Cap-Independent Circular mRNA Translation Efficiency
Abstract
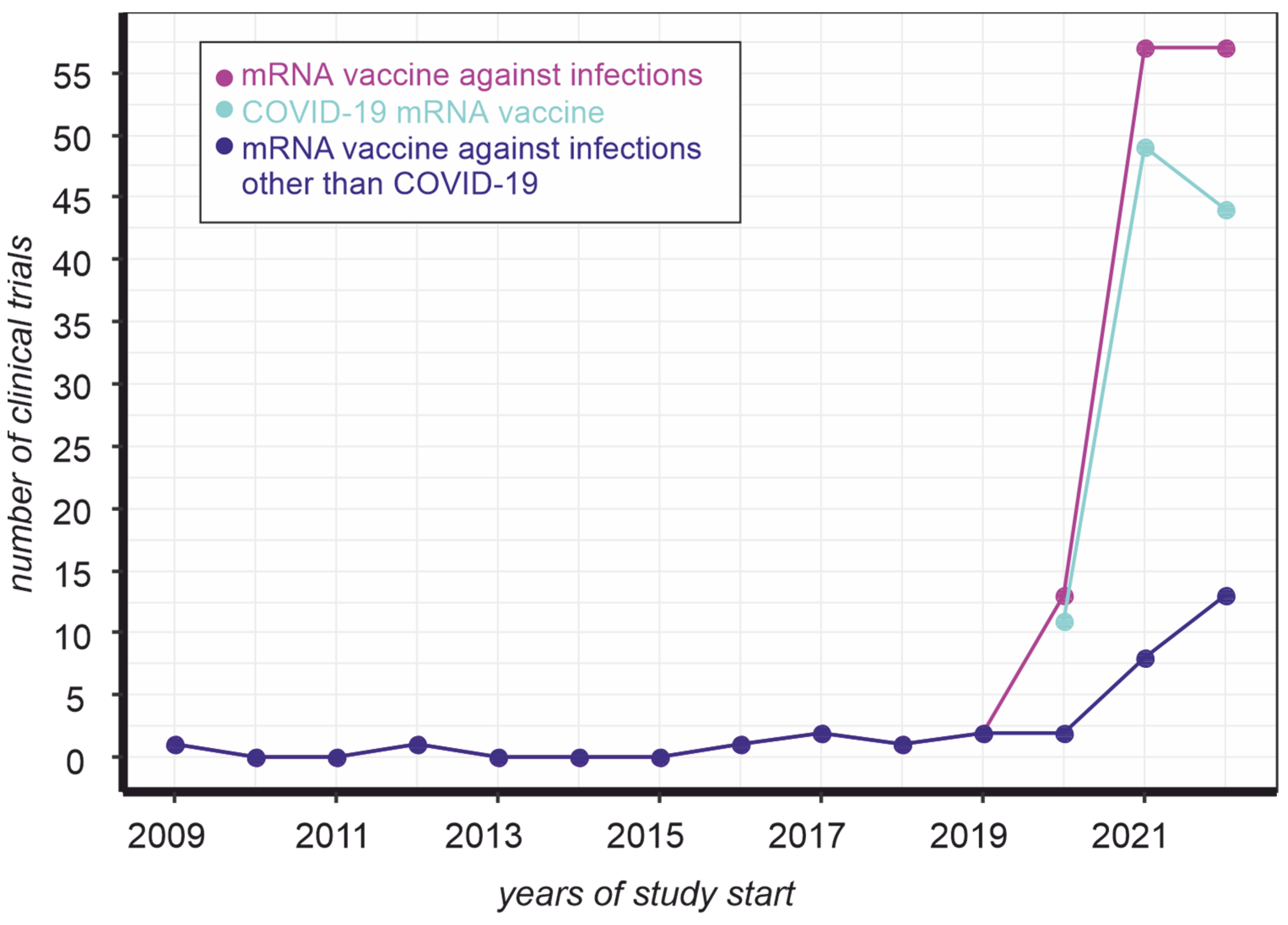
:1. Introduction
2. Translation Initiation Mechanisms
2.1. Efficiency of IRES-Dependent Translation Initiation
2.2. m6A and Translation Initiation
2.3. Endogenous IRES-like Elements in Eukaryotic Genome
2.4. Translation Enhancing Elements
2.5. Cap-Independent Translation Enhancers
2.6. R2 Elements
3. Circular RNA as Delivery Vehicle for Protein Synthesis
Future Directions
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodrigues, C.M.; Plotkin, S.A. Impact of vaccines; health, economic and social perspectives. Front. Microbiol. 2020, 11, 1526. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7371956/ (accessed on 17 January 2023). [CrossRef] [PubMed]
- Crommelin, D.J.; Anchordoquy, T.J.; Volkin, D.B.; Jiskoot, W.; Mastrobattista, E. Addressing the Cold Reality of mRNA Vaccine Stability. J. Pharm. Sci. 2020, 110, 997–1001. [Google Scholar] [CrossRef]
- Sharova, L.V.; Sharov, A.A.; Nedorezov, T.; Piao, Y.; Shaik, N.; Ko, M.S. Database for mRNA Half-Life of 19 977 Genes Obtained by DNA Microarray Analysis of Pluripotent and Differentiating Mouse Embryonic Stem Cells. DNA Res. 2009, 16, 45–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deutscher, M.P.; Li, Z. Exoribonucleases and their multiple roles in RNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 2000, 66, 67–105. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2012, 19, 141–157. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Wang, S.K.; Belk, J.A.; Amaya, L.; Li, Z.; Cardenas, A.; Abe, B.T.; Chen, C.K.; Wender, P.A.; Chang, H.Y. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 2022, 40, 1–10. [Google Scholar] [CrossRef]
- Meganck, R.M.; Liu, J.; Hale, A.E.; Simon, K.E.; Fanous, M.M.; Vincent, H.A.; Wilusz, J.E.; Moorman, N.J.; Marzluff, W.F.; Asokan, A. Engineering highly efficient backsplicing and translation of synthetic circRNAs. Mol. Ther. Nucleic Acids 2021, 23, 821–834. [Google Scholar] [CrossRef]
- Wesselhoeft, R.A.; Kowalski, P.S.; Anderson, D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.H.; Kim, S.; Lee, S.-W. Pros and Cons of In Vitro Methods for Circular RNA Preparation. Int. J. Mol. Sci. 2022, 23, 13247. [Google Scholar] [CrossRef]
- Hinnebusch, A.G. Structural Insights into the Mechanism of Scanning and Start Codon Recognition in Eukaryotic Translation Initiation. Trends Biochem. Sci. 2017, 42, 589–611. [Google Scholar] [CrossRef] [PubMed]
- Sonneveld, S.; Verhagen, B.M.; Tanenbaum, M.E. Heterogeneity in mRNA Translation. Trends Cell Biol. 2020, 30, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Eliseev, B.; Yeramala, L.; Leitner, A.; Karuppasamy, M.; Raimondeau, E.; Huard, K.; Alkalaeva, E.; Aebersold, R.; Schaffitzel, C. Structure of a human cap-dependent 48S translation pre-initiation complex. Nucleic Acids Res. 2018, 46, 2678–2689. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, Z. IRES-mediated cap-independent translation, a path leading to hidden proteome. J. Mol. Cell Biol. 2019, 11, 911–919. [Google Scholar] [CrossRef] [Green Version]
- Kieft, J.S. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 2008, 33, 274–283. [Google Scholar] [CrossRef] [Green Version]
- Kozak, M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 1987, 7, 3438–3445. [Google Scholar] [CrossRef]
- Peabody, D.S.; Berg, P. Termination-reinitiation occurs in the translation of mammalian cell mRNAs. Mol. Cell. Biol. 1986, 6, 2695–2703. [Google Scholar] [CrossRef]
- Al-Allaf, F.A.; Abduljaleel, Z.; Athar, M.; Taher, M.M.; Khan, W.; Mehmet, H.; Colakogullari, M.; Apostolidou, S.; Bigger, B.; Waddington, S.; et al. Modifying inter-cistronic sequence significantly enhances IRES dependent second gene expression in bicistronic vector: Construction of optimised cassette for gene therapy of familial hypercholesterolemia. Non-Coding RNA Res. 2019, 4, 1–14. [Google Scholar] [CrossRef]
- Mizuguchi, H.; Xu, Z.; Ishii-Watabe, A.; Uchida, E.; Hayakawa, T. IRES-Dependent Second Gene Expression Is Significantly Lower Than Cap-Dependent First Gene Expression in a Bicistronic Vector. Mol. Ther. 2000, 1, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Borman, A.M.; Le Mercier, P.; Girard, M.; Kean, K.M. Comparison of Picornaviral IRES-Driven Internal Initiation of Translation in Cultured Cells of Different Origins. Nucleic Acids Res. 1997, 25, 925–932. [Google Scholar] [CrossRef] [Green Version]
- Seephetdee, C.; Bhukhai, K.; Buasri, N.; Leelukkanaveera, P.; Lerdwattanasombat, P.; Manopwisedjaroen, S.; Phueakphud, N.; Kuhaudomlarp, S.; Olmedillas, E.; Saphire, E.O.; et al. A circular mRNA vaccine prototype producing VFLIP-X spike confers a broad neutralization of SARS-CoV-2 variants by mouse sera. Antivir. Res. 2022, 204, 105370. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Yi, Z.; Shen, Y.; Lin, L.; Chen, F.; Xu, Y.; Wu, Z.; Tang, H.; Zhang, X.; Tian, F.; et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 2022, 185, 1728–1744. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Yang, Y.; Chen, C.; Wang, Z. Pervasive translation of circular RNAs driven by short IRES-like elements. Nat. Commun. 2022, 13, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Whetter, L.E.; Day, S.P.; Elroy-Stein, O.; Brown, E.A.; Lemon, S.M. Low efficiency of the 5’ nontranslated region of hepatitis A virus RNA in directing cap-independent translation in permissive monkey kidney cells. J. Virol. 1994, 68, 5253–5263. [Google Scholar] [CrossRef] [Green Version]
- Chappell, S.A.; Edelman, G.M.; Mauro, V.P. Biochemical and functional analysis of a 9-nt RNA sequence that affects translation efficiency in eukaryotic cells. Proc. Natl. Acad. Sci. USA 2004, 101, 9590–9594. [Google Scholar] [CrossRef] [Green Version]
- Douin, V.; Bornes, S.; Creancier, L.; Rochaix, P.; Favre, G.; Prats, A.C.; Couderc, B. Use and comparison of different internal ribosomal entry sites (IRES) in tricistronic retroviral vectors. BMC Biotechnol. 2004, 4, 16. [Google Scholar] [CrossRef] [Green Version]
- Licursi, M.; Christian, S.L.; Pongnopparat, T.; Hirasawa, K. In vitro and in vivo comparison of viral and cellular internal ribosome entry sites for bicistronic vector expression. Gene Ther. 2011, 18, 631–636. [Google Scholar] [CrossRef] [Green Version]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5′ UTR m6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostyusheva, A.; Brezgin, S.; Glebe, D.; Kostyushev, D.; Chulanov, V. Host-cell interactions in HBV infection and pathogenesis: The emerging role of m6A modification. Emerg. Microbes Infect. 2021, 10, 2264–2275. [Google Scholar] [CrossRef]
- Yang, B.; Wang, J.Q.; Tan, Y.; Yuan, R.; Chen, Z.S.; Zou, C. RNA methylation and cancer treatment. Pharmacol. Res. 2021, 174, 105937. [Google Scholar] [CrossRef]
- Sakharov, P.A.; Smolin, E.A.; Lyabin, D.N.; Agalarov, S.C. ATP-Independent Initiation during Cap-Independent Translation of m6A-Modified mRNA. Int. J. Mol. Sci. 2021, 22, 3662. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N 6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinha, T.; Panigrahi, C.; Das, D.; Chandra Panda, A. Circular RNA translation, a path to hidden proteome. Wiley Interdiscip. Rev. RNA 2021, 13, e1685. [Google Scholar] [CrossRef] [PubMed]
- Van Nostrand, E.L.; Freese, P.; Pratt, G.A.; Wang, X.; Wei, X.; Xiao, R.; Blue, S.M.; Chen, J.Y.; Cody, N.A.; Dominguez, D.; et al. A large-scale binding and functional map of human RNA-binding proteins. Nature 2020, 583, 711–719. [Google Scholar] [CrossRef]
- Rubtsova, M.P.; Sizova, D.V.; Dmitriev, S.E.; Ivanov, D.S.; Prassolov, V.S.; Shatsky, I.N. Distinctive Properties of the 5′-Untranslated Region of Human Hsp70 mRNA. J. Biol. Chem. 2003, 278, 22350–22356. [Google Scholar] [CrossRef] [Green Version]
- Chappell, S.A.; Edelman, G.M.; Mauro, V.P. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl. Acad. Sci. USA 2000, 97, 1536–1541. [Google Scholar] [CrossRef] [Green Version]
- Spriggs, K.A.; Stoneley, M.; Bushell, M.; Willis, A.E. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol. Cell 2008, 100, 27–38. [Google Scholar] [CrossRef]
- Weingarten-Gabbay, S.; Elias-Kirma, S.; Nir, R.; Gritsenko, A.A.; Stern-Ginossar, N.; Yakhini, Z.; Weinberger, A.; Segal, E. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 2016, 351, aad4939. [Google Scholar] [CrossRef]
- Wellensiek, B.P.; Larsen, A.C.; Stephens, B.; Kukurba, K.; Waern, K.; Briones, N.; Liu, L.; Snyder, M.; Jacobs, B.L.; Kumar, S.; et al. Genome-wide profiling of human cap-independent translation-enhancing elements. Nat. Methods 2013, 10, 747–750. [Google Scholar] [CrossRef] [Green Version]
- Wellensiek, B.P.; Larsen, A.C.; Flores, J.; Jacobs, B.L.; Chaput, J.C. A leader sequence capable of enhancing RNA expression and protein synthesis in mammalian cells. Protein Sci. 2013, 22, 1392–1398. [Google Scholar] [CrossRef] [Green Version]
- Richard, H.B., Jr.; Minder, S.; Sidhu, A.; Juba, A.N.; Jancovich, J.K.; Jacobs, B.L.; Wellensiek, B.P. Optimization of translation enhancing element use to increase protein expression in a vaccinia virus system. J. Gen. Virol. 2021, 102, 001624. [Google Scholar] [CrossRef] [PubMed]
- Haizel, S.A.; Bhardwaj, U.; Gonzalez, R.L.; Mitra, S.; Goss, D.J. 5′-UTR recruitment of the translation initiation factor eIF4GI or DAP5 drives cap-independent translation of a subset of human mRNAs. J. Biol. Chem. 2020, 295, 11693–11706. [Google Scholar] [CrossRef] [PubMed]
- Terenin, I.M.; Andreev, D.E.; Dmitriev, S.E.; Shatsky, I.N. A novel mechanism of eukaryotic translation initiation that is neither m7G-cap-, nor IRES-dependent. Nucleic Acids Res. 2012, 41, 1807–1816. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Simon, A.E. Differential use of 3’CITEs by the subgenomic RNA of Pea enation mosaic virus 2. Virology 2017, 510, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, B.L.; White, K.A. 3′ Cap-independent translation enhancers of positive-strand RNA plant viruses. Curr. Opin. Virol. 2011, 1, 373–380. [Google Scholar] [CrossRef]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef] [Green Version]
- Moss, W.N.; Eickbush, D.G.; Lopez, M.J.; Eickbush, T.H.; Turner, D.H. The R2 retrotransposon RNA families. RNA Biol. 2011, 8, 714–718. [Google Scholar] [CrossRef] [Green Version]
- Eickbush, T.H.; Eickbush, D.G. Integration, Regulation, and Long-Term Stability of R2 Retrotransposons. Mobile DNA III 2015, 14627, 1125–1146. [Google Scholar] [CrossRef] [Green Version]
- Ruminski, D.J.; Webb, C.H.T.; Riccitelli, N.J.; Lupták, A. Processing and Translation Initiation of Non-long Terminal Repeat Retrotransposons by Hepatitis Delta Virus (HDV)-like Self-cleaving Ribozymes. J. Biol. Chem. 2011, 286, 41286–41295. [Google Scholar] [CrossRef] [Green Version]
- Rakotondrafara, A.M. Oscillating kissing stem-loop interactions mediate 5’ scanning-dependent translation by a viral 3’-cap-independent translation element. RNA 2006, 12, 1893–1906. [Google Scholar] [CrossRef] [Green Version]
- Fabian, M.R.; White, K.A. 5′-3′ RNA-RNA Interaction Facilitates Cap- and Poly(A) Tail-independent Translation of Tomato Bushy Stunt Virus mRNA. J. Biol. Chem. 2004, 279, 28862–28872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abe, N.; Matsumoto, K.; Nishihara, M.; Nakano, Y.; Shibata, A.; Maruyama, H.; Shuto, S.; Matsuda, A.; Yoshida, M.; Ito, Y.; et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci. Rep. 2015, 5, 16435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costello, A.; Lao, N.T.; Barron, N.; Clynes, M. Continuous translation of circularized mRNA improves recombinant protein titer. Metab. Eng. 2019, 52, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, P.; Zhao, D.; Zhu, L.; Tang, J.; Leng, W.; Su, J.; Liu, Y.; Bi, C.; Zhang, X. Engineering Circularized mRNAs for the Production of Spider Silk Proteins. Appl. Environ. Microbiol. 2022, 88, e00028-22. [Google Scholar] [CrossRef]
- Vicens, Q.; Paukstelis, P.J.; Westhof, E.; Lambowitz, A.M.; Cech, T.R. Toward predicting self-splicing and protein-facilitated splicing of group I introns. RNA 2008, 14, 2013–2029. [Google Scholar] [CrossRef] [Green Version]
- Haimov, O.; Sinvani, H.; Dikstein, R. Cap-dependent, scanning-free translation initiation mechanisms. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2015, 1849, 1313–1318. [Google Scholar] [CrossRef]
- de la Parra, C.; Ernlund, A.; Alard, A.; Ruggles, K.; Ueberheide, B.; Schneider, R.J. A widespread alternate form of cap-dependent mRNA translation initiation. Nat. Commun. 2018, 9, 3068. [Google Scholar] [CrossRef] [Green Version]
- Shatsky, I.N.; Terenin, I.M.; Smirnova, V.V.; Andreev, D.E. Cap-Independent Translation: What’s in a Name? Trends Biochem. Sci. 2018, 43, 882–895. [Google Scholar] [CrossRef]
- Truniger, V.; Miras, M.; Aranda, M.A. Structural and Functional Diversity of Plant Virus 3′-Cap-Independent Translation Enhancers (3′-CITEs). Front. Plant Sci. 2017, 8, 2047. [Google Scholar] [CrossRef] [Green Version]
- Coots, R.A.; Liu, X.M.; Mao, Y.; Dong, L.; Zhou, J.; Wan, J.; Zhang, X.; Qian, S.B. m6A Facilitates eIF4F-Independent mRNA Translation. Mol. Cell 2017, 68, 504–514.e7. [Google Scholar] [CrossRef]
- Abe, N.; Hiroshima, M.; Maruyama, H.; Nakashima, Y.; Nakano, Y.; Matsuda, A.; Sako, Y.; Ito, Y.; Abe, H. Rolling Circle Amplification in a Prokaryotic Translation System Using Small Circular RNA. Angew. Chem. Int. Ed. 2013, 52, 7004–7008. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Merits, A.; Wu, Y.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Zhao, X.; Yang, Q.; Wu, Y.; et al. cis-Acting Sequences and Secondary Structures in Untranslated Regions of Duck Tembusu Virus RNA Are Important for Cap-Independent Translation and Viral Proliferation. J. Virol. 2020, 94, e00906-20. [Google Scholar] [CrossRef] [PubMed]

| IRES | Cell Line | Method of Comparison | Results | Source |
|---|---|---|---|---|
| EMCV, CVB3 | HEK293, HeLa, A549, Min6 | Gaussia luciferase | The efficiency of IRESs varies according to cell type; CVB3 IRES was superior in all cell types | [9] |
| EMCV, Poliovirus, KSHV, and HCV | HEK293; U87; Huh7; 293T | GFP | Poliovirus IRES resulted in maximal expression in HEK293 | [7] |
| EMCV and HAV | Monkey kidney cells (BT7-H) | antibiotic resistance (bacterial chloramphenicol acetyltransferase) | EMCV IRES was more efficient than translation directed by the HAV IRES | [24] |
| A synthetic construct with five concatenated copies of the 9-nt Gtx IRES | mouse N2a cells | Photinus luciferase | The efficiency of the translation increases, if several copies of the 9-nt Gtx IRES are included in the construct | [25] |
| EMCV, c-myc, FGF-2, and HTLV-1 | B16.F10, TS/A, NIH-3T3, ψCRIP, 293T, and primary cultures of human melanoma cells | Immunostaining and flow cytometry | The efficiency of translation initiation depends on the type of cells induced and the presence of other genetic elements in the vector. | [26] |
| Five viral (FMDV, HCV, EMCV, PV, HRV) and eight cellular IRES elements (Rbm3, NRF, Apaf-1, BIP, VCIP, AQP-4, c-myc, CAT-1) | murine fibroblast cell line (NIH 3T3), mouse embryonic fibroblasts (MEF), human hepatoma cell line (Huh 7) and human lung fibroblasts (MRC-5) | Firefly and Renilla luciferase | Vascular endothelial growth factor and type 1 collagen-inducible protein (VCIP) IRES induced the highest firefly luciferase expression rate in all tested cell lines | [27] |
| Type of mRNA Translation Initiation | Mechanism of Initiation | Description of the Mechanism | Features | Applicability to circRNA Translation | References |
|---|---|---|---|---|---|
| Cap-dependent | eIF4E-dependent, with scanning | eIF4E binds the cap structure and the eIF4F complex, then the cap-binding complex recruits the 40S subunit, the initiation complex scans the mRNA until it reaches the start codon, and then the 60S ribosomal subunit joins this complex | The most common, canonical way of translation of mRNAs in higher eukaryotes | Not applicable | [11,12] |
| eIF4E-dependent, scanning free | There are several variants of the scanning free mechanism: - mRNA with short 5′-UTR (leaderless mRNA) translation; - translation initiator of short 5′ UTR mediated translation; - Histone H4 translation; - Ribosome shunting | Supposed to be a non-efficient process leading to leaky scanning. At the same time, several reports show efficient and accurate translation of short 5′ UTR mRNAs that are evidently translated differently from the well-known canonical scanning mechanism. | Not applicable | [54] | |
| eIF4E-independent | DAP5, homolog of eIF4G, which lacks eIF4E binding, forms complexes with eIF3d | About 20% of capped mRNAs are translated this way, during physiological conditions of mTOR inhibition and eIF4E depletion | Not applicable | [55] | |
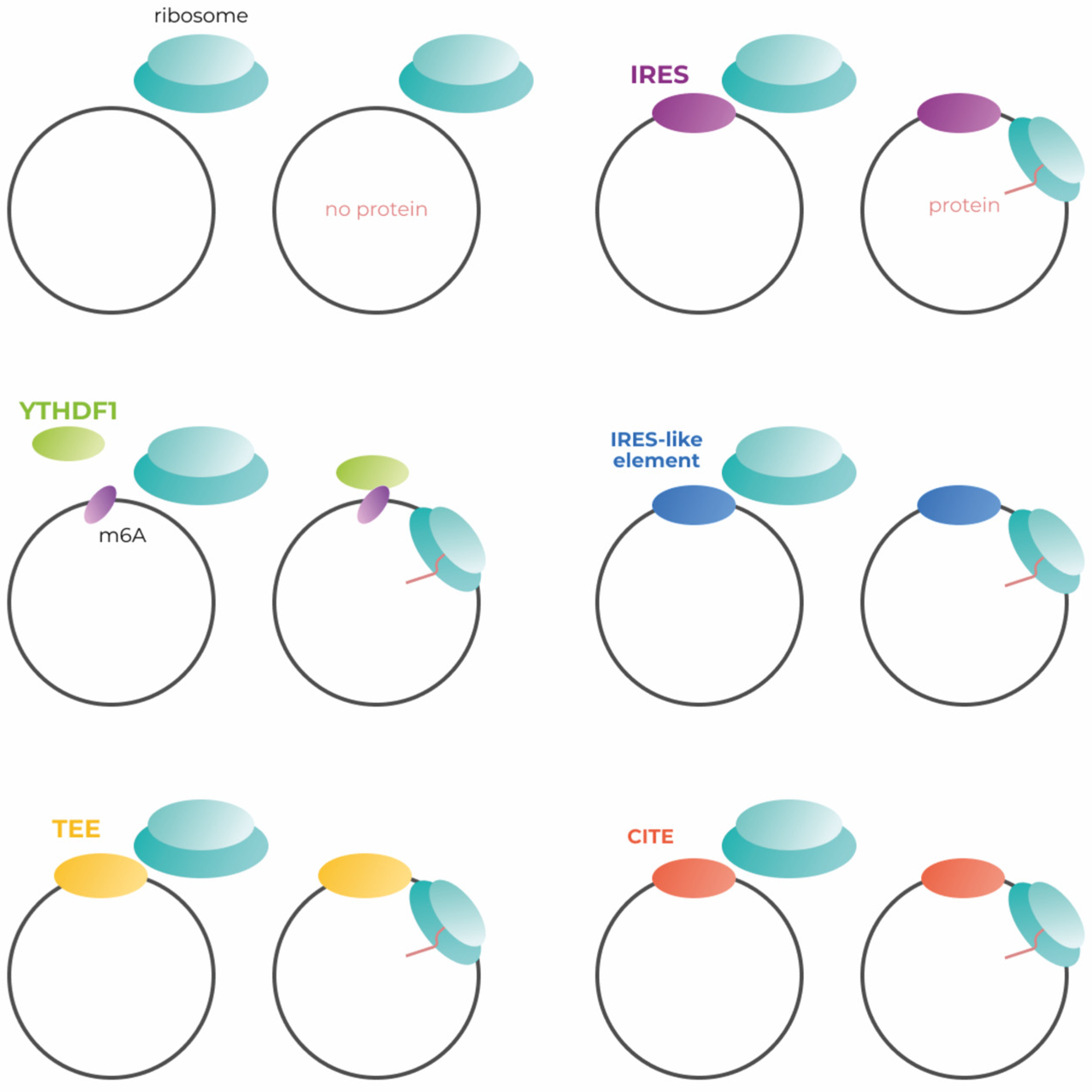
| Cap-independent | IRES-mediated initiation | IRES interacts with the 43S pre-initiation complex by direct binding via specific structural elements formed with the RNA, indirectly via ITAFs and cellular eIFs or by homology pairing of 5′UTR mRNA motifs with 18S rRNA | The translation efficiency generally is lower than cap-dependent one, but in some cases, it was shown to be equally effective, for example in the case of EMCV | Applicable, data available | [19] |
| CITE-mediated | CITEs can be located both within 5′ and 3′ UTRs and bind eIF4E and/or eIF4G subunits of eIF4F. Some CITEs can also directly bind ribosomal subunits, or the ribosomes themselves, without being dependent on elF4F | Translation efficiency of CITEs varies depending on their nature (5′ or 3′ type) and the translated sequence. 3′ CITEs were shown to be an effective substitution of cap-dependent translation | No available data, but theoretically applicable | [56,57] | |
| m6A-mediated | m6A modification within 5`UTR of mRNA can recruit a 40S ribosomal subunit through direct eIF3 binding | m6A-mediated translation co-exists with eIF4F-mediated translation for a great deal of transcripts, thus fully capped mRNAs can undergo m6A-mediated translation providing selectivity of mRNA translation in response to environmental and physiological conditions | Applicable, data available | [58] | |
| Mediated by IRES-like structures | Short hexamer sequences in endogenous circRNA are capable of translation initiation | This mechanism is less effective than viral IRESs, but each sequence less than 50 nt may contain a short IRES-like element, thus most human circRNAs might have a potential for translation. However, these structures are not conserved, hardly classified, and hardly predictable | Applicable, data available | [22,33,35] | |
| Rolling circle amplification translation | Translation initiation of circRNAs carrying only Kozak sequence (without 5′-cap, poly(A), IRES, stop codon) is possible. The ribosome continuously circles the circRNA molecule, which leads to the production of a long repeating peptide | CircRNAs can be efficiently translated by a rolling circle amplification mechanism in a cell-free E. coli translation system and in human cells | Applicable, data available | [50,59] | |
| R2-mediated | R2 element contains conservative structures (pseudoknots) which can be recognized by the translational machinery | Efficiency depends on the retrotransposon sequence and its structure, some of the R2 elements were shown to be 35 times more effective than HCV IRES | No available data, but theoretically applicable | [60,61] | |
| Mediated by cis-Acting Sequences and Secondary Structures in 5′ and 3′UTR | The precise mechanism is still unknown, but initiation of translation depends on both secondary structures and primary sequences within UTRs. 5′ and 3′ UTRs of uncapped RNA of Flaviviruses must be free and present in cis | This mechanism was shown for DTMUV, TMUV, DENV2, ZIKA, and JEV, but it is probably common for all Flaviviruses | No available data, most likely not applicable | [62] | |
| TEE-mediated | Mechanism is unclear | TEE-mediated initiation was described only for the vaccinia virus (VACV) | No available data | [39,40,41] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deviatkin, A.A.; Simonov, R.A.; Trutneva, K.A.; Maznina, A.A.; Soroka, A.B.; Kogan, A.A.; Feoktistova, S.G.; Khavina, E.M.; Mityaeva, O.N.; Volchkov, P.Y. Cap-Independent Circular mRNA Translation Efficiency. Vaccines 2023, 11, 238. https://doi.org/10.3390/vaccines11020238
Deviatkin AA, Simonov RA, Trutneva KA, Maznina AA, Soroka AB, Kogan AA, Feoktistova SG, Khavina EM, Mityaeva ON, Volchkov PY. Cap-Independent Circular mRNA Translation Efficiency. Vaccines. 2023; 11(2):238. https://doi.org/10.3390/vaccines11020238
Chicago/Turabian StyleDeviatkin, Andrei A., Ruslan A. Simonov, Kseniya A. Trutneva, Anna A. Maznina, Anastasiia B. Soroka, Anna A. Kogan, Sofya G. Feoktistova, Elena M. Khavina, Olga N. Mityaeva, and Pavel Y. Volchkov. 2023. "Cap-Independent Circular mRNA Translation Efficiency" Vaccines 11, no. 2: 238. https://doi.org/10.3390/vaccines11020238
APA StyleDeviatkin, A. A., Simonov, R. A., Trutneva, K. A., Maznina, A. A., Soroka, A. B., Kogan, A. A., Feoktistova, S. G., Khavina, E. M., Mityaeva, O. N., & Volchkov, P. Y. (2023). Cap-Independent Circular mRNA Translation Efficiency. Vaccines, 11(2), 238. https://doi.org/10.3390/vaccines11020238






