Capsid-Specific Antibody Responses of Domestic Pigs Immunized with Low-Virulent African Swine Fever Virus
Abstract
:1. Introduction
2. Materials and Methods
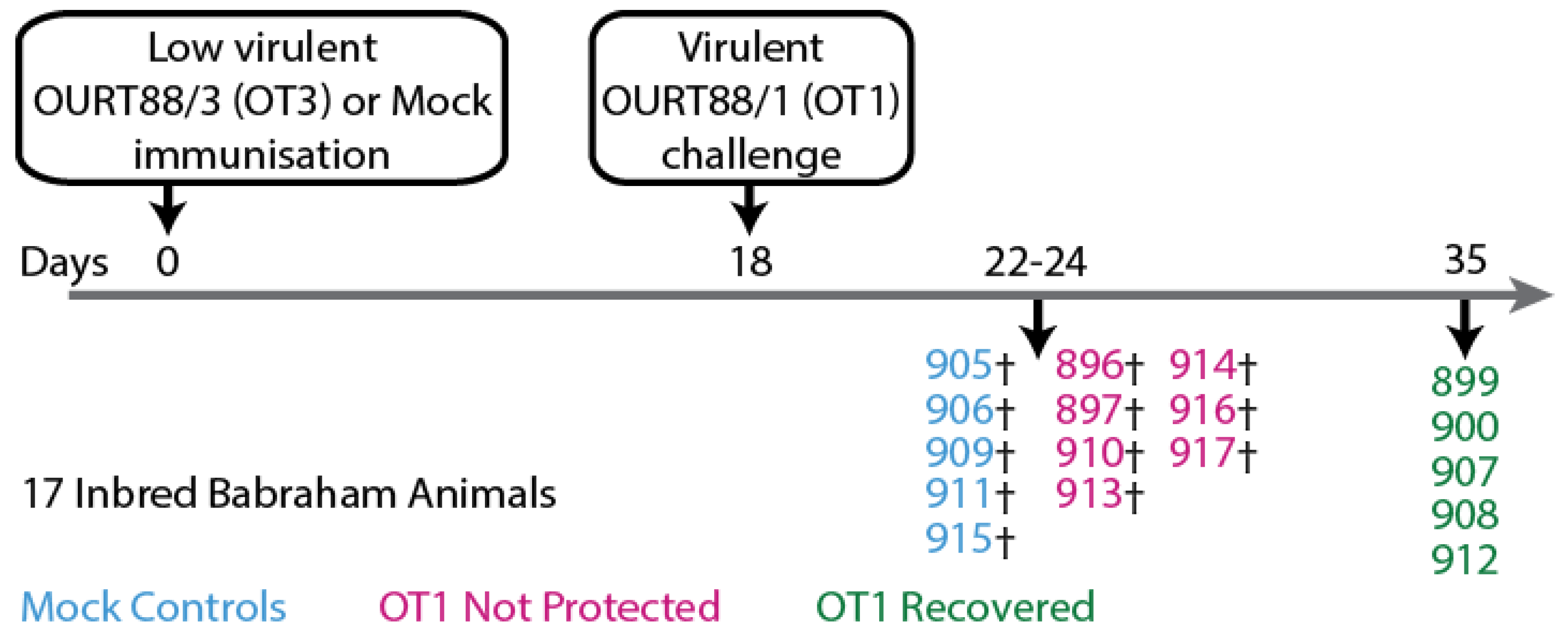
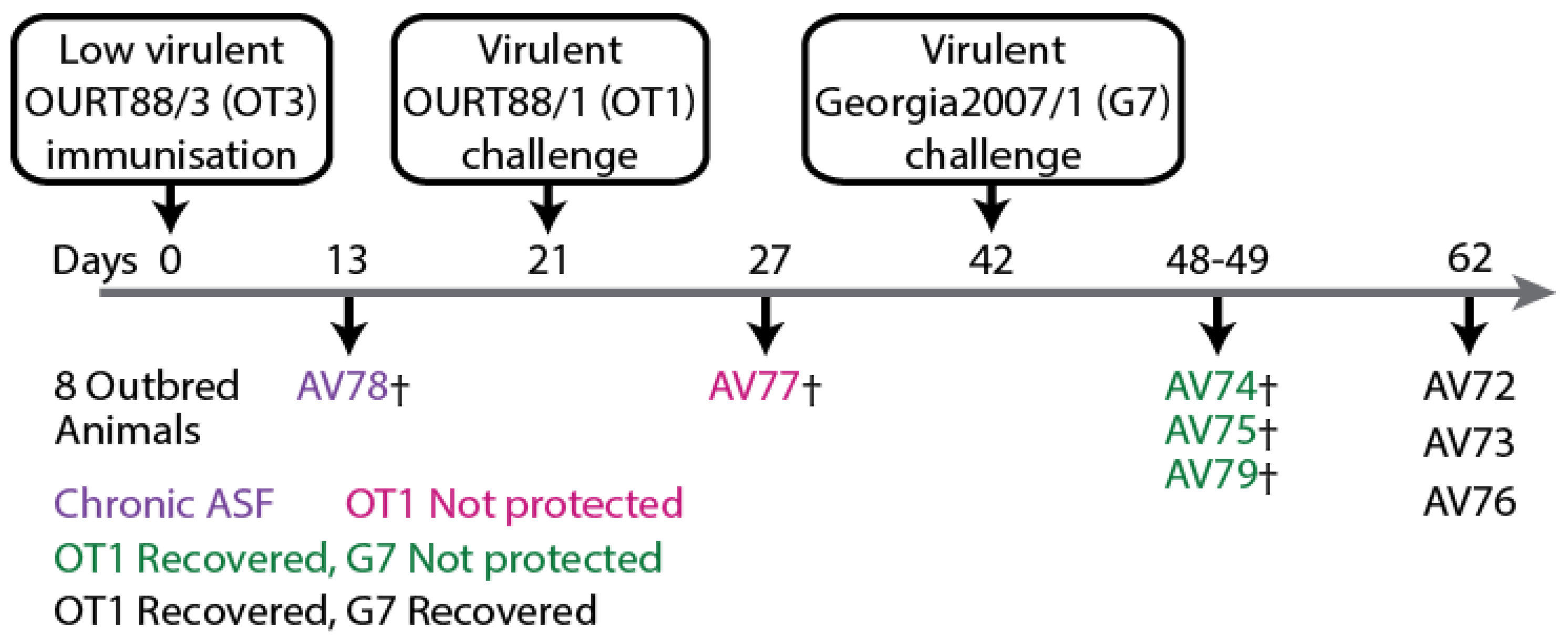
2.1. Sera Samples from Animal Experiments and Ethics Statement
2.2. Plasmids
2.3. Cell Culture and Transfections
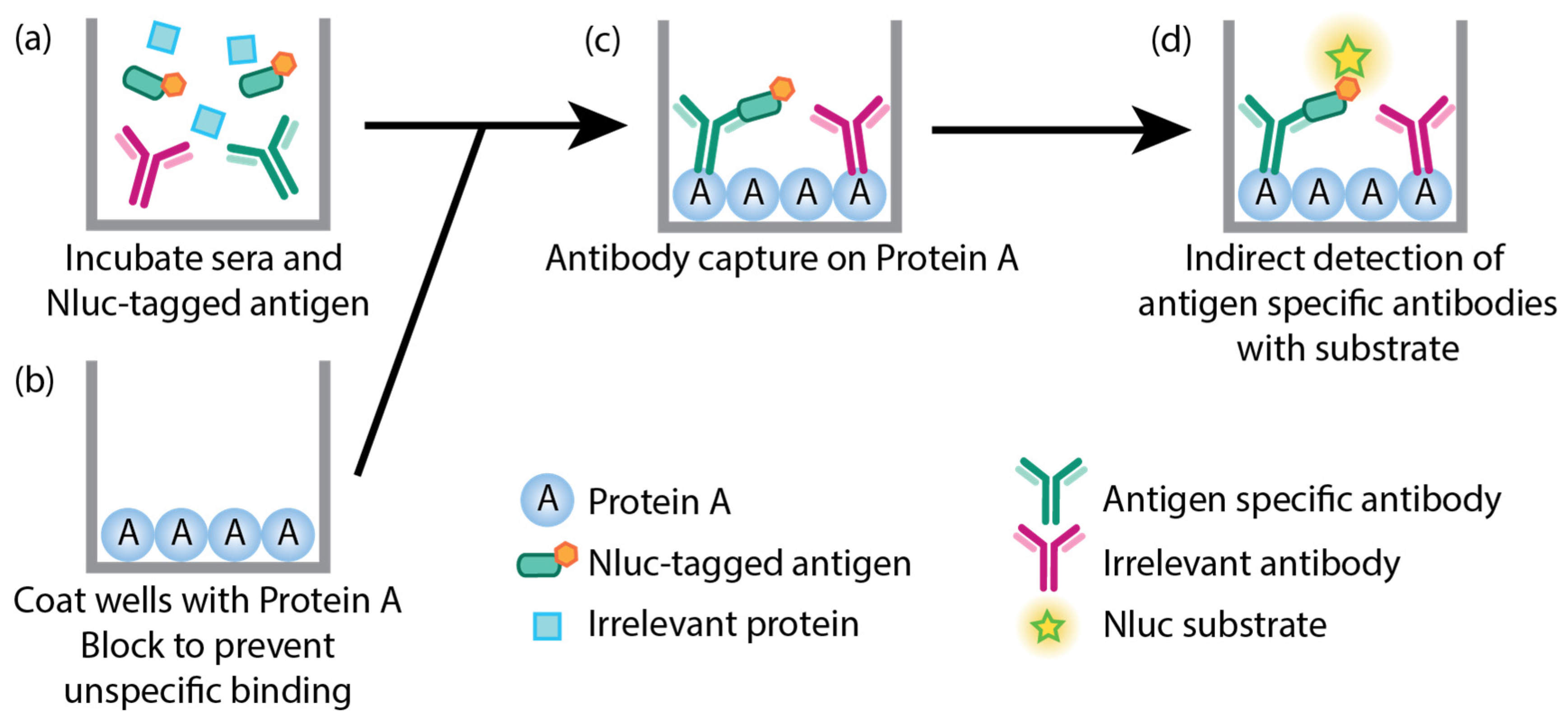
2.4. LACA
2.5. Statistics
3. Results
3.1. Antigen-Specific Antibody Responses of Inbred Babraham Pigs
3.2. Antigen-Specific Antibody Respones of Outbred Domestic Pigs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alejo, A.; Matamoros, T.; Guerra, M.; Andrés, G. A Proteomic Atlas of the African Swine Fever Virus Particle. J. Virol. 2018, 92, 23. [Google Scholar] [CrossRef]
- WOAH. World Organisation for Animal Health: African Swine Fever. Available online: https://www.woah.org/en/disease/african-swine-fever/ (accessed on 1 February 2023).
- Iacolina, L.; Penrith, M.L.; Bellini, S.; Chenais, E.; Jori, F.; Montoya, M.; Ståhl, K.; Gavier-Widén, D. Understanding and Combatting African Swine Fever: A European Perspective; Wageningen Academic Publishers: Wageningen, The Netherlands, 2021; pp. 161–182. [Google Scholar]
- Dixon, L.K.; Chapman, D.A.; Netherton, C.L.; Upton, C. African swine fever virus replication and genomics. Virus Res. 2013, 173, 3–14. [Google Scholar] [CrossRef]
- Cackett, G.; Matelska, D.; Sýkora, M.; Portugal, R.; Malecki, M.; Bähler, J.; Dixon, L.; Werner, F. The African Swine Fever Virus Transcriptome. J. Virol. 2020, 94, 9. [Google Scholar] [CrossRef]
- Bosch-Camós, L.; Alonso, U.; Esteve-Codina, A.; Chang, C.-Y.; Martín-Mur, B.; Accensi, F.; Muñoz, M.; Navas, M.J.; Dabad, M.; Vidal, E.; et al. Cross-protection against African swine fever virus upon intranasal vaccination is associated with an adaptive-innate immune crosstalk. PLoS Pathog. 2022, 18, e1010931. [Google Scholar] [CrossRef]
- Borca, M.V.; Ramirez-Medina, E.; Silva, E.; Vuono, E.; Rai, A.; Pruitt, S.; Holinka, L.G.; Velazquez-Salinas, L.; Zhu, J.; Gladue, D.P. Development of a Highly Effective African Swine Fever Virus Vaccine by Deletion of the I177L Gene Results in Sterile Immunity against the Current Epidemic Eurasia Strain. J. Virol. 2020, 94, 7. [Google Scholar] [CrossRef]
- Borca, M.V.; Ramirez-Medina, E.; Silva, E.; Vuono, E.; Rai, A.; Pruitt, S.; Espinoza, N.; Velazquez-Salinas, L.; Gay, C.G.; Gladue, D.P. ASFV-G-∆I177L as an Effective Oral Nasal Vaccine against the Eurasia Strain of Africa Swine Fever. Viruses 2021, 13, 765. [Google Scholar] [CrossRef] [PubMed]
- Gladue, D.P.; Borca, M.V. Recombinant ASF Live Attenuated Virus Strains as Experimental Vaccine Candidates. Viruses 2022, 14, 878. [Google Scholar] [CrossRef]
- Rathakrishnan, A.; Connell, S.; Petrovan, V.; Moffat, K.; Goatley, L.C.; Jabbar, T.; Sanchez-Cordon, P.J.; Reis, A.L.; Dixon, L.K. Differential Effect of Deleting Members of African Swine Fever Virus Multigene Families 360 and 505 from the Genotype II Georgia 2007/1 Isolate on Virus Replication, Virulence, and Induction of Protection. J. Virol. 2022, 96, e0189921. [Google Scholar] [CrossRef]
- Netherton, C.L.; Goatley, L.C.; Reis, A.L.; Portugal, R.; Nash, R.H.; Morgan, S.B.; Gault, L.; Nieto, R.; Norlin, V.; Gallardo, C.; et al. Identification and immunogenicity of African swine fever virus antigens. Front. Immunol. 2019, 10, 1318. [Google Scholar] [CrossRef] [PubMed]
- Argilaguet, J.M.; Pérez-Martín, E.; Nofrarías, M.; Gallardo, C.; Accensi, F.; Lacasta, A.; Mora, M.; Ballester, M.; Galindo-Cardiel, I.; López-Soria, S.; et al. DNA Vaccination Partially Protects against African Swine Fever Virus Lethal Challenge in the Absence of Antibodies. PLoS ONE 2012, 7, e40942. [Google Scholar] [CrossRef] [PubMed]
- Bosch-Camós, L.; López, E.; Collado, J.; Navas, M.J.; Blanco-Fuertes, M.; Pina-Pedrero, S.; Accensi, F.; Salas, M.L.; Mundt, E.; Nikolin, V.; et al. M448R and MGF505-7R: Two African Swine Fever Virus Antigens Commonly Recognized by ASFV-Specific T-Cells and with Protective Potential. Vaccines 2021, 9, 508. [Google Scholar] [CrossRef]
- Goatley, L.C.; Reis, A.L.; Portugal, R.; Goldswain, H.; Shimmon, G.L.; Hargreaves, Z.; Ho, C.-S.; Montoya, M.; Sánchez-Cordón, P.J.; Taylor, G.; et al. A pool of eight virally vectored African swine fever antigens protect pigs against fatal disease. Vaccines 2020, 8, 234. [Google Scholar] [CrossRef]
- Takamatsu, H.-H.; Denyer, M.S.; Lacasta, A.; Stirling, C.M.; Argilaguet, J.M.; Netherton, C.L.; Oura, C.A.; Martins, C.; Rodríguez, F. Cellular immunity in ASFV responses. Virus Res. 2013, 173, 110–121. [Google Scholar] [CrossRef]
- Wardley, R.; Norley, S.; Wilkinson, P.; Williams, S. The role of antibody in protection against African swine fever virus. Vet. Immunol. Immunopathol. 1985, 9, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Onisk, D.; Borca, M.; Kutish, S.; Kramer, E.; Irusta, P.; Rock, D. Passively transferred African swine fever virus antibodies protect swine against lethal infection. Virology 1994, 198, 350–354. [Google Scholar] [CrossRef]
- Silva, E.B.; Krug, P.W.; Ramirez-Medina, E.; Valladares, A.; Rai, A.; Espinoza, N.; Gladue, D.P.; Borca, M.V. The Presence of Virus Neutralizing Antibodies Is Highly Associated with Protection against Virulent Challenge in Domestic Pigs Immunized with ASFV live Attenuated Vaccine Candidates. Pathogens 2022, 11, 1311. [Google Scholar] [CrossRef]
- Escribano, J.M.; Galindo, I.; Alonso, C. Antibody-mediated neutralization of African swine fever virus: Myths and facts. Virus Res. 2013, 173, 101–109. [Google Scholar] [CrossRef]
- Canter, J.A.; Aponte, T.; Ramirez-Medina, E.; Pruitt, S.; Gladue, D.P.; Borca, M.V.; Zhu, J.J. Serum Neutralizing and Enhancing Effects on African Swine Fever Virus Infectivity in Adherent Pig PBMC. Viruses 2022, 14, 1249. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Shao, J.-J.; Zhang, G.-L.; Ge, S.-D.; Chang, Y.-Y.; Xiao, L.; Chang, H.-Y. Development of an indirect ELISA to specifically detect antibodies against African swine fever virus: Bioinformatics approaches. Virol. J. 2021, 18, 97. [Google Scholar] [CrossRef]
- Giménez-Lirola, L.G.; Mur, L.; Rivera, B.; Mogler, M.; Sun, Y.; Lizano, S.; Goodell, C.; Harris, D.H.; Rowland, R.R.; Gallardo, C.; et al. Detection of African Swine Fever Virus Antibodies in Serum and Oral Fluid Specimens Using a Recombinant Protein 30 (p30) Dual Matrix Indirect ELISA. PLoS ONE 2016, 11, e0161230. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, Q.; Liu, Y.; Wang, M.; Zhang, L.; Han, L.; Chu, X.; Ding, G.; Li, Y.; Hou, Y.; et al. Indirect ELISA Using Multi–Antigenic Dominants of p30, p54 and p72 Recombinant Proteins to Detect Antibodies against African Swine Fever Virus in Pigs. Viruses 2022, 14, 2660. [Google Scholar] [CrossRef]
- Lv, C.; Zhao, Y.; Jiang, L.; Zhao, L.; Wu, C.; Hui, X.; Hu, X.; Shao, Z.; Xia, X.; Sun, X.; et al. Development of a Dual ELISA for the Detection of CD2v-Unexpressed Lower-Virulence Mutational ASFV. Life 2021, 11, 1214. [Google Scholar] [CrossRef]
- Nah, J.-J.; Kwon, O.-K.; Choi, J.-D.; Jang, S.-H.; Lee, H.J.; Ahn, D.-G.; Lee, K.; Kang, B.; Kang, H.-E.; Shin, Y.-K. Development of an indirect ELISA against African swine fever virus using two recombinant antigens, partial p22 and p30. J. Virol. Methods 2022, 309, 114611. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Cao, C.; Yang, Z.; Jia, W. Identification of a conservative site in the African swine fever virus p54 protein and its preliminary application in a serological assay. J. Vet. Sci. 2022, 23, e55. [Google Scholar] [CrossRef]
- Wang, Y.; Kang, W.; Yang, W.; Zhang, J.; Li, D.; Zheng, H. Structure of African Swine Fever Virus and Associated Molecular Mechanisms Underlying Infection and Immunosuppression: A Review. Front. Immunol. 2021, 12, 715582. [Google Scholar] [CrossRef] [PubMed]
- Luong, H.Q.; Lai, H.T.; Do, L.D.; Ha, B.X.; Nguyen, G.V.; Vu, H.L. Differential antibody responses in sows and finishing pigs naturally infected with African swine fever virus under field conditions. Virus Res. 2021, 307, 198621. [Google Scholar] [CrossRef]
- Luong, H.Q.; Lai, H.T.L.; Vu, H.L.X. Evaluation of Antibody Response Directed against Porcine Reproductive and Respiratory Syndrome Virus Structural Proteins. Vaccines 2020, 8, 533. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, G.; Yang, D.; Zhao, B.; Zhao, Y.; Liu, Y.; Cai, X.; Nan, Y.; Zhou, E.-M.; Wu, C. Development of luciferase-linked antibody capture assay based on luciferase immunoprecipitation systems for antibody detection of porcine reproductive and respiratory syndrome virus. BMC Biotechnol. 2018, 18, 73. [Google Scholar] [CrossRef]
- Burbelo, P.D.; Lebovitz, E.E.; Notkins, A.L. Luciferase immunoprecipitation systems for measuring antibodies in autoimmune and infectious diseases. Transl. Res. 2015, 165, 325–335. [Google Scholar] [CrossRef]
- Liu, S.; Luo, Y.; Wang, Y.; Li, S.; Zhao, Z.; Bi, Y.; Sun, J.; Peng, R.; Song, H.; Zhu, D.; et al. Cryo-EM Structure of the African Swine Fever Virus. Cell Host Microbe 2019, 26, 836–843.e3. [Google Scholar] [CrossRef]
- Wang, N.; Zhao, D.; Wang, J.; Zhang, Y.; Wang, M.; Gao, Y.; Li, F.; Wang, J.; Bu, Z.; Rao, Z.; et al. Architecture of African swine fever virus and implications for viral assembly. Science 2019, 366, 640–644. [Google Scholar] [CrossRef]
- Andrés, G.; Charro, D.; Matamoros, T.; Dillard, R.S.; Abrescia, N.G.A. The cryo-EM structure of African swine fever virus unravels a unique architecture comprising two icosahedral protein capsids and two lipoprotein membranes. J. Biol. Chem. 2020, 295, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Ma, B.; Qian, N.; Zhang, F.; Tan, X.; Lei, J.; Xiang, Y. Structure of the African swine fever virus major capsid protein p72. Cell Res. 2019, 29, 953–955. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, M.C.; de la Torre Reoyo, A.; Fernández-Pinero, J.; Iglesias, I.; Muñoz, M.J.; Arias, M.L. African swine fever: A global view of the current challenge. Porc. Health Manag. 2015, 1, 21. [Google Scholar] [CrossRef]
- Bastos, A.D.S.; Penrith, M.-L.; Crucière, C.; Edrich, J.L.; Hutchings, G.; Roger, F.; Couacy-Hymann, E.; Thomson, G.R. Genotyping field strains of African swine fever virus by partial p72 gene characterisation. Arch. Virol. 2003, 148, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Cobbold, C.; Windsor, M.; Wileman, T. A virally encoded chaperone specialized for folding of the major capsid protein of African swine fever virus. J. Virol. 2001, 75, 7221–7229. [Google Scholar] [CrossRef]
- Epifano, C.; Krijnse-Locker, J.; Salas, M.L.; Rodríguez, J.M.; Salas, J. The African swine fever virus nonstructural protein pB602L is required for formation of the icosahedral capsid of the virus particle. J. Virol. 2006, 80, 12260–12270. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Castañeda, B.; Reis, A.L.; Corteyn, A.; Parkhouse, R.M.E.; Kollnberger, S. Expression, cellular localization and antibody responses of the African swine fever virus genes B602L and K205R. Arch. Virol. 2008, 153, 2303–2306. [Google Scholar] [CrossRef]
- Kollnberger, S.D.; Gutierrez-Castañeda, B.; Foster-Cuevas, M.; Corteyn, A.; Parkhouse, R.M.E. Identification of the principal serological immunodeterminants of African swine fever virus by screening a virus cDNA library with antibody. J. Gen. Virol. 2002, 83, 1331–1342. [Google Scholar] [CrossRef]
- Suárez, C.; Gutiérrez-Berzal, J.; Andrés, G.; Salas, M.L.; Rodríguez, J.M. African swine fever virus protein p17 is essential for the progression of viral membrane precursors toward icosahedral intermediates. J. Virol. 2010, 84, 7484–7499. [Google Scholar] [CrossRef]
- Jancovich, J.K.; Chapman, D.; Hansen, D.T.; Robida, M.D.; Loskutov, A.; Craciunescu, F.; Borovkov, A.; Kibler, K.; Goatley, L.; King, K.; et al. Immunization of Pigs by DNA Prime and Recombinant Vaccinia Virus Boost to Identify and Rank African Swine Fever Virus Immunogenic and Protective Proteins. J. Virol. 2018, 92, 8. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Xia, N.; Zhang, J.; Cao, Q.; Jiang, S.; Luo, J.; Wang, H.; Chen, N.; Zhang, Q.; Meurens, F.; et al. African Swine Fever Virus Structural Protein p17 Inhibits cGAS-STING Signaling Pathway Through Interacting with STING. Front. Immunol. 2022, 13, 941579. [Google Scholar] [CrossRef]
- Epifano, C.; Krijnse-Locker, J.; Salas, M.L.; Salas, J.; Rodríguez, J.M. Generation of filamentous instead of icosahedral particles by repression of African swine fever virus structural protein pB438L. J. Virol. 2006, 80, 11456–11466. [Google Scholar] [CrossRef]
- Lokhandwala, S.; Waghela, S.D.; Bray, J.; Sangewar, N.; Charendoff, C.; Martin, C.L.; Hassan, W.S.; Koynarski, T.; Gabbert, L.; Burrage, T.G.; et al. Adenovirus-vectored novel African Swine Fever Virus antigens elicit robust immune responses in swine. PLoS ONE 2017, 12, e0177007. [Google Scholar] [CrossRef] [PubMed]
- Andrés, G.; García-Escudero, R.; Viñuela, E.; Salas, M.L.; Rodríguez, J.M. African swine fever virus structural protein pE120R is essential for virus transport from assembly sites to plasma membrane but not for infectivity. J. Virol. 2001, 75, 6758–6768. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Pomares, L.; Simon-Mateo, C.; Lopez-Otin, C.; Viñuela, E. Characterization of the african swine fever virus structural protein p14.5: A DNA binding protein. Virology 1997, 229, 201–211. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, Z.; Feng, T.; Ma, Z.; Xue, Q.; Wu, P.; Li, P.; Li, S.; Yang, F.; Cao, W.; et al. African Swine Fever Virus E120R Protein Inhibits Interferon Beta Production by Interacting with IRF3 To Block Its Activation. J. Virol. 2021, 95, e0082421. [Google Scholar] [CrossRef]
- Goatley, L.C.; Nash, R.H.; Andrews, C.; Hargreaves, Z.; Tng, P.; Reis, A.L.; Graham, S.P.; Netherton, C.L. Cellular and Humoral Immune Responses after Immunisation with Low Virulent African Swine Fever Virus in the Large White Inbred Babraham Line and Outbred Domestic Pigs. Viruses 2022, 14, 1487. [Google Scholar] [CrossRef]
- Zhao, Y.; Ren, J.; Padilla-Parra, S.; Fry, E.E.; Stuart, D.I. Lysosome sorting of beta-glucocerebrosidase by LIMP-2 is targeted by the mannose 6-phosphate receptor. Nat. Commun. 2014, 5, 4321. [Google Scholar] [CrossRef]
- England, C.G.; Ehlerding, E.B.; Cai, W. NanoLuc: A Small Luciferase Is Brightening Up the Field of Bioluminescence. Bioconjug. Chem. 2016, 27, 1175–1187. [Google Scholar] [CrossRef]
- Pronobis, M.I.; Deuitch, N.; Peifer, M. The Miraprep: A Protocol that Uses a Miniprep Kit and Provides Maxiprep Yields. PLoS ONE 2016, 11, e0160509. [Google Scholar] [CrossRef]
- Cobbold, C.; Wileman, T. The major structural protein of African swine fever virus, p73, is packaged into large structures, indicative of viral capsid or matrix precursors, on the endoplasmic reticulum. J. Virol. 1998, 72, 5215–5223. [Google Scholar] [CrossRef]
- Petrovan, V.; Yuan, F.; Li, Y.; Shang, P.; Murgia, M.V.; Misra, S.; Rowland, R.R.; Fang, Y. Development and characterization of monoclonal antibodies against p30 protein of African swine fever virus. Virus Res. 2019, 269, 197632. [Google Scholar] [CrossRef]
- Gómez-Puertas, P.; Rodríguez, F.; Oviedo, J.M.; Brun, A.; Alonso, C.; Escribano, J.M. The African swine fever virus proteins p54 and p30 are involved in two distinct steps of virus attachment and both contribute to the antibody-mediated protective immune response. Virology 1998, 243, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Alcaraz, C.; De Diego, M.; Pastor, M.J.; Escribano, J.M. Comparison of a radioimmunoprecipitation assay to immunoblotting and ELISA for detection of antibody to African swine fever virus. J. Vet. Diagn. Investig. 1990, 2, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Barderas, M.G.; Rodríguez, F.; Gómez-Puertas, P.; Avilés, M.; Beitia, F.; Alonso, C.; Escribano, J.M. Antigenic and immunogenic properties of a chimera of two immunodominant African swine fever virus proteins. Arch. Virol. 2001, 146, 1681–1691. [Google Scholar] [CrossRef]
- Reis, A.L.; Parkhouse, R.M.E.; Penedos, A.R.; Martins, C.; Leitão, A. Systematic analysis of longitudinal serological responses of pigs infected experimentally with African swine fever virus. J. Gen. Virol. 2007, 88, 2426–2434. [Google Scholar] [CrossRef]
- Liu, W.; Li, H.; Liu, B.; Lv, T.; Yang, C.; Chen, S.; Feng, L.; Lai, L.; Duan, Z.; Chen, X.; et al. A new vaccination regimen using adenovirus-vectored vaccine confers effective protection against African swine fever virus in swine. Emerg. Microbes Infect. 2023, 12, 2233643. [Google Scholar] [CrossRef]
- Schwartz, J.C.; Hemmink, J.D.; Graham, S.P.; Tchilian, E.; Charleston, B.; Hammer, S.E.; Ho, C.-S.; Hammond, J.A. The major histocompatibility complex homozygous inbred Babraham pig as a resource for veterinary and translational medicine. Hla 2018, 92, 40–43. [Google Scholar] [CrossRef] [PubMed]
- Pappas, L.; Foglierini, M.; Piccoli, L.; Kallewaard, N.L.; Turrini, F.; Silacci, C.; Fernandez-Rodriguez, B.; Agatic, G.; Giacchetto-Sasselli, I.; Pellicciotta, G.; et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature 2014, 516, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Lucas, A.H. IGH V3-23*01 and its allele V3-23*03 differ in their capacity to form the canonical human antibody combining site specific for the capsular polysaccharide of Haemophilus influenzae type b. Immunogenetics 2003, 55, 336–338. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Sánchez, E.G.; Pérez-Núñez, D.; Nogal, M.; de Leon, P.; Carrascosa, Á.L.; Nieto, R.; Soler, A.; Arias, M.L.; Revilla, Y. African swine fever virus (ASFV) protection mediated by NH/P68 and NH/P68 recombinant live-attenuated viruses. Vaccine 2018, 36, 2694–2704. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Filgueira, D.M.; González-Camacho, F.; Gallardo, C.; Resino-Talaván, P.; Blanco, E.; Gómez-Casado, E.; Alonso, C.; Escribano, J.M. Optimization and validation of recombinant serological tests for african swine fever diagnosis based on detection of the p30 protein produced in trichoplusia ni larvae. J. Clin. Microbiol. 2006, 44, 3114–3121. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Blanco, E.; Rodríguez, J.M.; Carrascosa, A.L.; Sánchez-Vizcaíno, J.M. Antigenic properties and diagnostic potential of African swine fever virus protein pp62 expressed in insect cells. J. Clin. Microbiol. 2006, 44, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Duong, H.D.; Appiah-Kwarteng, C.; Takashima, Y.; Aye, K.M.; Nagayasu, E.; Yoshida, A. A novel luciferase-linked antibody capture assay (LACA) for the diagnosis of Toxoplasma gondii infection in chickens. Parasitol. Int. 2020, 77, 102125. [Google Scholar] [CrossRef]
- Paudyal, B.; Mwangi, W.; Rijal, P.; Schwartz, J.C.; Noble, A.; Shaw, A.; Sealy, J.E.; Placido, M.B.-D.; Graham, S.P.; Townsend, A.; et al. Fc-Mediated Functions of Porcine IgG Subclasses. Front. Immunol. 2022, 13, 903755. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tng, P.Y.L.; Al-Adwani, L.; Pauletto, E.; Hui, J.Y.K.; Netherton, C.L. Capsid-Specific Antibody Responses of Domestic Pigs Immunized with Low-Virulent African Swine Fever Virus. Vaccines 2023, 11, 1577. https://doi.org/10.3390/vaccines11101577
Tng PYL, Al-Adwani L, Pauletto E, Hui JYK, Netherton CL. Capsid-Specific Antibody Responses of Domestic Pigs Immunized with Low-Virulent African Swine Fever Virus. Vaccines. 2023; 11(10):1577. https://doi.org/10.3390/vaccines11101577
Chicago/Turabian StyleTng, Priscilla Y. L., Laila Al-Adwani, Egle Pauletto, Joshua Y. K. Hui, and Christopher L. Netherton. 2023. "Capsid-Specific Antibody Responses of Domestic Pigs Immunized with Low-Virulent African Swine Fever Virus" Vaccines 11, no. 10: 1577. https://doi.org/10.3390/vaccines11101577
APA StyleTng, P. Y. L., Al-Adwani, L., Pauletto, E., Hui, J. Y. K., & Netherton, C. L. (2023). Capsid-Specific Antibody Responses of Domestic Pigs Immunized with Low-Virulent African Swine Fever Virus. Vaccines, 11(10), 1577. https://doi.org/10.3390/vaccines11101577




