Generation of a DSF-Guided Refolded Bacterially Expressed Hemagglutinin Ectodomain of Influenza Virus A/Puerto Rico/8/1934 H1N1 as a Model for Influenza Vaccine Antigens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Construction of Expression Vector
2.3. Recombinant Protein Expression in E. coli
2.4. Inclusion Body Purification and Solubilization
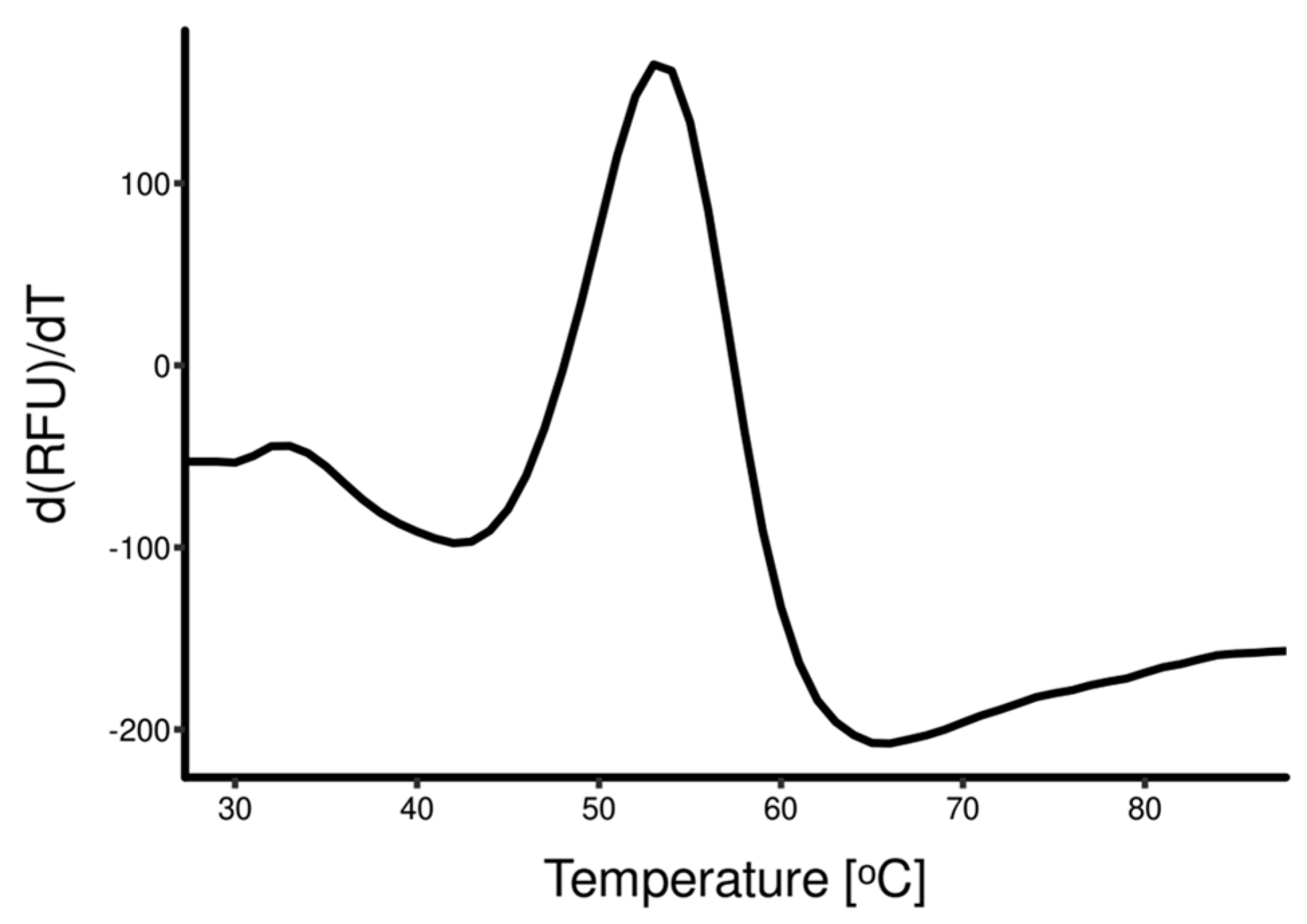
2.5. Differential Scanning Fluorimetry Guided Refolding
2.6. Protein Purification and Characterization
2.7. Hemagglutination Assay
2.8. Mice Immunization and Challenge Infection with Influenza Virus A H1N1
2.9. Humoral Immune Response Evaluation
2.10. Hemagglutination Inhibition Assay
3. Results
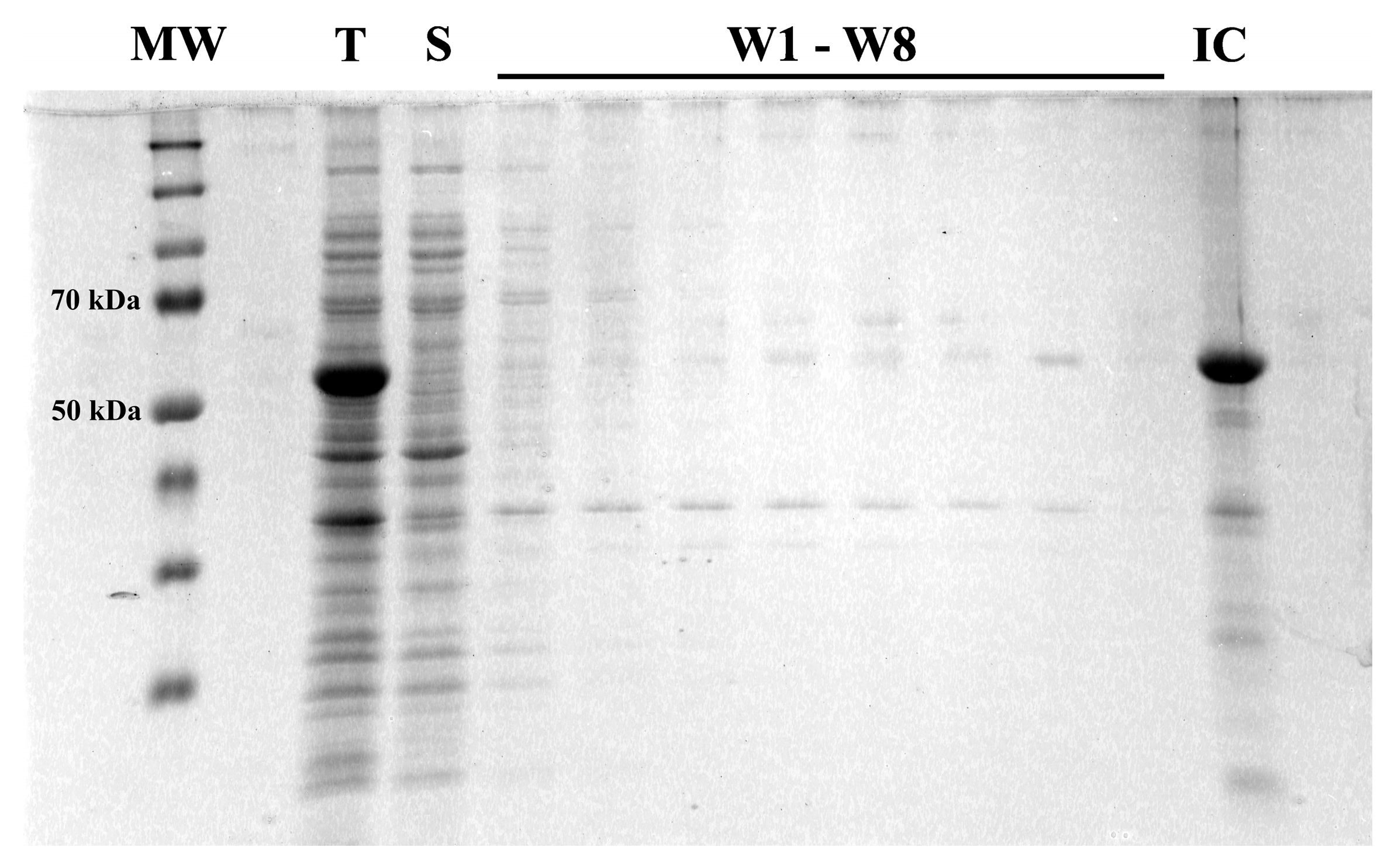
3.1. Hemagglutinin Extracellular Domain Was Expressed in Bacterial Host
3.2. Rapid Dilution Protein Refolding Optimization Using DSF
3.3. Refolded Hemagglutinin Consists of a Mix of Soluble Monomers and Oligomers with Biological Activity
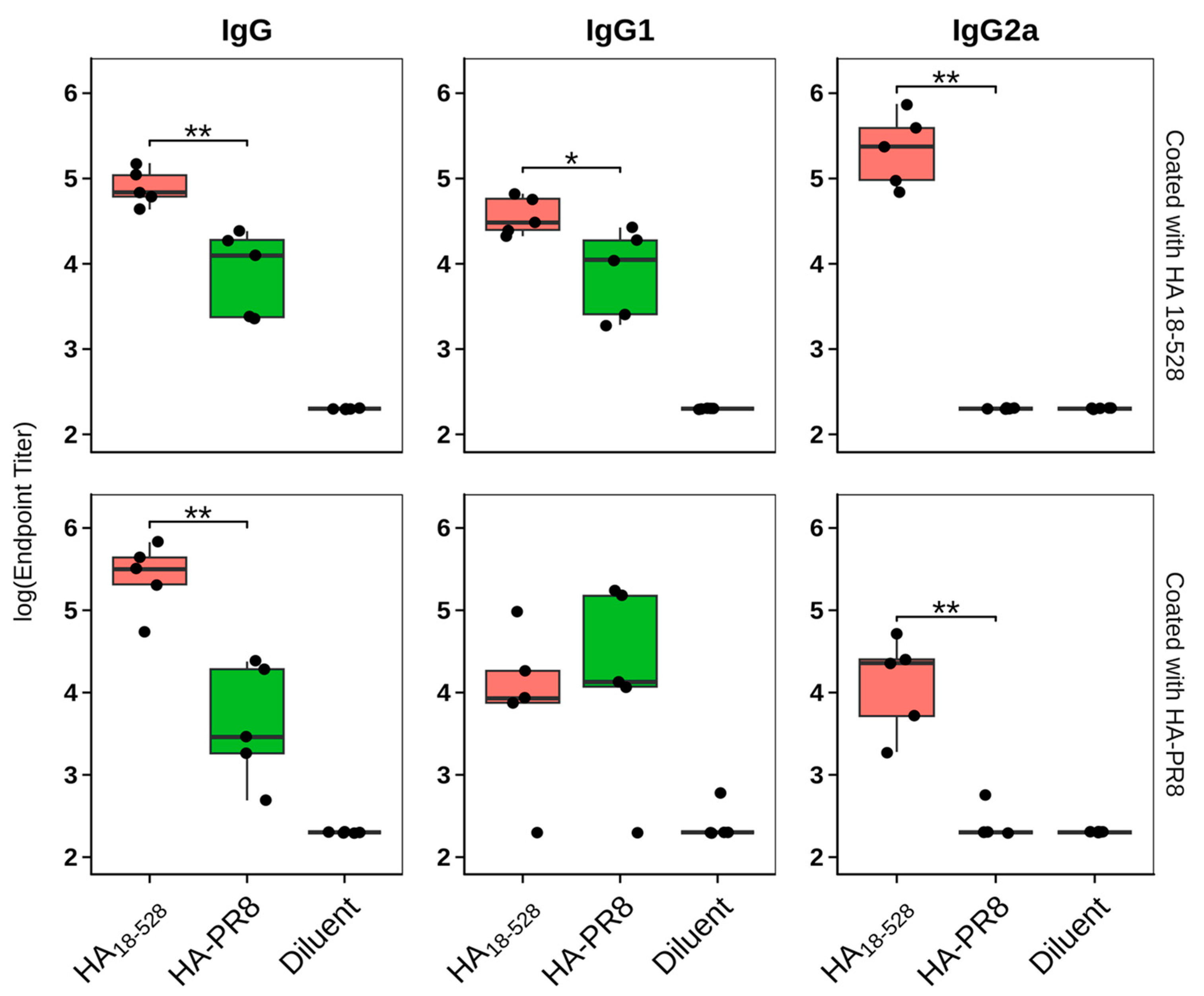
3.4. Immunization of Mice with HA18–528 Generates Neutralizing Antibodies Coupled with a Balanced Th1/Th2 Immune Response and Protects against Viral Challenge
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Tanner, A.R.; Dorey, R.B.; Brendish, N.J.; Clark, T.W. Influenza vaccination: Protecting the most vulnerable. Eur. Respir. Rev. 2021, 30, 200258. [Google Scholar] [CrossRef] [PubMed]
- Iuliano, A.D.; Roguski, K.M.; Chang, H.H.; Muscatello, D.J.; Palekar, R.; Tempia, S.; Cohen, C.; Gran, J.M.; Schanzer, D.; Cowling, B.J.; et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet 2018, 391, 1285–1300. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control Prevention 2022–2023, U.S. Flu Season: Preliminary In-Season Burden Estimates. Available online: https://www.cdc.gov/flu/about/burden/preliminary-in-season-estimates.htm (accessed on 8 August 2023).
- Zumofen, M.H.B.; Frimpter, J.; Hansen, S.A. Impact of Influenza and Influenza-Like Illness on Work Productivity Outcomes: A Systematic Literature Review. PharmacoEconomics 2022, 41, 253–273. [Google Scholar] [CrossRef]
- Uyeki, T.M.; Hui, D.S.; Zambon, M.; Wentworth, D.E.; Monto, A.S. Influenza. Lancet 2022, 400, 693–706. [Google Scholar] [CrossRef]
- Dey, P.; Ahuja, A.; Panwar, J.; Choudhary, P.; Rani, S.; Kaur, M.; Sharma, A.; Kaur, J.; Yadav, A.K.; Sood, V.; et al. Immune Control of Avian Influenza Virus Infection and Its Vaccine Development. Vaccines 2023, 11, 593. [Google Scholar] [CrossRef] [PubMed]
- Dawood, F.S.; Iuliano, A.D.; Reed, C.; Meltzer, M.I.; Shay, D.K.; Cheng, P.-Y.; Bandaranayake, D.; Breiman, R.F.; Brooks, W.A.; Buchy, P.; et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: A modelling study. Lancet Infect. Dis. 2012, 12, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Morens, D.M. 1918 Influenza: The Mother of All Pandemics. Emerg. Infect. Dis. 2006, 12, 15–22. [Google Scholar] [CrossRef]
- Kilbourne, E.D. Influenza Pandemics of the 20th Century. Emerg. Infect. Dis. 2006, 12, 9–14. [Google Scholar] [CrossRef]
- Duwe, S.C.; Schmidt, B.; Gärtner, B.C.; Timm, J.; Adams, O.; Fickenscher, H.; Schmidtke, M. Prophylaxis and treatment of influenza: Options, antiviral susceptibility, and existing recommendations. GMS Infect. Dis. 2021, 9, Doc02. [Google Scholar] [CrossRef]
- Javanian, M.; Barary, M.; Ghebrehewet, S.; Koppolu, V.; Vasigala, V.; Ebrahimpour, S. A brief review of influenza virus infection. J. Med. Virol. 2021, 93, 4638–4646. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.R.; Liu, Y.M.; Tseng, Y.C.; Ma, C. Better influenza vaccines: An industry perspective. J. Biomed. Sci. 2020, 27, 33. [Google Scholar] [CrossRef] [PubMed]
- Stavaru, C.; Onu, A.; Lupulescu, E.; Tucureanu, C.; Rasid, O.; Vlase, E.; Coman, C.; Caras, I.; Ghiorghisor, A.; Berbecila, L.; et al. Technology transfer of oil-in-water emulsion adjuvant manufacturing for pandemic influenza vaccine production in Romania: Preclinical evaluation of split virion inactivated H5N1 vaccine with adjuvant. Hum. Vaccines Immunother. 2016, 12, 1009–1026. [Google Scholar] [CrossRef]
- Widjaja, L.; Ilyushina, N.; Webster, R.G.; Webby, R.J. Molecular changes associated with adaptation of human influenza A virus in embryonated chicken eggs. Virology 2006, 350, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Chambers, B.S.; Parkhouse, K.; Ross, T.M.; Alby, K.; Hensley, S.E. Identification of Hemagglutinin Residues Responsible for H3N2 Antigenic Drift during the 2014–2015 Influenza Season. Cell Rep. 2015, 12, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Trombetta, C.M.; Gianchecchi, E.; Montomoli, E. Influenza vaccines: Evaluation of the safety profile. Hum. Vaccines Immunother. 2018, 14, 657–670. [Google Scholar] [CrossRef]
- Cox, M.M.J.; Izikson, R.; Post, P.; Dunkle, L. Safety, efficacy, and immunogenicity of Flublok in the prevention of seasonal influenza in adults. Ther. Adv. Vaccines 2015, 3, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef]
- Legastelois, I.; Buffin, S.; Peubez, I.; Mignon, C.; Sodoyer, R.; Werle, B. Non-conventional expression systems for the production of vaccine proteins and immunotherapeutic molecules. Hum. Vaccines Immunother. 2017, 13, 947–961. [Google Scholar] [CrossRef]
- Selas Castiñeiras, T.; Williams, S.G.; Hitchcock, A.G.; Smith, D.C. E. coli strain engineering for the production of advanced biopharmaceutical products. FEMS Microbiol. Lett. 2018, 365, fny162. [Google Scholar] [CrossRef]
- Nabiel, A.; Yosua, Y.; Sriwidodo, S.; Maksum, I.P. Overview of refolding methods on misfolded recombinant proteins from Escherichia coli inclusion bodies. J. App Biol. Biotech. 2023, 11, 47–52. [Google Scholar] [CrossRef]
- Vigerust, D.J.; Shepherd, V.L. Virus glycosylation: Role in virulence and immune interactions. Trends Microbiol. 2007, 15, 211–218. [Google Scholar] [CrossRef]
- Cheng, N.; Liu, M.; Li, W.; Sun, B.; Liu, D.; Wang, G.; Shi, J.; Li, L. Protein post-translational modification in SARS-CoV-2 and host interaction. Front. Immunol. 2023, 13, 1068449. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z. Hetero-Stagger Cloning: Efficient and Rapid Cloning of PCR Products. Nucleic Acids Res. 1996, 24, 2458–2459. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Neu, H.C.; Heppel, L.A. The Release of Enzymes from Escherichia coli by Osmotic Shock and during the Formation of Spheroplasts. J. Biol. Chem. 1965, 240, 3685–3692. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Van Oosterwijk, N.; Ali, A.M.; Adawy, A.; Anindya, A.L.; Dömling, A.S.; Groves, M.R. A systematic protein refolding screen method using the DGR approach reveals that time and secondary TSA are essential variables. Sci. Rep. 2017, 7, 9355. [Google Scholar] [CrossRef]
- Wittig, I.; Braun, H.P.; Schägger, H. Blue native PAGE. Nat. Protoc. 2006, 1, 418–428. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis [Internet]; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 15 August 2023).
- Wiley, D.C.; Skehel, J.J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 1987, 56, 365–394. [Google Scholar] [CrossRef] [PubMed]
- Subbiah, J.; Oh, J.; Kim, K.H.; Shin, C.H.; Park, B.R.; Bhatnagar, N.; Seong, B.L.; Wang, B.Z. A chimeric thermostable M2e and H3 stalk-based universal influenza A virus vaccine. NPJ Vaccines 2022, 7, 68. [Google Scholar] [CrossRef] [PubMed]
- Farahmand, B.; Akbari, A.; Fotouhi Chahouki, F.; Mehrbod, P.; Jalili, N. Cloning, expression and purification of hemagglutinin conserved domain (HA2) of influenza A virus, to be used in broad-spectrum subunit vaccine cocktails. Vaccine Res. 2016, 3, 69–75. [Google Scholar] [CrossRef]
- Song, L.; Nakaar, V.; Kavita, U.; Price, A.; Huleatt, J.; Tang, J.; Jacobs, A.; Liu, G.; Huang, Y.; Desai, P.; et al. Efficacious Recombinant Influenza Vaccines Produced by High Yield Bacterial Expression: A Solution to Global Pandemic and Seasonal Needs. PLoS ONE 2008, 3, e2257. [Google Scholar] [CrossRef] [PubMed]
- Sączyńska, V.; Romanik-Chruścielewska, A.; Florys, K.; Cecuda-Adamczewska, V.; Łukasiewicz, N.; Sokołowska, I.; Kęsik-Brodacka, M.; Płucienniczak, G. Prime-Boost Vaccination with a Novel Hemagglutinin Protein Produced in Bacteria Induces Neutralizing Antibody Responses Against H5-Subtype Influenza Viruses in Commercial Chickens. Front. Immunol. 2019, 10, 2006. [Google Scholar] [CrossRef] [PubMed]
- Sączyńska, V. Influenza virus hemagglutinin as a vaccine antigen produced in bacteria. Acta Biochim. Pol. 2014, 61, 561–572. [Google Scholar] [CrossRef] [PubMed]
- García-Fruitós, E.; González-Montalbán, N.; Morell, M.; Vera, A.; Ferraz, R.M.; Arís, A.; Ventura, S. Aggregation as bacterial inclusion bodies does not imply inactivation of enzymes and fluorescent proteins. Microb. Cell Fact. 2005, 4, 27. [Google Scholar] [CrossRef]
- Niesen, F.H.; Berglund, H.; Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007, 2, 2212–2221. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Yamamoto, E.; Mannen, T.; Nagamune, T.; Nagamune, T. Protein refolding using chemical refolding additives. Biotechnol. J. 2013, 8, 17–31. [Google Scholar] [CrossRef]
- Russell, C.J. Orthomyxoviruses: Structure of Antigens. In Reference Module in Biomedical Sciences; Elsevier: Berlin/Heidelberg, Germany, 2016. [Google Scholar] [CrossRef]
- Khurana, S.; Verma, S.; Verma, N.; Crevar, C.J.; Carter, D.M.; Manischewitz, J.; King, L.R.; Ross, T.M.; Golding, H. Properly Folded Bacterially Expressed H1N1 Hemagglutinin Globular Head and Ectodomain Vaccines Protect Ferrets against H1N1 Pandemic Influenza Virus. PLoS ONE 2010, 5, e11548. [Google Scholar] [CrossRef]
- Ekiert, D.C.; Bhabha, G.; Elsliger, M.A.; Friesen, R.H.E.; Jongeneelen, M.; Throsby, M.; Goudsmit, J.; Wilson, I.A. Antibody Recognition of a Highly Conserved Influenza Virus Epitope. Science 2009, 324, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Huber, V.C.; McKeon, R.M.; Brackin, M.N.; Miller, L.A.; Keating, R.; Brown, S.A.; Makarova, N.; Perez, D.R.; MacDonald, G.H.; McCullers, J.A. Distinct Contributions of Vaccine-Induced Immunoglobulin G1 (IgG1) and IgG2a Antibodies to Protective Immunity against Influenza. Clin. Vaccine Immunol. 2006, 13, 981–990. [Google Scholar] [CrossRef]
- Heinimäki, S.; Lampinen, V.; Tamminen, K.; Hankaniemi, M.M.; Malm, M.; Hytönen, V.P.; Blazevic, V. Antigenicity and immunogenicity of HA2 and M2e influenza virus antigens conjugated to norovirus-like, VP1 capsid-based particles by the SpyTag/SpyCatcher technology. Virology 2022, 566, 89–97. [Google Scholar] [CrossRef]




| Number of Amino Acids | Molecular Weight [kDa] | pI | |
|---|---|---|---|
| HA-PR8 | 556 | 62.6 | 7.26 |
| HA18–528 | 519 | 58.7 | 7.02 |
| HA18–528 | HA-PR8 | Diluent | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse ID | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 |
| HI Titer | 1/40 | 1/40 | - | 1/160 | 1/80 | - | - | - | - | - | - | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tofan, V.-C.; Ermeneanu, A.-L.; Caraș, I.; Lenghel, A.; Ionescu, I.-E.; Țucureanu, C.; Gal, C.; Stăvaru, C.-G.; Onu, A. Generation of a DSF-Guided Refolded Bacterially Expressed Hemagglutinin Ectodomain of Influenza Virus A/Puerto Rico/8/1934 H1N1 as a Model for Influenza Vaccine Antigens. Vaccines 2023, 11, 1520. https://doi.org/10.3390/vaccines11101520
Tofan V-C, Ermeneanu A-L, Caraș I, Lenghel A, Ionescu I-E, Țucureanu C, Gal C, Stăvaru C-G, Onu A. Generation of a DSF-Guided Refolded Bacterially Expressed Hemagglutinin Ectodomain of Influenza Virus A/Puerto Rico/8/1934 H1N1 as a Model for Influenza Vaccine Antigens. Vaccines. 2023; 11(10):1520. https://doi.org/10.3390/vaccines11101520
Chicago/Turabian StyleTofan, Vlad-Constantin, Andreea-Laura Ermeneanu, Iuliana Caraș, Alina Lenghel, Irina-Elena Ionescu, Cătălin Țucureanu, Claudiu Gal, Crina-Georgeta Stăvaru, and Adrian Onu. 2023. "Generation of a DSF-Guided Refolded Bacterially Expressed Hemagglutinin Ectodomain of Influenza Virus A/Puerto Rico/8/1934 H1N1 as a Model for Influenza Vaccine Antigens" Vaccines 11, no. 10: 1520. https://doi.org/10.3390/vaccines11101520






