Long Term Assessment of Anti-SARS-CoV-2 Immunogenicity after mRNA Vaccine in Persons Living with HIV
Abstract
:1. Introduction
2. Materials and Methods
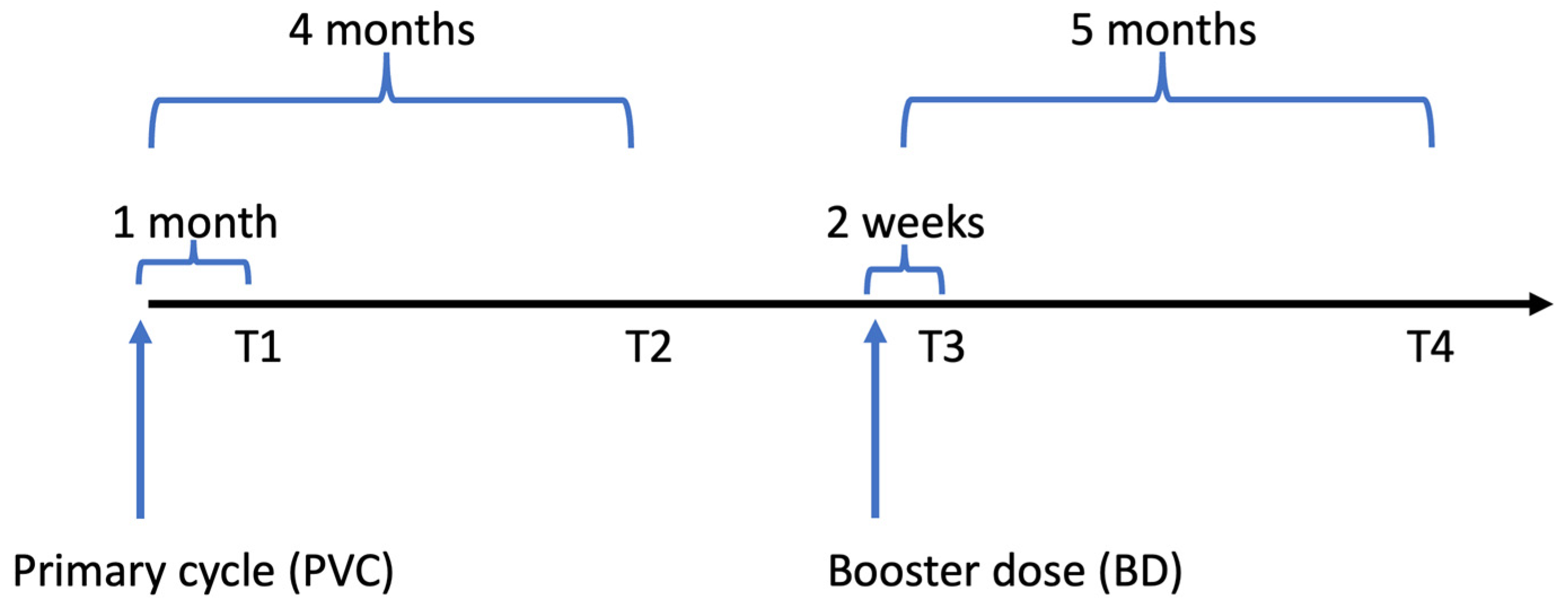
2.1. Study Design and Population
2.2. Laboratory Procedures
2.3. Statistical Analyses
3. Results
3.1. Study Population
3.2. Neutralization Activity
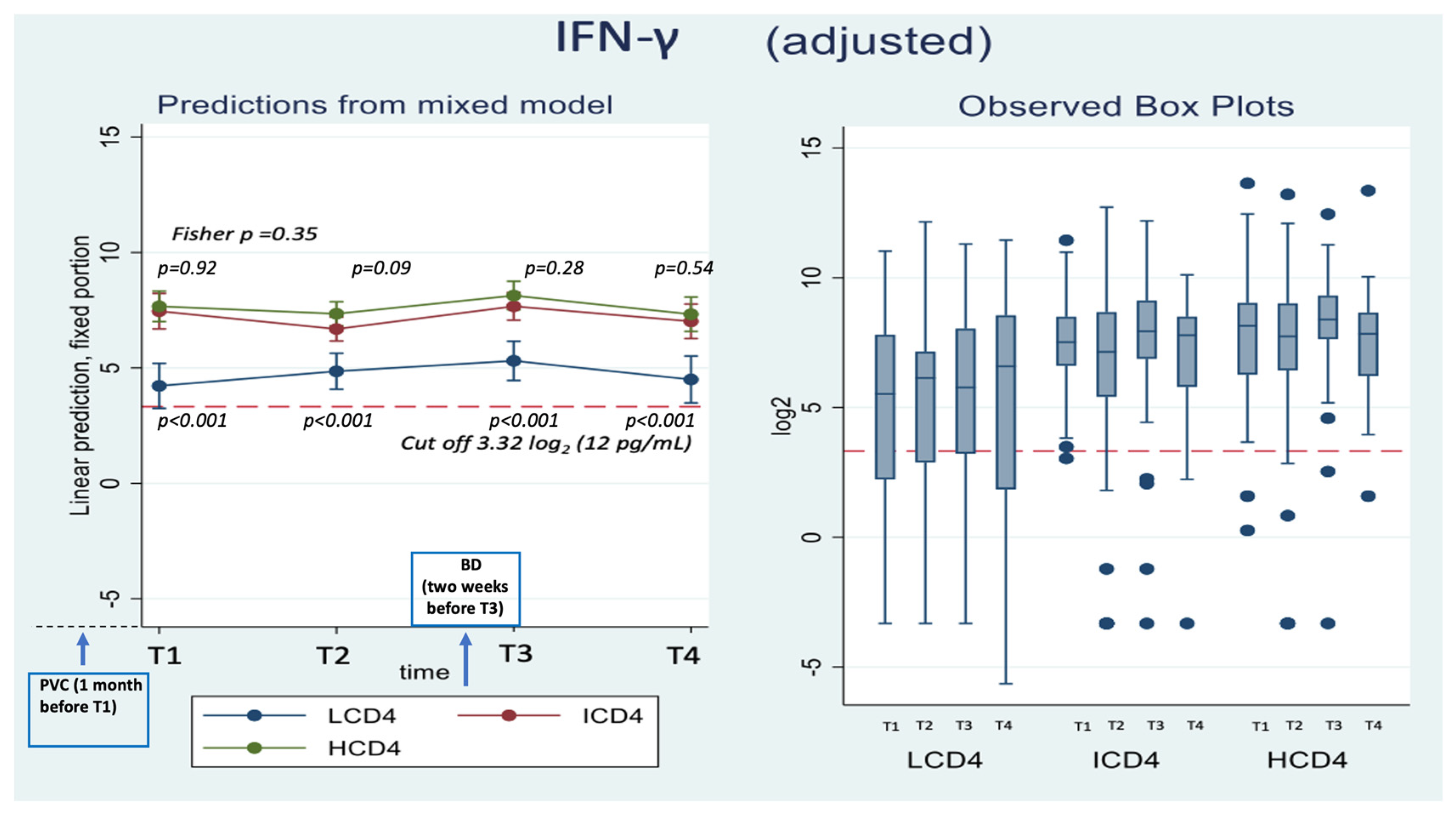
3.3. IFN-γ
3.4. Correlation between WD614G nAbs and IFN-γ Response Both at Time T1 and T3, Overall and after Stratification for CD4 Count Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martin, G.E.; Sen, D.R.; Pace, M.; Robinson, N.; Meyerowitz, J.; Adland, E.; Thornhill, J.P.; Jones, M.; Ogbe, A.; Parolini, L.; et al. Epigenetic features of HIV-induced T-cell exhaustion persist despite early antiretroviral therapy. Front. Immunol. 2021, 12, 1458. [Google Scholar] [CrossRef] [PubMed]
- Hileman, C.O.; Funderburg, N.T. Inflammation, immune activation, and antiretroviral therapy in HIV. Curr. HIV/AIDS Rep. 2017, 14, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Nakanjako, D.; Ssewanyana, I.; Mayanja-Kizza, H.; Kiragga, A.; Colebunders, R.; Manabe, Y.C.; Nabatanzi, R.; Kamya, M.R.; Cao, H. High T-cell immune activation and immune exhaustion among individuals with suboptimal CD4 recovery after 4 years of antiretroviral therapy in an African cohort. BMC Infect. Dis. 2011, 11, 43. [Google Scholar] [CrossRef] [PubMed]
- Almeida, C.-A.M.; Price, P.; French, M.A.; Evans, C.; Humphrey, J.H.; Ntozini, R.; Prendergast, A.J.; Brunt, S.J.; Cysique, L.A.; Lee, S.; et al. Immune activation in patients infected with HIV type 1 and maintaining suppression of viral replication by highly active antiretroviral therapy. AIDS Res. Hum. Retrovir. 2002, 18, 1351–1355. [Google Scholar] [CrossRef] [PubMed]
- Collie, S.; Champion, J.; Moultrie, H.; Bekker, L.-G.; Gray, G. Effectiveness of BNT162b2 vaccine against omicron variant in South Africa. N. Engl. J. Med. 2022, 386, 494–496. [Google Scholar] [CrossRef] [PubMed]
- Kalimuddin, S.; Tham, C.Y.L.; Qui, M.; de Alwis, R.; Sim, J.X.Y.; Lim, J.M.E.; Tan, H.-C.; Syenina, A.; Zhang, S.L.; Le Bert, N.; et al. Early T cell and binding antibody responses are associated with COVID-19 RNA vaccine efficacy onset. Med 2021, 2, 682–688.e4. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Cai, C.; Grifoni, A.; Müller, T.R.; Niessl, J.; Olofsson, A.; Humbert, M.; Hansson, L.; Österborg, A.; Bergman, P.; et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med. 2022, 28, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Tarke, A.; Coelho, C.H.; Zhang, Z.; Dan, J.M.; Yu, E.D.; Methot, N.; Bloom, N.I.; Goodwin, B.; Philips, E.; Mallal, S.; et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell 2022, 185, 847–859.e11. [Google Scholar] [CrossRef]
- Tarke, A.; Coelho, C.H.; Zhang, Z.; Dan, J.M.; Yu, E.D.; Methot, N.; Bloom, N.I.; Goodwin, B.; Phillips, E.; Mallal, S.; et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature 2022, 603, 488–492. [Google Scholar]
- Liu, J.; Chandrashekar, A.; Sellers, D.; Barrett, J.; Jacob-Dolan, C.; Lifton, M.; McMahan, K.; Sciacca, M.; Van Wyk, H.; Wu, C.; et al. Vaccines elicit highly conserved cellular immunity to SARS-CoV-2 Omicron. Nature 2022, 603, 493–496. [Google Scholar] [CrossRef]
- Gilbert, P.B.; Donis, R.O.; Koup, R.A.; Fong, Y.; Plotkin, S.A.; Follmann, D. A COVID-19 Milestone Attained—A Correlate of Protection for Vaccines. N. Engl. J. Med. 2022, 387, 2203–2206. [Google Scholar] [CrossRef] [PubMed]
- Widge, A.T.; Rouphael, N.G.; Jackson, L.A.; Anderson, E.J.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. N. Engl. J. Med. 2020, 384, 80–82. [Google Scholar] [CrossRef] [PubMed]
- Mateus, J.; Dan, J.M.; Zhang, Z.; Moderbacher, C.R.; Lammers, M.; Goodwin, B.; Sette, A.; Crotty, S.; Weiskopf, D. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science 2021, 374, eabj9853. [Google Scholar] [CrossRef] [PubMed]
- Pegu, A.; O’Connell, S.E.; Schmidt, S.D.; O’Dell, S.; Talana, C.A.; Lai, L.; Albert, J.; Anderson, E.; Bennett, H.; Corbett, K.S.; et al. Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science 2021, 373, 1372–1377. [Google Scholar] [CrossRef] [PubMed]
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880. [Google Scholar] [CrossRef] [PubMed]
- Goel, R.R.; Painter, M.M.; Apostolidis, S.A.; Mathew, D.; Meng, W.; Rosenfeld, A.M.; Lunfgreen, K.A.; Reynaldi, A.D.; Khoury, D.S.; Pattekar, A.; et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science 2021, 374, abm0829. [Google Scholar] [CrossRef] [PubMed]
- Agrati, C.; Castilletti, C.; Goletti, D.; Sacchi, A.; Bordoni, V.; Mariotti, D.; Notari, S.; Matusali, G.; Meschi, S.; Petrone, L.; et al. Persistent spike-specific T cell immunity despite antibody reduction after 3 months from SARS-CoV-2 BNT162b2- mRNA vaccine. Sci. Rep. 2022, 12, 6687. [Google Scholar] [CrossRef]
- Woldemeskel, B.A.; Karaba, A.H.; Garliss, C.C.; Beck, E.J.; Aytenfisu, T.Y.; Johnston, T.S.; Laeyendecker, O.; Cox, A.L.; Blankson, J.N. Decay of coronavirus disease 2019 mRNA vaccine-induced immunity in people with HIV. AIDS 2022, 36, 1315–1317. [Google Scholar] [CrossRef]
- Ogbe, A.; Pace, M.; Bittaye, M.; Tipoe, T.; Adele, S.; Alagaratnam, J.; Aley, P.K.; Ansari, M.A.; Bara, A.; Broadhead, S.; et al. Durability of ChAdOx1 nCoV-19 vaccination in people living with HIV. JCI Insight 2022, 7, e157031. [Google Scholar] [CrossRef]
- Pérez-Alós, L.; Armenteros, J.J.A.; Madsen, J.R.; Hansen, C.B.; Jarlhelt, I.; Hamm, S.R.; Heftdal, L.D.; Pries-Heje, M.M.; Møller, D.L.; Fogh, K.; et al. Modeling of waning immunity after SARS-CoV-2 vaccination and influencing factors. Nat. Commun. 2022, 13, 1614. [Google Scholar] [CrossRef]
- Antinori, A.; Cicalini, S.; Meschi, S.; Bordoni, V.; Lorenzini, P.; Vergori, A.; Lanini, S.; De Pascale, L.; Matusali, G.; Mariotti, D.; et al. Humoral and cellular immune response elicited by mRNA vaccination against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in people living with human immunodefciency virus receiving antiretroviral therapy based on current CD4 T-lymphocyte count. Clin. Infect. Dis. 2022, 75, e552–e563. [Google Scholar] [CrossRef] [PubMed]
- Vergori, A.; Cozzi-Lepri, A.; Matusali, G.; Colavita, F.; Cicalini, S.; Gallì, P.; Garbuglia, A.R.; Fusto, M.; Puro, V.; Maggi, F.; et al. HIV-VAC Study Group SARS-CoV-2 Omicron Variant Neutralization after Third Dose Vaccination in PLWH. Viruses 2022, 14, 1710. [Google Scholar] [CrossRef] [PubMed]
- Matusali, G.; Sberna, G.; Meschi, S.; Gramigna, G.; Colavita, F.; Lapa, D.; Francalancia, M.; Bettini, A.; Capobianchi, M.R.; Puro, V.; et al. Differential Dynamics of SARS-CoV-2 Binding and Functional Antibodies upon BNT162b2 Vaccine: A 6-Month Follow-Up. Viruses 2022, 14, 312. [Google Scholar] [CrossRef] [PubMed]
- Martini, F.; D’Offizi, G.; Girardi, E.; Vitale, A.; Cimini, E.; Mollicone, B.; Vlassi, C.; Aiuti, F.; Other Members of 2005 Italian CD4 Quality Control Network. The 2005 Italian Quality Control Study for the evaluation of CD4 cells in centers involved in the treatment of HIV type 1 patients. AIDS Res. Hum. Retrovir. 2007, 23, 777–781. [Google Scholar] [CrossRef] [PubMed]
- Nault, L.; Marchitto, L.; Goyette, G.; Tremblay-Sher, D.; Fortin, C.; Martel-Laferrière, V.; Trottier, B.; Richard, J.; Durand, M.; Kaufmann, D.; et al. COVID-19 vaccine immunogenicity in people living with HIV-1. Vaccine 2022, 40, 3633–3637. [Google Scholar] [CrossRef] [PubMed]
- Sisteré-Oró, M.; Andrade, N.; Wortmann, D.D.; Du, J.; Garcia-Giralt, N.; González-Cao, M.; Güerri-Fernández, R.; Meyerhans, A. Anti-SARS-CoV-2 specifc immunity in HIV immunological non-responders after mRNA-based COVID-19 vaccination. Front. Immunol. 2022, 13, 994173. [Google Scholar] [CrossRef] [PubMed]
- Tau, L.; Turner, D.; Adler, A.; Marom, R.; Ahsanov, S.; Matus, N.; Levi, I.; Gerber, G.; Lev, S.; Ziv-Baran, T.; et al. SARS-CoV-2 humoral and cellular immune responses of patients with HIV after vaccination with BNT162b2 mRNA COVID-19 vaccine in the Tel-Aviv Medical Center. Open Forum Infect. Dis. 2022, 9, ofac089. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, M.A.; Peluso, M.J.; Lynch, K.L.; Yun, C.; Glidden, D.V.; Henrich, T.J.; Deeks, S.G.; Gandhi, M. Differences in post-mRNA vaccination severeacute respiratory syndrome coronavirus 2 (SARS-CoV-2) immunoglobulin G (IgG) concentrations and surrogate virus neutralization test response by human immunodefciency virus (HIV) status and type of vaccine: A matched case-control observational study. Clin. Infect. Dis. 2022, 75, e916–e919. [Google Scholar] [CrossRef]
- Benet, S.; Blanch-Lombarte, O.; Ainsua-Enrich, E.; Pedreño-Lopez, N.; Muñoz-Basagoiti, J.; Raïch-Regué, D.; Perez-Zsolt, D.; Peña, R.; Jiménez, E.; de la Concepción, M.L.R.; et al. Limited humoral and specific T-cell responses after SARS-CoV-2 vaccination in PLWH with poor immune reconstitution. J. Infect. Dis. 2022. [CrossRef]
- Hensley, K.S.; Jongkees, M.J.; Geers, D.; GeurtsvanKessel, C.H.; Mueller, Y.M.; Dalm, V.A.S.H.; Papageorgiou, G.; Steggink, H.; Gorska, A.; Bogers, S.; et al. Immunogenicity and reactogenicity of SARS-CoV-2 vaccines in people living with HIV in the Netherlands: A nationwide prospective cohort study. PLoS Med. 2022, 19, e1003979. [Google Scholar] [CrossRef]
- Corma-Gómez, A.; Fernández-Fuertes, M.; García, E.; Fuentes-López, A.; Gómez-Ayerbe, C.; Rivero-Juárez, A.; Domínguez, C.; Santos, M.; Viñuela, L.; Palacios, R.; et al. Severe immunosuppression is related to poorer immunogenicity to SARS-CoV-2 vaccines among people living with HIV. Clin. Microbiol. Infect. 2022, 28, 1492–1498. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-R.; Jiang, Y.-W.; Li, F.-X.; Liu, D.; Lin, T.-F.; Zhao, Z.-Y.; Wei, C.; Jin, Q.-Y.; Li, X.-M.; Jia, Y.-X.; et al. Efficacy of SARS-CoV-2 vaccines and the dose-response relationship with three major antibodies: A systematic review and meta-analysis of randomised controlled trials. Lancet Microbe 2023, 4, e236–e246. [Google Scholar] [CrossRef] [PubMed]
- Chemaitelly, H.; Ayoub, H.H.; AlMukdad, S.; Coyle, P.; Tang, P.; Yassine, H.M.; Al-Khatib, H.A.; Smatti, M.K.; Hasan, M.R.; Al-Kanaani, Z.; et al. Duration of mRNA vaccine protection against SARS-CoV-2 omicron BA.1 and BA.2 subvariants in Qatar. Nat. Commun. 2022, 13, 3082. [Google Scholar] [CrossRef] [PubMed]
- Abu-Raddad, L.J.; Chemaitelly, H.; Ayoub, H.H.; Yassine, H.M.; Benslimane, F.M.; Al Khatib, H.A.; Tang, P.; Hasan, M.R.; Coyle, P.; Al Kanaani, Z.; et al. Association of prior SARS-CoV-2 infection with risk of breakthrough infection following mRNA vaccination in Qatar. JAMA 2021, 326, 1930–1939. [Google Scholar] [CrossRef] [PubMed]
- Qu, P.; Faraone, J.N.; Evans, J.P.; Zheng, Y.-M.; Yu, L.; Ma, Q.; Carlin, C.; Lozanski, G.; Saif, L.J.; Oltz, E.M.; et al. Durability of Booster mRNA Vaccine against SARS-CoV-2 BA.2.12.1, BA.4, and BA.5 Subvariants. N. Engl. J. Med. 2022, 387, 1329–1331. [Google Scholar] [CrossRef] [PubMed]
- Tuan, J.J.; Zapata, H.; Barakat, L.; Andrews, L.; Behnegar, A.; Kim, Y.W.; Kayani, J.; Mutic, S.; Ryall, L.; Turcotte, B.; et al. Long-term quantitative assessment of anti-SARS-CoV-2 spike protein immunogenicity (QUASI) after COVID-19 vaccination in older people living with HIV (PWH). BMC Infect. Dis. 2022, 22, 744. [Google Scholar] [CrossRef] [PubMed]
- Santoro, A.; Capri, A.; Petrone, D.; Colavita, F.; Meschi, S.; Matusali, G.; Mizzoni, K.; Notari, S.; Agrati, C.; Goletti, D.; et al. SARS-CoV-2 Breakthrough Infections According to the Immune Response Elicited after mRNA Third Dose Vaccination in COVID-19-Naïve Hospital Personnel. Biomedicines 2023, 11, 1247. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zeng, Q.; Deng, C.; Li, M.; Li, L.; Liu, D.; Liu, M.; Ruan, X.; Mei, J.; Mo, R.; et al. Robust induction of B cell and T cell responses by a third dose of inactivated SARS-CoV-2 vaccine. Cell Discov. 2022, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Tartof, S.Y.; Slezak, J.M.; Fischer, H.; Hong, V.; Ackerson, B.K.; Ranasinghe, O.N.; Frankland, T.B.; Ogun, O.A.; Zamparo, J.M.; Gray, S.; et al. Effectiveness of mRNA BNT162b2 COVID-19 vaccine up to 6 months in a large integrated health system in the USA: A retrospective cohort study. Lancet 2021, 398, 1407–1416. [Google Scholar] [CrossRef]

| CD4 Count at T1 $ (Cells/mm3) | |||||
|---|---|---|---|---|---|
| Characteristics | LCD4 0–200/mm3 | ICD4 201–500/mm3 | HCD4 >500/mm3 | p-Value * | Total |
| N = 56 | N = 120 | N = 138 | N = 314 | ||
| Age, years | 0.208 | ||||
| Median (IQR) | 58 (53, 63) | 56 (50, 62) | 56 (48, 60) | 56 (50, 61) | |
| Female, n (%) | 17 (30.4) | 16 (13.3) | 22 (15.9) | 0.018 | 55 (17.5) |
| Caucasian, n (%) | 37 (66.1) | 77 (64.2) | 115 (83.3) | 0.001 | 229 (72.9) |
| Nadir CD4 count, cells/mm3 | <0.001 | ||||
| Median (IQR) | 32 (7, 70) | 48 (23, 129) | 166 (52, 298) | 74 (26, 180) | |
| Time from HIV diagnosis, years | 0.007 | ||||
| Median (IQR) | 17 (2, 24) | 6 (2, 13) | 10 (5, 15) | 9 (4, 21) | |
| Time from AIDS diagnosis, years | 0.069 | ||||
| Median (IQR) | 2 (1, 2) | 6 (3, 7) | 5 (5, 10) | 5 (4, 8) | |
| AIDS, n (%) | 14 (25.9) | 32 (30.2) | 27 (21.3) | 0.297 | 73 (25.4) |
| Year of starting ART | 0.029 | ||||
| Median (IQR) | 2009 (2000, 2020) | 2016 (2010, 2019) | 2013 (2010, 2016) | 2014 (2009, 2018) | |
| VL≤ 50 at T1, n (%) | 44 (78.6) | 116 (97.5) | 137 (99.3) | <0.001 | 297 (94.9) |
| Cancer, n (%) | 2 (3.6) | 6 (5.0) | 12 (8.7) | 0.308 | 20 (6.4) |
| BMI, median (IQR) | 22 (21, 24) | 24 (21, 26) | 23 (22, 26) | 0. 109 | 23 (21, 26) |
| Autoimmune disease, n (%) | 0 (0.0) | 1 (0.9) | 0 (0.0) | 0.426 | 1 (0.3) |
| Cardiopathy, n (%) | 0 (0.0) | 0 (0.0) | 1 (0.8) | 0.533 | 1 (0.3) |
| CKD, n (%) | 8 (14.8) | 9 (8.5) | 9 (7.1) | 0.246 | 26 (9.1) |
| COPD, n (%) | 3 (5.6) | 6 (5.7) | 4 (3.1) | 0.606 | 13 (4.5) |
| MI, n (%) | 1 (1.9%) | 1 (0.9%) | 1 (0.8%) | 0.806 | 3 (1.0) |
| Hypertension, n (%) | 8 (14.8) | 17 (16.0) | 11 (8.7) | 0.205 | 36 (12.5) |
| Mild liver disease, n (%) | 12 (22.2) | 23 (21.7) | 29 (22.8) | 0.979 | 64 (22.3) |
| Severe liver disease, n (%) | 4 (7.4) | 3 (2.8) | 1 (0.8) | 0.047 | 8 (2.8) |
| No. of comorbidities & | 0.189 | ||||
| Median (IQR) | 1 (1, 2) | 1 (1, 2) | 1 (1, 1) | 1 (1, 2) | |
| Time from T1 to T3, days | 0.472 | ||||
| Median (IQR) | 178 (175, 183) | 174 (162, 187) | 175 (167, 199) | 175 (166, 186) | |
| Time from T3 to T4, days | 0.110 | ||||
| Median (IQR) | 126 (122, 130) | 119 (103, 133) | 119 (109, 130) | 120 (108, 130) | |
| (A) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Adjusted Means | Adjusted Difference in Means | ||||||||
| T1 95% CI | T2 95% CI | T3 95% CI | T4 95% CI | T1 95% CI p-Value | T2 95% CI p-Value | T3 95% CI p-Value | T4 95% CI p-Value | p-Value * | |
| Nabs Whuhan | |||||||||
| CD4 group | 0.037 | ||||||||
| HCD4 | 6.5 (6.1, 6.9) | 5.3 (4.9, 5.6) | 8.8 (8.4, 9.2) | 6.6 (6.1, 7.1) | 0 | 0 | 0 | 0 | |
| ICD4 | 5.7 (5.2, 6.1) | 4.8 (4.4, 5.1) | 9.1 (8.7, 9.4) | 6.9 (6.4, 7.3) | −0.8 (−1.4, −0.2) | −0.5 (−1.0, −0.0) | 0.2 (−0.4, 0.8) | 0.2 (−0.4, 0.9) | |
| 0.008 | 0.045 | 0.460 | 0.498 | ||||||
| LCD4 | 3.9 (3.3, 4.5) | 3.2 (2.7, 3.7) | 7.5 (6.9, 8.0) | 5.4 (4.8, 6.1) | −2.6 (−3.3, −1.8) | −2.1 (−2.7, −1.5) | −1.4 (−2.1, −0.7) | −1.2 (−2.0, −0.3) | |
| <0.001 | <0.001 | <0.001 | 0.006 | ||||||
| (B) | |||||||||
| Adjusted & Mean Changes from T2 $ | Adjusted & Difference in Mean Changes from T2 $ | ||||||||
| Response | |||||||||
| T1 95% CI | T2 95% CI | T3 95% CI | T4 95% CI | T1 95% CI p-Value | T3 95% CI p-Value | T4 95% CI p-Value | p-Value * | ||
| Nabs Wuhan | |||||||||
| CD4 group | 0.011 | ||||||||
| HCD4 | 1.2 (0.9, 1.6) | REF | 3.5 (3.1, 3.8) | 1.4 (1.0, 1.9) | 0 | 0 | 0 | ||
| ICD4 | 1.2 (0.8, 1.6) | REF | 4.3 (4.0, 4.7) | 2.2 (1.9, 2.6) | −0.0 (−0.5, 0.5) | 0.9 (0.4, 1.4) | 0.8 (0.2, 1.4) | ||
| 0.887 | <0.001 | 0.006 | |||||||
| LCD4 | 0.9 (0.4, 1.4) | REF | 4.4 (3.9, 4.8) | 2.3 (1.7, 2.8) | −0.4 (−1.0, 0.2) | 0.9 (0.3, 1.5) | 0.8 (0.1, 1.5) | ||
| 0.239 | 0.002 | 0.021 | |||||||
| (C) | |||||||||
| Adjusted & Mean Changes from T3$$ | Adjusted & Difference in Mean Changes from T3 $$ | ||||||||
| Response | |||||||||
| T1 95% CI | T2 95% CI | T3 95% CI | T4 95% CI | T1 95% CI p-Value | T2 95% CI p-Value | T4 95% CI p-Value | p-Value * | ||
| Nabs Wuhan | |||||||||
| CD4 group | 0.024 | ||||||||
| HCD4 | −2.6 (−3.1, −2.1) | −3.5 (−3.9, −3.1) | REF | −2.3 (−2.8, −1.8) | 0 | 0 | 0 | ||
| ICD4 | −3.6 (−4.1, −3.1) | −4.3 (−4.7, −3.9) | REF | −2.2 (−2.6, −1.8) | −1.0 (−1.7, −0.3) | −0.8 (−1.3, −0.2) | 0.1 (−0.5, 0.7) | ||
| 0.003 | 0.004 | 0.736 | |||||||
| LCD4 | −3.7 (−4.2, −3.1) | −4.3 (−4.8, −3.7) | REF | −2.1 (−2.8, −1.5) | −1.1 (−1.9, −0.3) | 0.2 (−0.3, 0.7) | 0.2 (−0.6, 1.0) | ||
| 0.008 | 0.534 | 0.684 | |||||||
| (A) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Adjusted Means | Adjusted Difference in Means | ||||||||
| Response | |||||||||
| T1 95% CI | T2 95% CI | T3 95% CI | T4 95% CI | T1 95% CI | T2 95% CI | T3 95% CI p-Value | T4 95% CI p-Value | p-Value * | |
| IFN-γ | |||||||||
| CD4 group | 0.34 | ||||||||
| HCD4 | 7.6 (6.9, 8.3) | 7.3 (6.8, 7.9) | 8.2 (7.5, 8.8) | 7.5 (6.7, 8.3) | 0 | 0 | 0 | 0 | |
| ICD4 | 7.6 (6.8, 8.4) | 6.7 (6.2, 7.2) | 7.7 (7.1, 8.3) | 7.1 (6.3, 7.9) | 0.1 (−1.0, 1.1) | −0.7 (−1.4, 0.1) | −0.5 (−1.4, 0.4) | −0.4 (−1.5, 0.8) | |
| 0.917 | 0.089 | 0.280 | 0.543 | ||||||
| LCD4 | 4.2 (3.2, 5.2) | 4.9 (4.1, 5.6) | 5.3 (4.5, 6.2) | 4.8 (3.7, 5.9) | −3.3 (−4.6, −2.1) | −2.5 (−3.5, −1.5) | −2.9 (−3.9, −1.8) | -2.7 (−4.0, −1.3) | |
| <0.001 | <0.001 | <.001 | <0.001 | ||||||
| (B) | |||||||||
| Adjusted & Mean Changes from T2 $ | Adjusted & Difference in Mean Changes from T2 $ | ||||||||
| Response | |||||||||
| T1 95% CI | T2 95% CI | T3 95% CI | T4 95% CI | T1 95% CI p-Value | T3 95% CI p-Value | T4 95% CI p-Value | p-Value * | ||
| IFN-γ | |||||||||
| CD4 group | 0.023 | ||||||||
| HCD4 | 0.0 (−0.5, 0.6) | REF | 0.8 (0.3, 1.3) | −0.2 (−0.8, 0.4) | 0 | 0 | 0 | ||
| ICD4 | 1.2 (0.5, 1.8) | REF | 1.0 (0.6, 1.5) | 0.3 (−0.3, 0.9) | 1.1 (0.3, 2.0) | 0.2 (−0.5, 0.9) | 0.5 (−0.4, 1.4) | ||
| 0.011 | 0.559 | 0.271 | |||||||
| LCD4 | −1.0 (−1.8, −0.2) | REF | 0.5 (−0.1, 1.2) | −0.7 (−1.5, 0.2) | −1.0 (−2.0, −0.0) | −0.3 (−1.2, 0.5) | −0.5 (−1.5, 0.6) | ||
| 0.040 | 0.450 | 0.376 | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vergori, A.; Cozzi-Lepri, A.; Matusali, G.; Cicalini, S.; Bordoni, V.; Meschi, S.; Mazzotta, V.; Colavita, F.; Fusto, M.; Cimini, E.; et al. Long Term Assessment of Anti-SARS-CoV-2 Immunogenicity after mRNA Vaccine in Persons Living with HIV. Vaccines 2023, 11, 1739. https://doi.org/10.3390/vaccines11121739
Vergori A, Cozzi-Lepri A, Matusali G, Cicalini S, Bordoni V, Meschi S, Mazzotta V, Colavita F, Fusto M, Cimini E, et al. Long Term Assessment of Anti-SARS-CoV-2 Immunogenicity after mRNA Vaccine in Persons Living with HIV. Vaccines. 2023; 11(12):1739. https://doi.org/10.3390/vaccines11121739
Chicago/Turabian StyleVergori, Alessandra, Alessandro Cozzi-Lepri, Giulia Matusali, Stefania Cicalini, Veronica Bordoni, Silvia Meschi, Valentina Mazzotta, Francesca Colavita, Marisa Fusto, Eleonora Cimini, and et al. 2023. "Long Term Assessment of Anti-SARS-CoV-2 Immunogenicity after mRNA Vaccine in Persons Living with HIV" Vaccines 11, no. 12: 1739. https://doi.org/10.3390/vaccines11121739






