immunization with an Inactivated COVID-19 Vaccine. Clin. Microbiol. Infect. 2022, 28, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Long, Q.-X.; Tang, X.-J.; Shi, Q.-L.; Li, Q.; Deng, H.-J.; Yuan, J.; Hu, J.-L.; Xu, W.; Zhang, Y.; Lv, F.-J.; et al. Clinical and Immunological Assessment of Asymptomatic SARS-CoV-2 Infections. Nat. Med. 2020, 26, 1200–1204. [Google Scholar] [CrossRef] [PubMed]
- Robbiani, D.F.; Gaebler, C.; Muecksch, F.; Lorenzi, J.C.C.; Wang, Z.; Cho, A.; Agudelo, M.; Barnes, C.O.; Gazumyan, A.; Finkin, S.; et al. Convergent Antibody Responses to SARS-CoV-2 in Convalescent Individuals. Nature 2020, 584, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Riley, T.P.; Chou, H.-T.; Hu, R.; Bzymek, K.P.; Correia, A.R.; Partin, A.C.; Li, D.; Gong, D.; Wang, Z.; Yu, X.; et al. Enhancing the Prefusion Conformational Stability of SARS-CoV-2 Spike Protein Through Structure-Guided Design. Front. Immunol. 2021, 12, 660198. [Google Scholar] [CrossRef]
- Carnell, G.W.; Ciazynska, K.A.; Wells, D.A.; Xiong, X.; Aguinam, E.T.; McLaughlin, S.H.; Mallery, D.; Ebrahimi, S.; Ceron-Gutierrez, L.; Asbach, B.; et al. SARS-CoV-2 Spike Protein Stabilized in the Closed State Induces Potent Neutralizing Responses. J. Virol. 2021, 95, e00203-21. [Google Scholar] [CrossRef]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schäfer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA Vaccine Design Enabled by Prototype Pathogen Preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef]
- Corbett, K.S.; Flynn, B.; Foulds, K.E.; Francica, J.R.; Boyoglu-Barnum, S.; Werner, A.P.; Flach, B.; O’Connell, S.; Bock, K.W.; Minai, M.; et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020, 383, 1544–1555. [Google Scholar] [CrossRef]
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020, 383, 2427–2438. [Google Scholar] [CrossRef]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Sahin, U.; Muik, A.; Derhovanessian, E.; Vogler, I.; Kranz, L.M.; Vormehr, M.; Baum, A.; Pascal, K.; Quandt, J.; Maurus, D.; et al. COVID-19 Vaccine BNT162b1 Elicits Human Antibody and TH1 T Cell Responses. Nature 2020, 586, 594–599. [Google Scholar] [CrossRef]
- Mateus, J.; Grifoni, A.; Tarke, A.; Sidney, J.; Ramirez, S.I.; Dan, J.M.; Burger, Z.C.; Rawlings, S.A.; Smith, D.M.; Phillips, E.; et al. Selective and Cross-Reactive SARS-CoV-2 T Cell Epitopes in Unexposed Humans. Science 2020, 370, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Juno, J.A.; Tan, H.-X.; Lee, W.S.; Reynaldi, A.; Kelly, H.G.; Wragg, K.; Esterbauer, R.; Kent, H.E.; Batten, C.J.; Mordant, F.L.; et al. Humoral and Circulating Follicular Helper T Cell Responses in Recovered Patients with COVID-19. Nat. Med. 2020, 26, 1428–1434. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.E.; Frenck, R.W.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Yu, J.; Tostanoski, L.H.; Peter, L.; Mercado, N.B.; McMahan, K.; Mahrokhian, S.H.; Nkolola, J.P.; Liu, J.; Li, Z.; Chandrashekar, A.; et al. DNA Vaccine Protection against SARS-CoV-2 in Rhesus Macaques. Science 2020, 369, 806–811. [Google Scholar] [CrossRef]
- Folegatti, P.M.; Ewer, K.J.; Aley, P.K.; Angus, B.; Becker, S.; Belij-Rammerstorfer, S.; Bellamy, D.; Bibi, S.; Bittaye, M.; Clutterbuck, E.A.; et al. Safety and Immunogenicity of the ChAdOx1 nCoV-19 Vaccine against SARS-CoV-2: A Preliminary Report of a Phase 1/2, Single-Blind, Randomised Controlled Trial. Lancet 2020, 396, 467–478. [Google Scholar] [CrossRef]
- Logunov, D.Y.; Dolzhikova, I.V.; Zubkova, O.V.; Tukhvatulin, A.I.; Shcheblyakov, D.V.; Dzharullaeva, A.S.; Grousova, D.M.; Erokhova, A.S.; Kovyrshina, A.V.; Botikov, A.G.; et al. Safety and Immunogenicity of an rAd26 and rAd5 Vector-Based Heterologous Prime-Boost COVID-19 Vaccine in Two Formulations: Two Open, Non-Randomised Phase 1/2 Studies from Russia. Lancet 2020, 396, 887–897. [Google Scholar] [CrossRef]
- Zhu, F.-C.; Li, Y.-H.; Guan, X.-H.; Hou, L.-H.; Wang, W.-J.; Li, J.-X.; Wu, S.-P.; Wang, B.-S.; Wang, Z.; Wang, L.; et al. Safety, Tolerability, and Immunogenicity of a Recombinant Adenovirus Type-5 Vectored COVID-19 Vaccine: A Dose-Escalation, Open-Label, Non-Randomised, First-in-Human Trial. Lancet 2020, 395, 1845–1854. [Google Scholar] [CrossRef]
- Kim, Y.C.; Dema, B.; Reyes-Sandoval, A. COVID-19 Vaccines: Breaking Record Times to First-in-Human Trials. NPJ Vaccines 2020, 5, 34. [Google Scholar] [CrossRef]
- Smith, T.R.F.; Patel, A.; Ramos, S.; Elwood, D.; Zhu, X.; Yan, J.; Gary, E.N.; Walker, S.N.; Schultheis, K.; Purwar, M.; et al. Immunogenicity of a DNA Vaccine Candidate for COVID-19. Nat. Commun. 2020, 11, 2601. [Google Scholar] [CrossRef]
- Park, J.W.; Lagniton, P.N.P.; Liu, Y.; Xu, R.-H. mRNA Vaccines for COVID-19: What, Why and How. Int. J. Biol. Sci. 2021, 17, 1446–1460. [Google Scholar] [CrossRef]
- Zhang, N.; Li, C.; Hu, Y.; Li, K.; Liang, J.; Wang, L.; Du, L.; Jiang, S. Current Development of COVID-19 Diagnostics, Vaccines and Therapeutics. Microbes Infect. 2020, 22, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, M.; Sharma, A.; Priyanka; Thakur, N.; Rajkhowa, T.K.; Choudhary, O.P. Delta Variant (B.1.617.2) of SARS-CoV-2: Mutations, Impact, Challenges and Possible Solutions. Hum. Vaccines Immunother. 2022, 18, 2068883. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Sidney, J.; Vita, R.; Peters, B.; Crotty, S.; Weiskopf, D.; Sette, A. SARS-CoV-2 Human T Cell Epitopes: Adaptive Immune Response against COVID-19. Cell Host Microbe 2021, 29, 1076–1092. [Google Scholar] [CrossRef]
- Tarke, A.; Sidney, J.; Methot, N.; Yu, E.D.; Zhang, Y.; Dan, J.M.; Goodwin, B.; Rubiro, P.; Sutherland, A.; Wang, E.; et al. Impact of SARS-CoV-2 Variants on the Total CD4+ and CD8+ T Cell Reactivity in Infected or Vaccinated Individuals. Cell Rep. Med. 2021, 2, 100355. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Y.; Wang, M.; Islam, M.S.; Liao, P.; Hu, Y.; Chen, X. Humoral and Cellular Immune Responses of COVID-19 Vaccines against SARS-Cov-2 Omicron Variant: A Systemic Review. Int. J. Biol. Sci. 2022, 18, 4629–4641. [Google Scholar] [CrossRef]
- Naranbhai, V.; Nathan, A.; Kaseke, C.; Berrios, C.; Khatri, A.; Choi, S.; Getz, M.A.; Tano-Menka, R.; Ofoman, O.; Gayton, A.; et al. T Cell Reactivity to the SARS-CoV-2 Omicron Variant Is Preserved in Most but Not All Individuals. Cell 2022, 185, 1041–1051.e6. [Google Scholar] [CrossRef] [PubMed]
- Woldemeskel, B.A.; Garliss, C.C.; Aytenfisu, T.Y.; Johnston, T.S.; Beck, E.J.; Dykema, A.G.; Frumento, N.; Wright, D.A.; Yang, A.H.; Damanakis, A.I.; et al. SARS-CoV-2–Specific Immune Responses in Boosted Vaccine Recipients with Breakthrough Infections during the Omicron Variant Surge. JCI Insight 2022, 7. [Google Scholar] [CrossRef]
- Keeton, R.; Tincho, M.B.; Ngomti, A.; Baguma, R.; Benede, N.; Suzuki, A.; Khan, K.; Cele, S.; Bernstein, M.; Karim, F.; et al. T Cell Responses to SARS-CoV-2 Spike Cross-Recognize Omicron. Nature 2022, 603, 488–492. [Google Scholar] [CrossRef]
- Atmar, R.L.; Lyke, K.E.; Deming, M.E.; Jackson, L.A.; Branche, A.R.; El Sahly, H.M.; Rostad, C.A.; Martin, J.M.; Johnston, C.; Rupp, R.E.; et al. Homologous and Heterologous Covid-19 Booster Vaccinations. N. Engl. J. Med. 2022, 386, 1046–1057. [Google Scholar] [CrossRef]
- Collier, A.Y.; Yu, J.; McMahan, K.; Liu, J.; Chandrashekar, A.; Maron, J.S.; Atyeo, C.; Martinez, D.R.; Ansel, J.L.; Aguayo, R.; et al. Differential Kinetics of Immune Responses Elicited by Covid-19 Vaccines. N. Engl. J. Med. 2021, 385, 2010–2012. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, A.; Yu, J.; McMahan, K.; Jacob-Dolan, C.; Liu, J.; He, X.; Hope, D.; Anioke, T.; Barrett, J.; Chung, B.; et al. Vaccine Protection against the SARS-CoV-2 Omicron Variant in Macaques. Cell 2022, 185, 1549–1555.e11. [Google Scholar] [CrossRef] [PubMed]
- Muik, A.; Lui, B.G.; Wallisch, A.-K.; Bacher, M.; Mühl, J.; Reinholz, J.; Ozhelvaci, O.; Beckmann, N.; Güimil Garcia, R.d.l.C.; Poran, A.; et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA Vaccine–Elicited Human Sera. Science 2022, 375, 678–680. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Cai, C.; Grifoni, A.; Müller, T.R.; Niessl, J.; Olofsson, A.; Humbert, M.; Hansson, L.; Österborg, A.; Bergman, P.; et al. Ancestral SARS-CoV-2-Specific T Cells Cross-Recognize the Omicron Variant. Nat. Med. 2022, 28, 472–476. [Google Scholar] [CrossRef]
- Cohen, H.; Rotem, S.; Elia, U.; Bilinsky, G.; Levy, I.; Chitlaru, T.; Bar-Haim, E. T Cell Response Following Anti-COVID-19 BNT162b2 Vaccination Is Maintained against the SARS-CoV-2 Omicron B.1.1.529 Variant of Concern. Viruses 2022, 14, 347. [Google Scholar] [CrossRef]
- Adamo, S.; Michler, J.; Zurbuchen, Y.; Cervia, C.; Taeschler, P.; Raeber, M.E.; Baghai Sain, S.; Nilsson, J.; Moor, A.E.; Boyman, O. Signature of Long-Lived Memory CD8+ T Cells in Acute SARS-CoV-2 Infection. Nature 2021, 602, 148–155. [Google Scholar] [CrossRef]
- Vega-Magaña, N.; Muñoz-Valle, J.F.; Peña-Rodríguez, M.; Viera-Segura, O.; Pereira-Suárez, A.L.; Hernández-Bello, J.; García-Chagollan, M. Specific T-Cell Immune Response to SARS-CoV-2 Spike Protein over Time in Naïve and SARS-CoV-2 Previously Infected Subjects Vaccinated with BTN162b2. Vaccines 2022, 10, 1117. [Google Scholar] [CrossRef]
- Vardhana, S.A.; Wolchok, J.D. The Many Faces of the Anti-COVID Immune Response. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef]
- Rondovic, G.; Djordjevic, D.; Udovicic, I.; Stanojevic, I.; Zeba, S.; Abazovic, T.; Vojvodic, D.; Abazovic, D.; Khan, W.; Surbatovic, M. From Cytokine Storm to Cytokine Breeze: Did Lessons Learned from Immunopathogenesis Improve Immunomodulatory Treatment of Moderate-to-Severe COVID-19? Biomedicines 2022, 10, 2620. [Google Scholar] [CrossRef]
- McLane, L.M.; Abdel-Hakeem, M.S.; Wherry, E.J. CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu. Rev. Immunol. 2019, 37, 457–495. [Google Scholar] [CrossRef]
- Eskandari-Malayeri, F.; Rezaei, M. Immune Checkpoint Inhibitors as Mediators for Immunosuppression by Cancer-Associated Fibroblasts: A Comprehensive Review. Front. Immunol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Sears, J.D.; Waldron, K.J.; Wei, J.; Chang, C. Targeting Metabolism to Reverse T-cell Exhaustion in Chronic Viral Infections. Immunology 2020, 162, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Brooks, D.G.; Trifilo, M.J.; Edelmann, K.H.; Teyton, L.; McGavern, D.B.; Oldstone, M.B.A. Interleukin-10 Determines Viral Clearance or Persistence in Vivo. Nat. Med. 2006, 12, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Ejrnaes, M.; Filippi, C.M.; Martinic, M.M.; Ling, E.M.; Togher, L.M.; Crotty, S.; von Herrath, M.G. Resolution of a Chronic Viral Infection after Interleukin-10 Receptor Blockade. J. Exp. Med. 2006, 203, 2461–2472. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Flies, D.B. Molecular Mechanisms of T Cell Co-Stimulation and Co-Inhibition. Nat. Rev. Immunol. 2013, 13, 227–242. [Google Scholar] [CrossRef]
- Lanier, O.L.; Pérez-Herrero, E.; Andrea, A.P.D.; Bahrami, K.; Lee, E.; Ward, D.M.; Ayala-Suárez, N.; Rodríguez-Méndez, S.M.; Peppas, N.A. Immunotherapy Approaches for Hematological Cancers. iScience 2022, 25, 105326. [Google Scholar] [CrossRef]
- Chiappelli, F. CoViD-19 Immunopathology & Immunotherapy. Bioinformation 2020, 16, 219–222. [Google Scholar] [CrossRef]
- Saeidi, A.; Zandi, K.; Cheok, Y.Y.; Saeidi, H.; Wong, W.F.; Lee, C.Y.Q.; Cheong, H.C.; Yong, Y.K.; Larsson, M.; Shankar, E.M. T-Cell Exhaustion in Chronic Infections: Reversing the State of Exhaustion and Reinvigorating Optimal Protective Immune Responses. Front. Immunol. 2018, 9, 2569. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Wang, X.-M.; Xing, X.; Xu, Z.; Zhang, C.; Song, J.-W.; Fan, X.; Xia, P.; Fu, J.-L.; Wang, S.-Y.; et al. Single-Cell Landscape of Immunological Responses in Patients with COVID-19. Nat. Immunol. 2020, 21, 1107–1118. [Google Scholar] [CrossRef]
- Zheng, H.-Y.; Zhang, M.; Yang, C.-X.; Zhang, N.; Wang, X.-C.; Yang, X.-P.; Dong, X.-Q.; Zheng, Y.-T. Elevated Exhaustion Levels and Reduced Functional Diversity of T Cells in Peripheral Blood May Predict Severe Progression in COVID-19 Patients. Cell. Mol. Immunol. 2020, 17, 541–543. [Google Scholar] [CrossRef]
- Toor, S.M.; Saleh, R.; Sasidharan Nair, V.; Taha, R.Z.; Elkord, E. T-cell Responses and Therapies against SARS-CoV-2 Infection. Immunology 2020, 162, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Demoliou, C.; Papaneophytou, C.; Nicolaidou, V. SARS-CoV-2 and HIV-1: So Different yet so Alike. Immune Response at the Cellular and Molecular Level. Int. J. Med. Sci. 2022, 19, 1787–1795. [Google Scholar] [CrossRef] [PubMed]
- De Biasi, S.; Meschiari, M.; Gibellini, L.; Bellinazzi, C.; Borella, R.; Fidanza, L.; Gozzi, L.; Iannone, A.; Lo Tartaro, D.; Mattioli, M.; et al. Marked T Cell Activation, Senescence, Exhaustion and Skewing towards TH17 in Patients with COVID-19 Pneumonia. Nat. Commun. 2020, 11, 3434. [Google Scholar] [CrossRef] [PubMed]
- Rha, M.-S.; Jeong, H.W.; Ko, J.-H.; Choi, S.J.; Seo, I.-H.; Lee, J.S.; Sa, M.; Kim, A.R.; Joo, E.-J.; Ahn, J.Y.; et al. PD-1-Expressing SARS-CoV-2-Specific CD8+ T Cells Are Not Exhausted, but Functional in Patients with COVID-19. Immunity 2021, 54, 44–52.e3. [Google Scholar] [CrossRef]
- Tan, J.; Li, Y. Immune Checkpoint Alterations and Their Blockade in COVID-19 Patients. Blood Sci. 2022, 4, 192–198. [Google Scholar] [CrossRef]
- Van Eeden, C.; Khan, L.; Osman, M.S.; Cohen Tervaert, J.W. Natural Killer Cell Dysfunction and Its Role in COVID-19. Int. J. Mol. Sci. 2020, 21, 6351. [Google Scholar] [CrossRef]
- Antonioli, L.; Fornai, M.; Pellegrini, C.; Blandizzi, C. NKG2A and COVID-19: Another Brick in the Wall. Cell. Mol. Immunol. 2020, 17, 672–674. [Google Scholar] [CrossRef]
- Taefehshokr, N.; Taefehshokr, S.; Hemmat, N.; Heit, B. Covid-19: Perspectives on Innate Immune Evasion. Front. Immunol. 2020, 11, 580641. [Google Scholar] [CrossRef]
- Mohammed, R.N.; Tamjidifar, R.; Rahman, H.S.; Adili, A.; Ghoreishizadeh, S.; Saeedi, H.; Thangavelu, L.; Shomali, N.; Aslaminabad, R.; Marofi, F.; et al. A Comprehensive Review about Immune Responses and Exhaustion during Coronavirus Disease (COVID-19). Cell Commun. Signal. 2022, 20, 79. [Google Scholar] [CrossRef]
- Soltani-Zangbar, M.S.; Parhizkar, F.; Abdollahi, M.; Shomali, N.; Aghebati-Maleki, L.; Shahmohammadi Farid, S.; Roshangar, L.; Mahmoodpoor, A.; Yousefi, M. Immune System-Related Soluble Mediators and COVID-19: Basic Mechanisms and Clinical Perspectives. Cell Commun. Signal. 2022, 20, 1–131. [Google Scholar] [CrossRef]
- Lo Presti, E.; Dieli, F.; Meraviglia, S. Lymphopenia in COVID-19: Γδ T Cells-Based Therapeutic Opportunities. Vaccines 2021, 9, 562. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and Clinical Characteristics of 99 Cases of 2019 Novel Coronavirus Pneumonia in Wuhan, China: A Descriptive Study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Patra, R.; Chandra Das, N.; Mukherjee, S. Targeting Human TLRs to Combat COVID-19: A Solution? J. Med. Virol. 2020, 93, 615–617. [Google Scholar] [CrossRef] [PubMed]
- Poulas, K.; Farsalinos, K.; Zanidis, C. Activation of TLR7 and Innate Immunity as an Efficient Method Against COVID-19 Pandemic: Imiquimod as a Potential Therapy. Front. Immunol. 2020, 11, 1373. [Google Scholar] [CrossRef] [PubMed]
- Angelopoulou, A.; Alexandris, N.; Konstantinou, E.; Mesiakaris, K.; Zanidis, C.; Farsalinos, K.; Poulas, K. Imiquimod—A Toll like Receptor 7 Agonist—Is an Ideal Option for Management of COVID 19. Environ. Res. 2020, 188, 109858. [Google Scholar] [CrossRef]
- Iqbal Yatoo, M.; Hamid, Z.; Rather, I.; Nazir, Q.U.A.; Bhat, R.A.; Ul Haq, A.; Magray, S.N.; Haq, Z.; Sah, R.; Tiwari, R.; et al. Immunotherapies and Immunomodulatory Approaches in Clinical Trials—A Mini Review. Hum. Vaccines Immunother. 2021, 17, 1897–1909. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Al-Ahmed, S.H.; Muhammad, J.; Khan, A.; Sule, A.A.; Tirupathi, R.; Mutair, A.A.; Alhumaid, S.; Al-Omari, A.; Dhawan, M.; et al. Role of Inflammatory Cytokines in COVID-19 Patients: A Review on Molecular Mechanisms, Immune Functions, Immunopathology and Immunomodulatory Drugs to Counter Cytokine Storm. Vaccines 2021, 9, 436. [Google Scholar] [CrossRef]
- Feldmann, M.; Maini, R.N.; Woody, J.N.; Holgate, S.T.; Winter, G.; Rowland, M.; Richards, D.; Hussell, T. Trials of Anti-Tumour Necrosis Factor Therapy for COVID-19 Are Urgently Needed. Lancet 2020, 395, 1407–1409. [Google Scholar] [CrossRef]
- Capochiani, E.; Frediani, B.; Iervasi, G.; Paolicchi, A.; Sani, S.; Roncucci, P.; Cuccaro, A.; Franchi, F.; Simonetti, F.; Carrara, D.; et al. Ruxolitinib Rapidly Reduces Acute Respiratory Distress Syndrome in COVID-19 Disease. Analysis of Data Collection From RESPIRE Protocol. Front. Med. 2020, 7, 466. [Google Scholar] [CrossRef]
- Gupta, P.K.; Godec, J.; Wolski, D.; Adland, E.; Yates, K.; Pauken, K.E.; Cosgrove, C.; Ledderose, C.; Junger, W.G.; Robson, S.C.; et al. CD39 Expression Identifies Terminally Exhausted CD8+ T Cells. PLOS Pathog. 2015, 11, e1005177. [Google Scholar] [CrossRef]
- Trautmann, L.; Janbazian, L.; Chomont, N.; Said, E.A.; Gimmig, S.; Bessette, B.; Boulassel, M.-R.; Delwart, E.; Sepulveda, H.; Balderas, R.S.; et al. Upregulation of PD-1 Expression on HIV-Specific CD8+ T Cells Leads to Reversible Immune Dysfunction. Nat. Med. 2006, 12, 1198–1202. [Google Scholar] [CrossRef] [PubMed]
- Zahm, C.D.; Colluru, V.T.; McIlwain, S.J.; Ong, I.M.; McNeel, D.G. TLR Stimulation during T-Cell Activation Lowers PD-1 Expression on CD8+ T Cells. Cancer Immunol. Res. 2018, 6, 1364–1374. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, H.-H.; Tu, B.; Zhou, M.-J.; Hu, W.; Fu, Y.-L.; Li, X.-Y.; Yang, T.; Song, J.-W.; Fan, X.; et al. Reversal of the CD8+ T-Cell Exhaustion Induced by Chronic HIV-1 Infection Through Combined Blockade of the Adenosine and PD-1 Pathways. Front. Immunol. 2021, 12, 687296. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Han, F.; Zhu, W. CD39—A Bright Target for Cancer Immunotherapy. Biomed. Pharmacother. 2022, 151, 113066. [Google Scholar] [CrossRef]
- Kaufmann, D.E.; Walker, B.D. PD-1 and CTLA-4 Inhibitory Cosignaling Pathways in HIV Infection and the Potential for Therapeutic Intervention. J. Immunol. 2009, 182, 5891–5897. [Google Scholar] [CrossRef]
- Isazadeh, A.; Hajazimian, S.; Garshasbi, H.; Shadman, B.; Baghbanzadeh, A.; Chavoshi, R.; Taefehshokr, S.; Farhoudi Sefidan Jadid, M.; Hajiasgharzadeh, K.; Baradaran, B. Resistance Mechanisms to Immune Checkpoints Blockade by Monoclonal Antibody Drugs in Cancer Immunotherapy: Focus on Myeloma. J. Cell. Physiol. 2020, 236, 791–805. [Google Scholar] [CrossRef]
- Vivarelli, S.; Falzone, L.; Torino, F.; Scandurra, G.; Russo, G.; Bordonaro, R.; Pappalardo, F.; Spandidos, D.; Raciti, G.; Libra, M. Immune-Checkpoint Inhibitors from Cancer to COVID-19: A Promising Avenue for the Treatment of Patients with COVID-19 (Review). Int. J. Oncol. 2020, 58, 145–157. [Google Scholar] [CrossRef]
- Whitfield, S.J.C.; Taylor, C.; Risdall, J.E.; Griffiths, G.D.; Jones, J.T.A.; Williamson, E.D.; Rijpkema, S.; Saraiva, L.; Vessillier, S.; Green, A.C.; et al. Interference of the T Cell and Antigen-Presenting Cell Costimulatory Pathway Using CTLA4-Ig (Abatacept) Prevents Staphylococcal Enterotoxin B Pathology. J. Immunol. 2017, 198, 3989–3998. [Google Scholar] [CrossRef]
- Bersanelli, M.; Scala, S.; Affanni, P.; Veronesi, L.; Colucci, M.E.; Banna, G.L.; Cortellini, A.; Liotta, F. Immunological Insights on Influenza Infection and Vaccination during Immune Checkpoint Blockade in Cancer Patients. Immunotherapy 2020, 12, 105–110. [Google Scholar] [CrossRef]
- Di Cosimo, S.; Malfettone, A.; Pérez-García, J.M.; Llombart-Cussac, A.; Miceli, R.; Curigliano, G.; Cortés, J. Immune Checkpoint Inhibitors: A Physiology-Driven Approach to the Treatment of Coronavirus Disease 2019. Eur. J. Cancer 2020, 135, 62–65. [Google Scholar] [CrossRef]
- Pezeshki, P.S.; Rezaei, N. Immune Checkpoint Inhibition in COVID-19: Risks and Benefits. Expert Opin. Biol. Ther. 2021, 21, 1173–1179. [Google Scholar] [CrossRef]
- Irvani, S.S.N.; Golmohammadi, M.; Pourhoseingholi, M.A.; Shokouhi, S.; Darazam, I.A. Effectiveness of Interferon Beta 1a, Compared to Interferon Beta 1b and the Usual Therapeutic Regimen to Treat Adults with Moderate to Severe COVID-19: Structured Summary of a Study Protocol for a Randomized Controlled Trial. Trials 2020, 21, 473. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, T.; Hato, T.; Okayama, T.; Ikeo, K.; Miyamoto, Y.; Iwanaga, N.; Suzuki, K.; Yoshida, M.; Yamashita, K.; Yamashita, S.; et al. Th1 Cytokine Endotype Discriminates and Predicts Severe Complications in COVID-19. Eur. Cytokine Netw. 2022, 33, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Yang, X.O. TH17 Responses in Cytokine Storm of COVID-19: An Emerging Target of JAK2 Inhibitor Fedratinib. J.Microbiol. Immunol. Infect. 2020, 53, 368–370. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, M.; Dhama, K.; Parmar, M.; Sharma, A.; Angural, S. Unravelling the Potentialities of Tocilizumab for the Development of a Potential Immunotherapeutic Regimen against COVID-19—A Narrative Review. J. Appl. Pharm. Sci. 2021, 11, 026–033. [Google Scholar] [CrossRef]
- Corcoran, A.E.; Riddell, A.; Krooshoop, D.; Venkitaraman, A.R. Impaired Immunoglobulin Gene Rearrangement in Mice Lacking the IL-7 Receptor. Nature 1998, 391, 904–907. [Google Scholar] [CrossRef] [PubMed]
- Terrazzini, N.; Mantegani, P.; Kern, F.; Fortis, C.; Mondino, A.; Caserta, S. Interleukin-7 Unveils Pathogen-Specific T Cells by Enhancing Antigen-Recall Responses. J. Infect. Dis. 2018, 217, 1997–2007. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Long, Z.; Jia, R.; Wang, M.; Zhu, D.; Liu, M.; Chen, S.; Zhao, X.; Yang, Q.; Wu, Y.; et al. The Broad Immunomodulatory Effects of IL-7 and Its Application In Vaccines. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef]
- Arenas-Ramirez, N.; Woytschak, J.; Boyman, O. Interleukin-2: Biology, Design and Application. Trends Immunol. 2015, 36, 763–777. [Google Scholar] [CrossRef]
- Pietrobon, V.; Todd, L.A.; Goswami, A.; Stefanson, O.; Yang, Z.; Marincola, F. Improving CAR T-Cell Persistence. Int. J. Mol. Sci. 2021, 22, 10828. [Google Scholar] [CrossRef]
- Mahmoudpour, S.H.; Jankowski, M.; Valerio, L.; Becker, C.; Espinola-Klein, C.; Konstantinides, S.; Quitzau, K.; Barco, S. Safety of Low-Dose Subcutaneous Recombinant Interleukin-2: Systematic Review and Meta-Analysis of Randomized Controlled Trials. Sci. Rep. 2019, 9, 7145. [Google Scholar] [CrossRef]
- Wang, D.Y.; Salem, J.-E.; Cohen, J.V.; Chandra, S.; Menzer, C.; Ye, F.; Zhao, S.; Das, S.; Beckermann, K.E.; Ha, L.; et al. Fatal Toxic Effects Associated With Immune Checkpoint Inhibitors. JAMA Oncol. 2018, 4, 1721. [Google Scholar] [CrossRef]
- Schmitt, C.A.; Tchkonia, T.; Niedernhofer, L.J.; Robbins, P.D.; Kirkland, J.L.; Lee, S. COVID-19 and Cellular Senescence. Nat. Rev. Immunol. 2022, 5, 1–13. [Google Scholar] [CrossRef]
- Leen, A.M.; Myers, G.D.; Sili, U.; Huls, M.H.; Weiss, H.; Leung, K.S.; Carrum, G.; Krance, R.A.; Chang, C.-C.; Molldrem, J.J.; et al. Monoculture-Derived T Lymphocytes Specific for Multiple Viruses Expand and Produce Clinically Relevant Effects in Immunocompromised Individuals. Nat. Med. 2006, 12, 1160–1166. [Google Scholar] [CrossRef]
- Sportès, C.; Hakim, F.T.; Memon, S.A.; Zhang, H.; Chua, K.S.; Brown, M.R.; Fleisher, T.A.; Krumlauf, M.C.; Babb, R.R.; Chow, C.K.; et al. Administration of rhIL-7 in Humans Increases in Vivo TCR Repertoire Diversity by Preferential Expansion of Naive T Cell Subsets. J. Exp. Med. 2008, 205, 1701–1714. [Google Scholar] [CrossRef]
- Unsinger, J.; McGlynn, M.; Kasten, K.R.; Hoekzema, A.S.; Watanabe, E.; Muenzer, J.T.; McDonough, J.S.; Tschoep, J.; Ferguson, T.A.; McDunn, J.E.; et al. IL-7 Promotes T Cell Viability, Trafficking, and Functionality and Improves Survival in Sepsis. J. Immunol. 2010, 184, 3768–3779. [Google Scholar] [CrossRef]
- Perales, M.-A.; Goldberg, J.D.; Yuan, J.; Koehne, G.; Lechner, L.; Papadopoulos, E.B.; Young, J.W.; Jakubowski, A.A.; Zaidi, B.; Gallardo, H.; et al. Recombinant Human Interleukin-7 (CYT107) Promotes T-Cell Recovery after Allogeneic Stem Cell Transplantation. Blood 2012, 120, 4882–4891. [Google Scholar] [CrossRef]
- Nanjappa, S.G.; Kim, E.H.; Suresh, M. Immunotherapeutic Effects of IL-7 during a Chronic Viral Infection in Mice. Blood 2011, 117, 5123–5132. [Google Scholar] [CrossRef]
- Nelson, B.H. IL-2, Regulatory T Cells, and Tolerance. J. Immunol. 2004, 172, 3983–3988. [Google Scholar] [CrossRef]
- Watson, J.; Mochizuki, D.; Gillis, S. T-Cell Growth Factors: Interleukin 2. Immunol. Today 1980, 1, 113–117. [Google Scholar] [CrossRef]
- Pham, M.N.; von Herrath, M.G.; Vela, J.L. Antigen-Specific Regulatory T Cells and Low Dose of IL-2 in Treatment of Type 1 Diabetes. Front. Immunol. 2016, 6, 651. [Google Scholar] [CrossRef]
- Ye, C.; Brand, D.; Zheng, S.G. Targeting IL-2: An Unexpected Effect in Treating Immunological Diseases. Signal Transduct. Target. Ther. 2018, 3, 2. [Google Scholar] [CrossRef]
- Schwarz, M.; Mzoughi, S.; Lozano-Ojalvo, D.; Tan, A.T.; Bertoletti, A.; Guccione, E. T Cell Immunity Is Key to the Pandemic Endgame: How to Measure and Monitor It. Curr. Res. Immunol. 2022, 3, 215–221. [Google Scholar] [CrossRef]
- Cox, J.; Ferrari, G.; Janetzki, S. Measurement of Cytokine Release at the Single Cell Level Using the ELISPOT Assay. Methods 2006, 38, 274–282. [Google Scholar] [CrossRef]
- Czerkinsky, C.C.; Nilsson, L.-Å.; Nygren, H.; Ouchterlony, Ö.; Tarkowski, A. A Solid-Phase Enzyme-Linked Immunospot (ELISPOT) Assay for Enumeration of Specific Antibody-Secreting Cells. J. Immunol. Methods 1983, 65, 109–121. [Google Scholar] [CrossRef]
- Gazagne, A.; Claret, E.; Wijdenes, J.; Yssel, H.; Bousquet, F.; Levy, E.; Vielh, P.; Scotte, F.; Goupil, T.L.; Fridman, W.H.; et al. A Fluorospot Assay to Detect Single T Lymphocytes Simultaneously Producing Multiple Cytokines. J. Immunol. Methods 2003, 283, 91–98. [Google Scholar] [CrossRef]
- Janetzki, S.; Rueger, M.; Dillenbeck, T. Stepping up ELISpot: Multi-Level Analysis in FluoroSpot Assays. Cells 2014, 3, 1102–1115. [Google Scholar] [CrossRef]
- Safont, G.; Latorre, I.; Villar-Hernández, R.; Stojanovic, Z.; Marín, A.; Pérez-Cano, C.; Lacoma, A.; Molina-Moya, B.; Solis, A.J.; Arméstar, F.; et al. Measuring T-Cell Responses against SARS-CoV-2 Is of Utility for Disease and Vaccination Management. J. Clin. Med. 2022, 11, 5103. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Sohail, M.S.; Quadeer, A.A.; McKay, M.R. Identification of Potential SARS-CoV-2 CD8+ T Cell Escape Mutants. Vaccines 2022, 10, 542. [Google Scholar] [CrossRef]

| Drug and Treatment Approach | ClinicalTrials.gov Identifier | Drug Candidates | Mechanism of Action | Therapeutic Benefits | Possible Side Effects | References |
|---|---|---|---|---|---|---|
| NCT04268537 | PD-1 blocking antibody and thymosin | Blockade of PD-1 | Reversal of T-cell exhaustion; Stimulate T-cells production | Immune-related adverse events and exaggerated activation of immune cells | [141,170,171] | |
| Inhibition of immune checkpoints | NCT04413838, NCT04356508 | Nivolumab | Blockade of PD-1 | Anti-PD1 | Immune-related adverse events | [141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170] |
| NCT04333914 NCT04333914 | GNS561, Monalizumab, Avdoralimab | Inhibition of autophagy inhibitor; blocking of NKG2A; anti-C5aR | Reversal of T-cell exhaustion; Restoration of T-cell numbers; Restored effector T-cell function | uncontrolled activation of immune cells | [140,170,171,172,173] | |
| Th1 activators | NCT04343768 | Ziferon | Activation of Th1 type T cells and release of | Improved symptoms | Uncontrolled activation of immune cells | [173,174,175] |
| IL-6 inhibitors | NCT04320615, NCT04317092 | Tocilizumab | Inhibit the binding of IL-6 with their receptors and alleviate the cytokine storm | Indirectly reduce the T cells’ exhaustion and lymphocytopenia | Efficacy and risk status of TCZ in patients at risk of other deadly infections, High cost, availability and | [170,176,177] |
| Th17 blockers | N.A. | Anti-IL-17, and anti-IL-22 | Inhibition of the cytokines such as IL-17 and IL-22 | Decreased production of IL-17, and IL-22, Activation of Th1 type cells, reduction in the viral load | N.A. | [141,176] |
| JAK2 inhibitor | N.A. | Fedratinib | Inhibit the excessive production of cytokines and chemokines | Reduce the inflammation | May suppress the immune response | [174,175,176] |
| Administration of recombinant IL-7 or IL-7 as vaccine adjuvant | NCT04407689, NCT04379076, NCT04426201 | CYT107 | Rearrangement of immunoglobulin (Ig) genes in immature B cell subsets regulated by IL-7. In order to maintain the diversity of T cells and the primary antibody repertoire, IL-7 also controls the T cell receptor (TCR) genes in precursor T cell subsets through the IL-7 receptor (IL-7R) signalling pathway. | Restored T-cell count and reversed lymphopenia, Enhanced TCR repertoire diversity and generation of memory CD8+ T cells, Improved trafficking of T cells to the infection site | N.A. | [141,178] |
| A low dose of recombinant IL-2 | NCT04357444 | ILT101 | IL-2 maintains homeostasis in the immune response through its influence on the Tregs and effector lymphocyte responses. | Controlled stimulation of Tregs to control excessive inflammation and proliferation of Tregs and other T-cell subsets, including effector cells | Suppression of unwarranted and other inflammatory cells, including those that are necessary | [141,178,179] |
| Adoptive T-cell transfer (ACT) | NCT04351659 | N.A. | SARS-CoV-2-reactive T cells from the patient are genetically modified ex vivo to kill virally infected cells before being reintroduced into the patient. | Overcome the T cell exhaustion. Improved specific antiviral T-cell responses against SARS-CoV-2 | Risk of relapse due to several factors, including poor T-cell expansion and lack of long-term persistence after adoptive transfer | [141,180] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhawan, M.; Rabaan, A.A.; Fawarah, M.M.A.; Almuthree, S.A.; Alsubki, R.A.; Alfaraj, A.H.; Mashraqi, M.M.; Alshamrani, S.A.; Abduljabbar, W.A.; Alwashmi, A.S.S.; et al. Updated Insights into the T Cell-Mediated Immune Response against SARS-CoV-2: A Step towards Efficient and Reliable Vaccines. Vaccines 2023, 11, 101. https://doi.org/10.3390/vaccines11010101
Dhawan M, Rabaan AA, Fawarah MMA, Almuthree SA, Alsubki RA, Alfaraj AH, Mashraqi MM, Alshamrani SA, Abduljabbar WA, Alwashmi ASS, et al. Updated Insights into the T Cell-Mediated Immune Response against SARS-CoV-2: A Step towards Efficient and Reliable Vaccines. Vaccines. 2023; 11(1):101. https://doi.org/10.3390/vaccines11010101
Chicago/Turabian StyleDhawan, Manish, Ali A. Rabaan, Mahmoud M. Al Fawarah, Souad A. Almuthree, Roua A. Alsubki, Amal H. Alfaraj, Mutaib M. Mashraqi, Saleh A. Alshamrani, Wesam A. Abduljabbar, Ameen S. S. Alwashmi, and et al. 2023. "Updated Insights into the T Cell-Mediated Immune Response against SARS-CoV-2: A Step towards Efficient and Reliable Vaccines" Vaccines 11, no. 1: 101. https://doi.org/10.3390/vaccines11010101
APA StyleDhawan, M., Rabaan, A. A., Fawarah, M. M. A., Almuthree, S. A., Alsubki, R. A., Alfaraj, A. H., Mashraqi, M. M., Alshamrani, S. A., Abduljabbar, W. A., Alwashmi, A. S. S., Ibrahim, F. A., Alsaleh, A. A., Khamis, F., Alsalman, J., Sharma, M., & Emran, T. B. (2023). Updated Insights into the T Cell-Mediated Immune Response against SARS-CoV-2: A Step towards Efficient and Reliable Vaccines. Vaccines, 11(1), 101. https://doi.org/10.3390/vaccines11010101









