A Competitive Panning Method Reveals an Anti-SARS-CoV-2 Nanobody Specific for an RBD-ACE2 Binding Site
Abstract
:1. Introduction
2. Materials and Methods
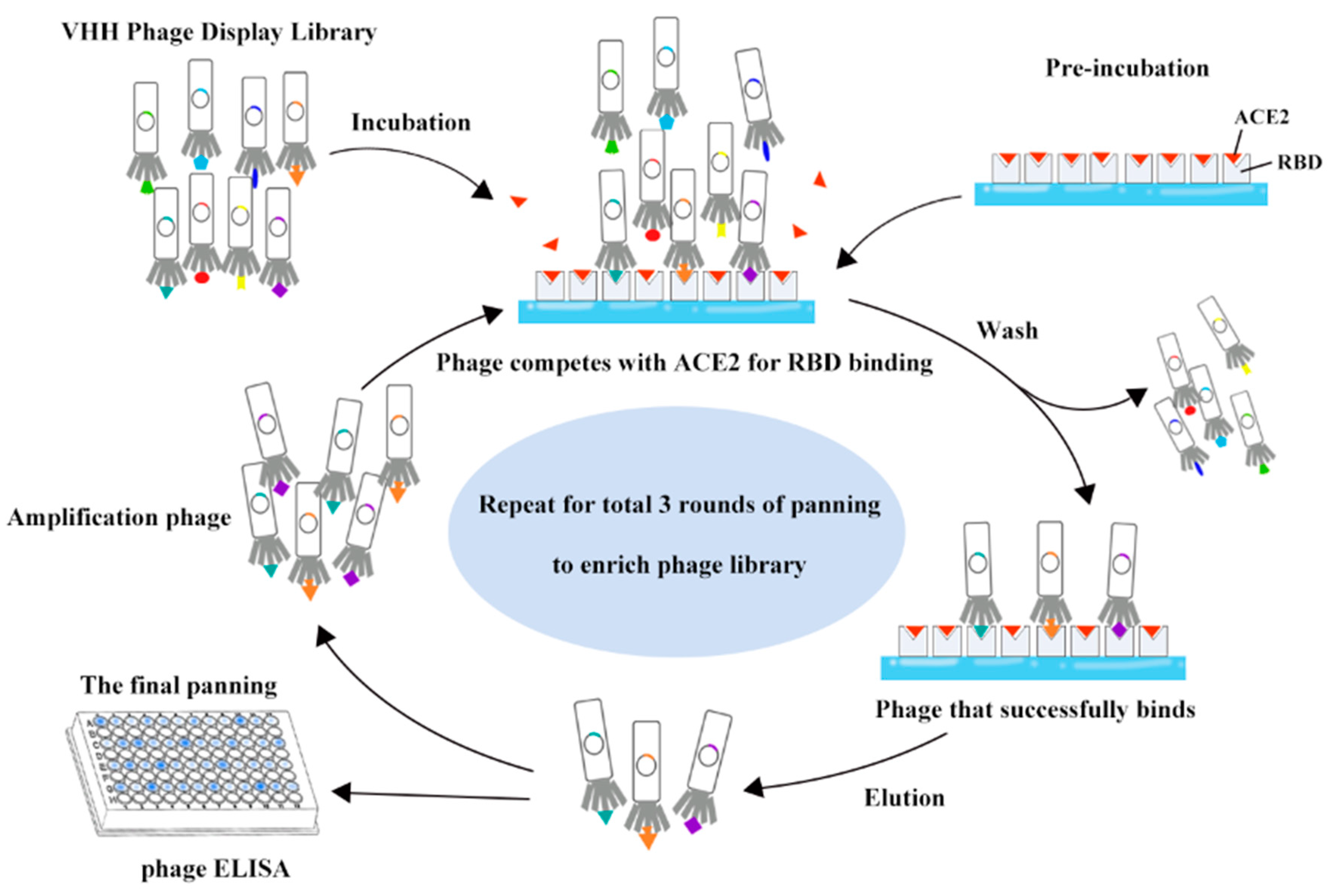
2.1. SARS-CoV-2 RBD-ACE2 Competitive Planning of Phage Libraries
2.2. Phage ELISA for Identification of Positive Clones
2.3. Competitive Phage ELISA
2.4. Prokaryotic Expression and Purification of the VHH Obtained by Screening
2.5. Validation of VHH Binding Specificity to the SARS-CoV-2 RBD by ELISA
2.6. Determination of the Binding Capacity of the VHH to the SARS-CoV-2 RBD (EC50)
2.7. Validation of VHH Competition with ACE2 for Binding to the SARS-CoV-2 RBD
2.8. Docking Simulation Studies and Analysis of the Complexes
2.9. Pseudovirus-Based Neutralization Assay
2.10. Binding Activity of VHH to Recombinant Phage Displaying the RBD of SARS-CoV-2 Mutants
2.11. Quantification and Statistical Analysis
3. Results
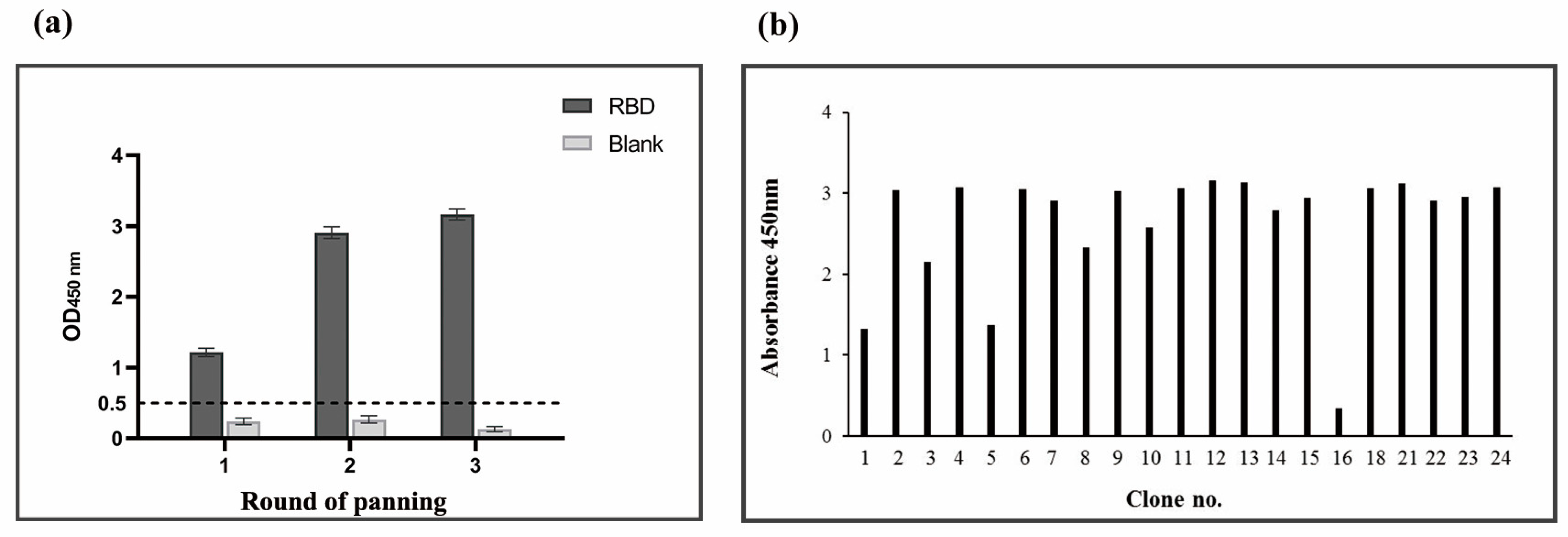
3.1. Enrichment of ACE2-Competitive Tight Binders against the SARS-CoV-2 RBD by Panning
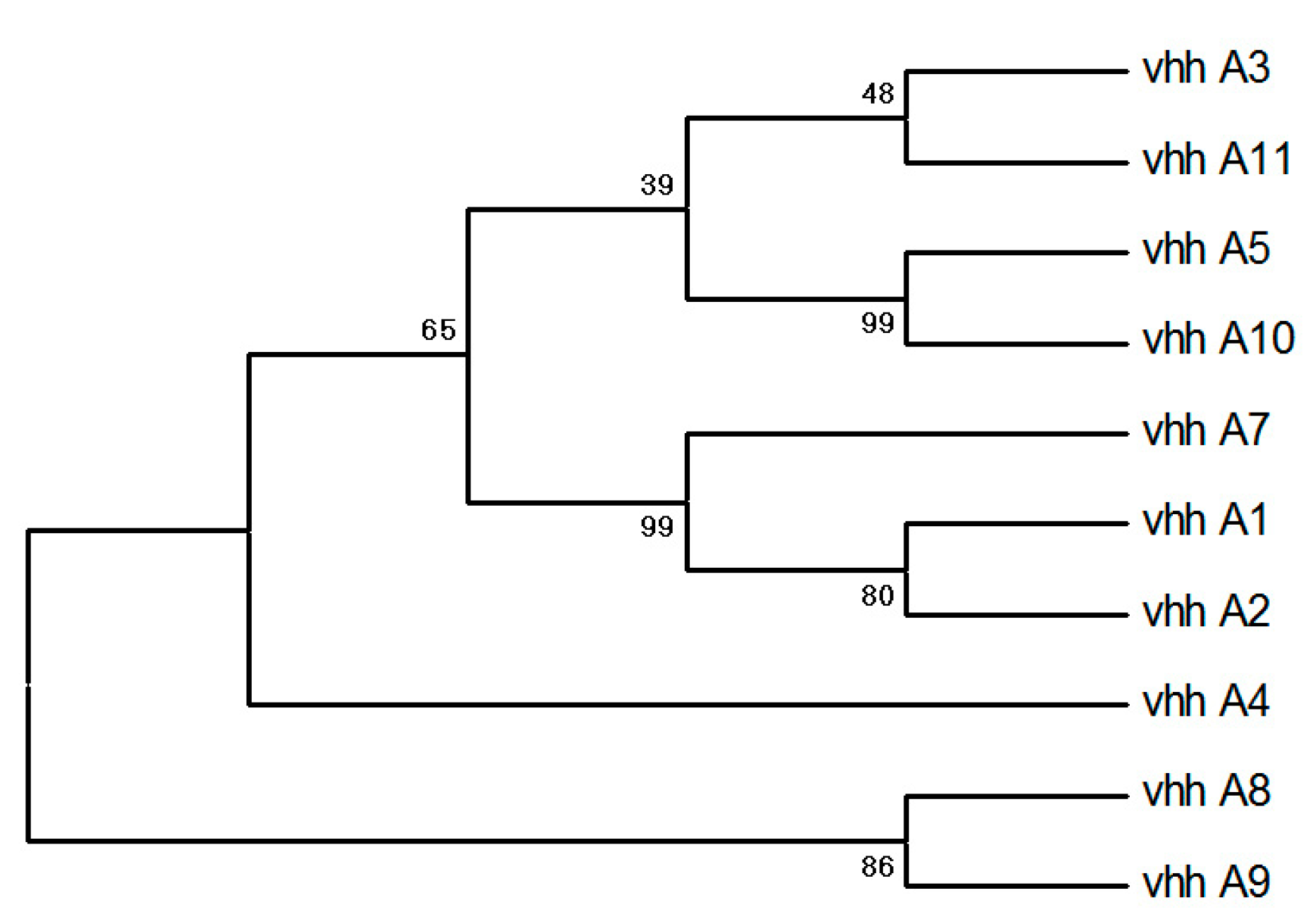
3.2. Selection of SARS-CoV-2 RBD-Specific Binders
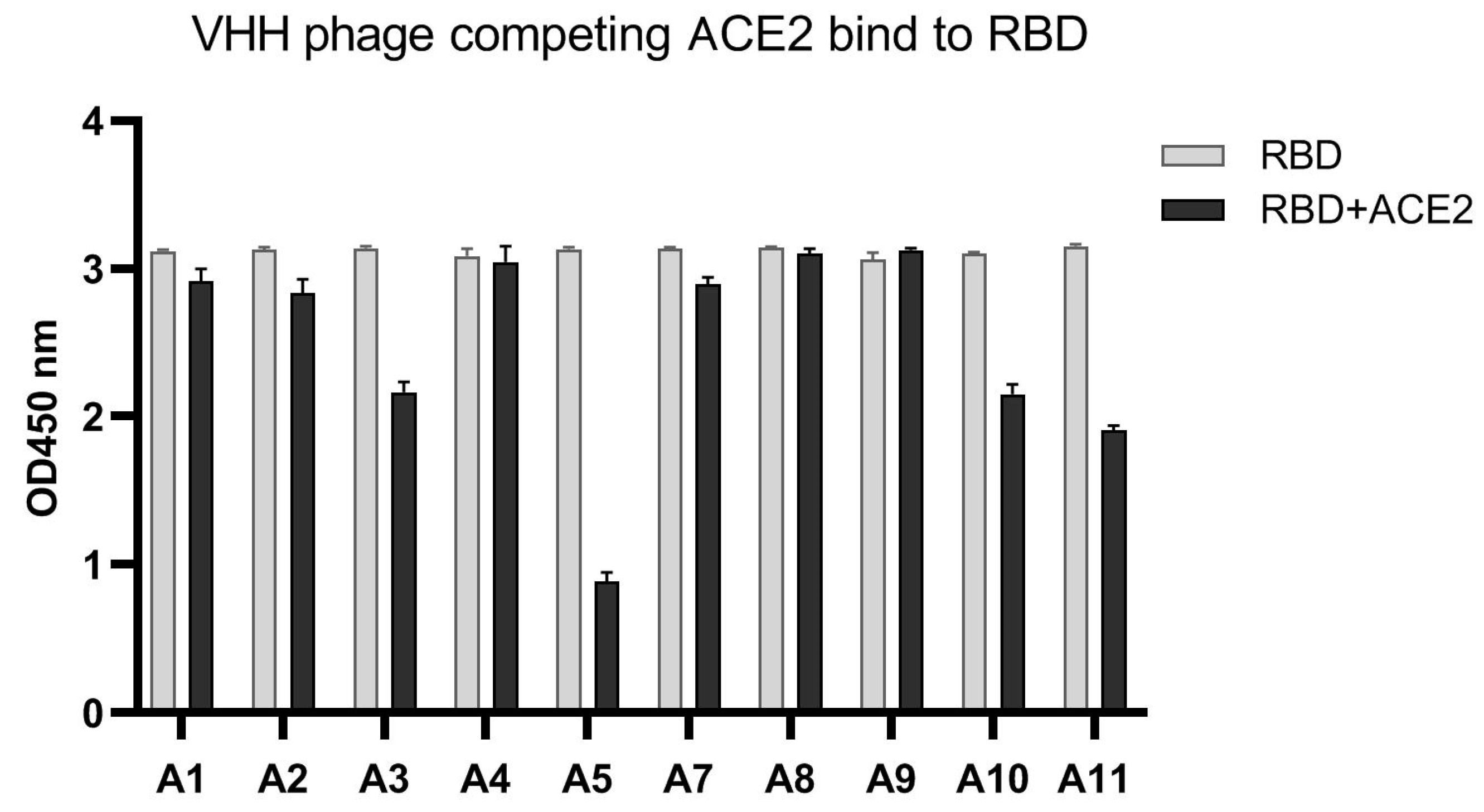
3.3. VHH-Phages with RBD-ACE2 Blocking Capability
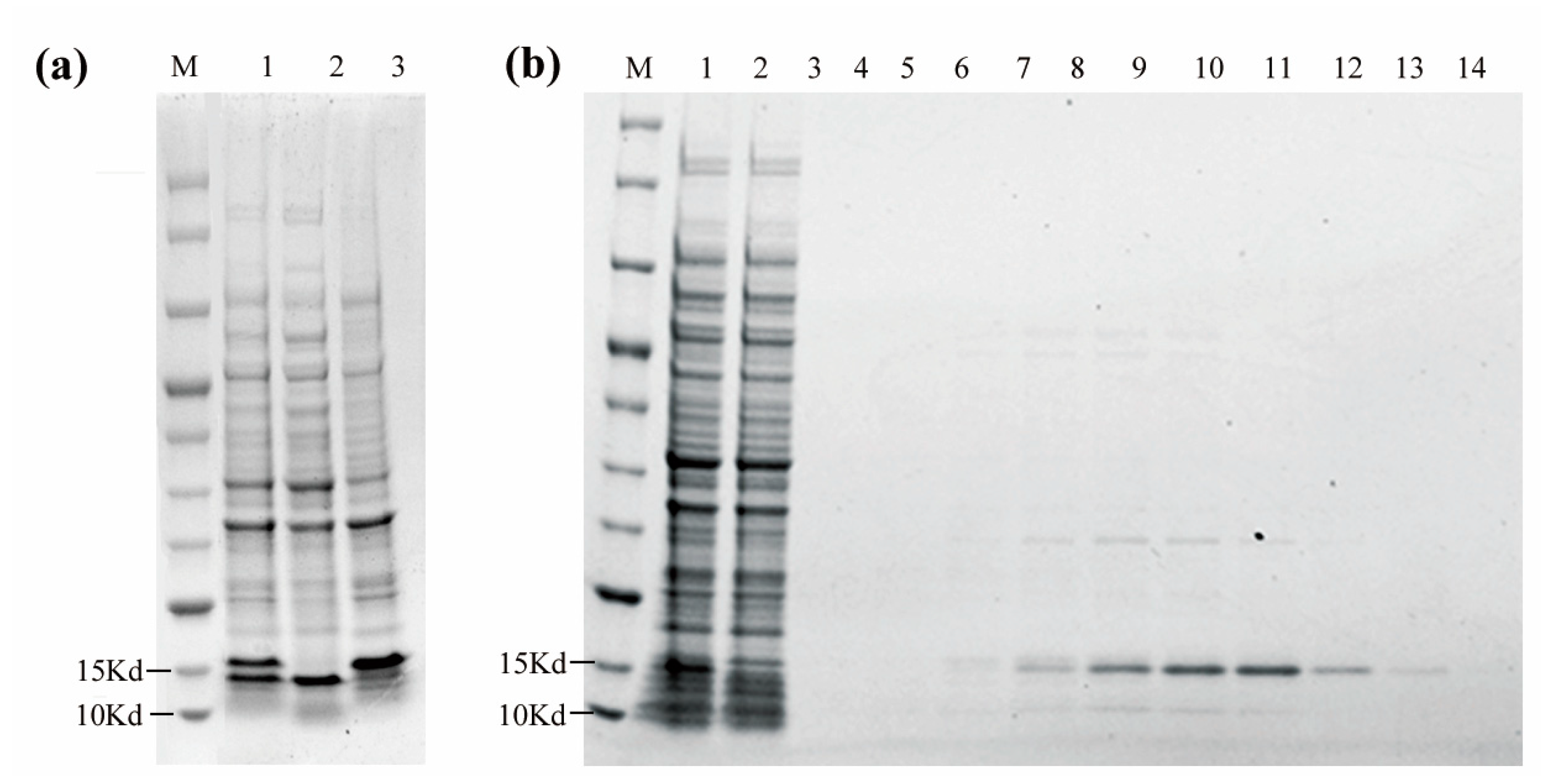
3.4. Expression and Purification of VHH5-05
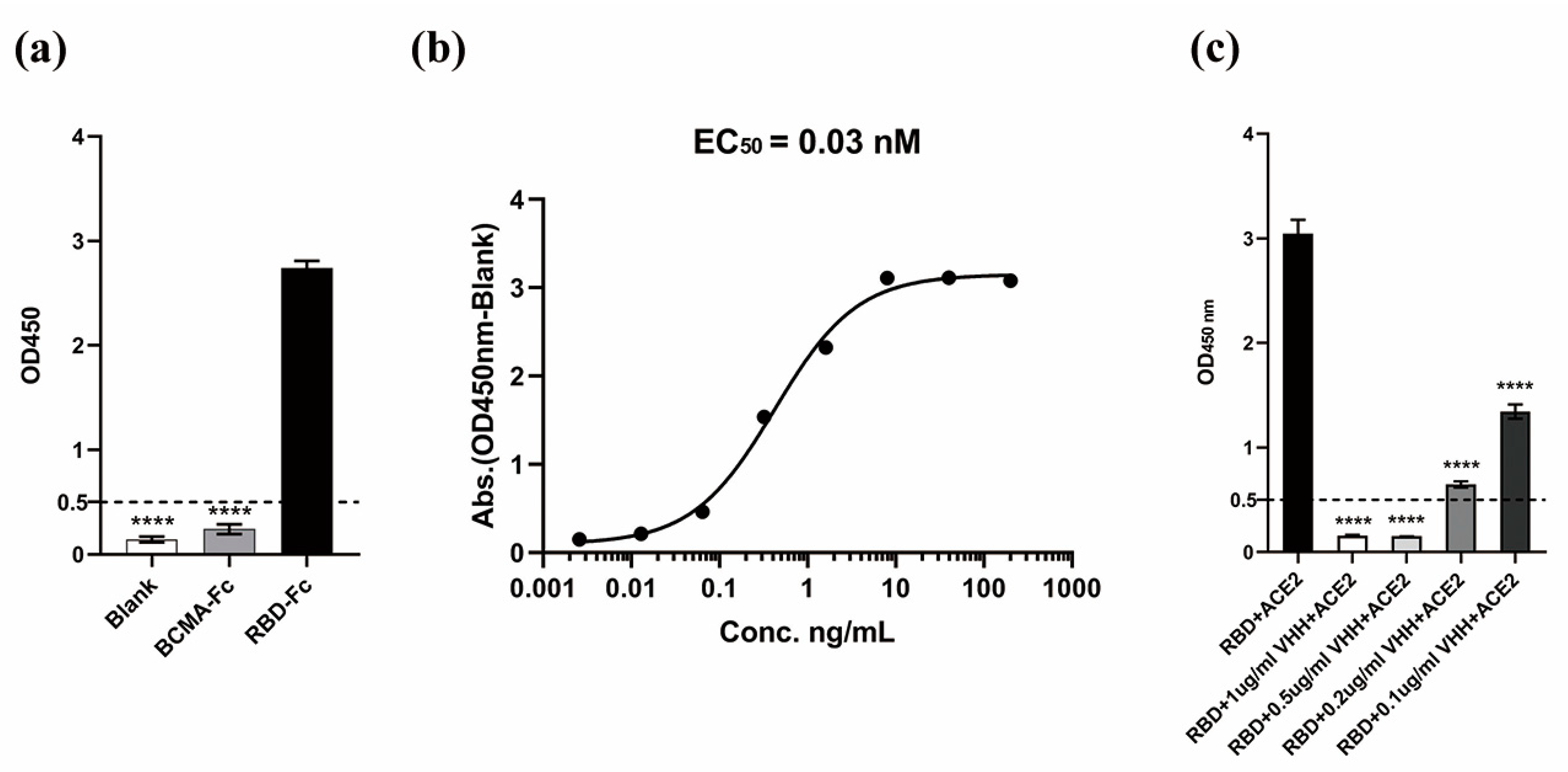
3.5. Binding Specificity and Affinity Determination of VHH5-05 to the SARS-CoV-2 RBD
3.6. VHH5-05 Competes with ACE2 for Binding to the SARS-CoV-2 RBD
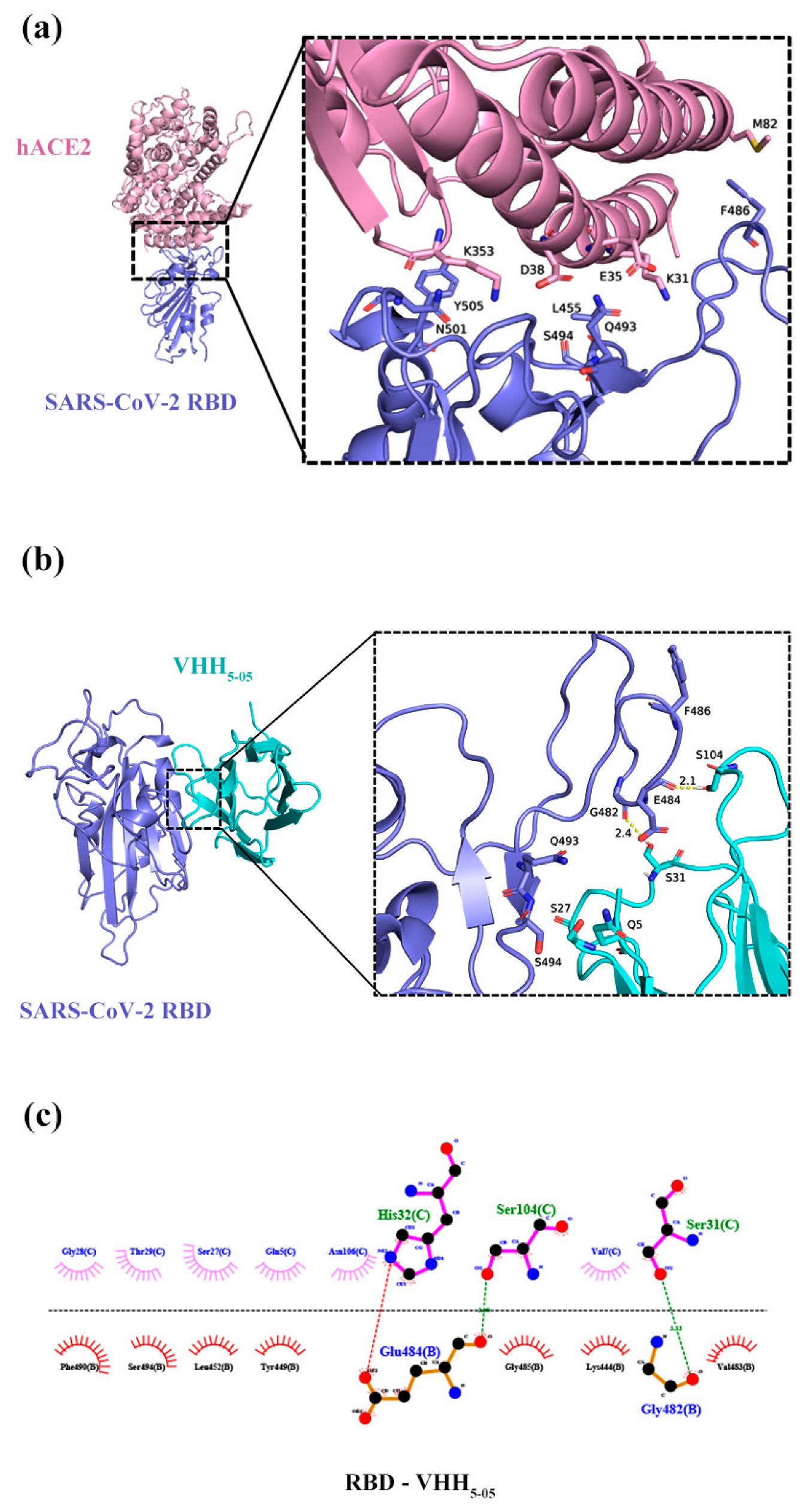
3.7. Prediction of VHH5-05 Binding Sites to RBD Using a Docking Simulation
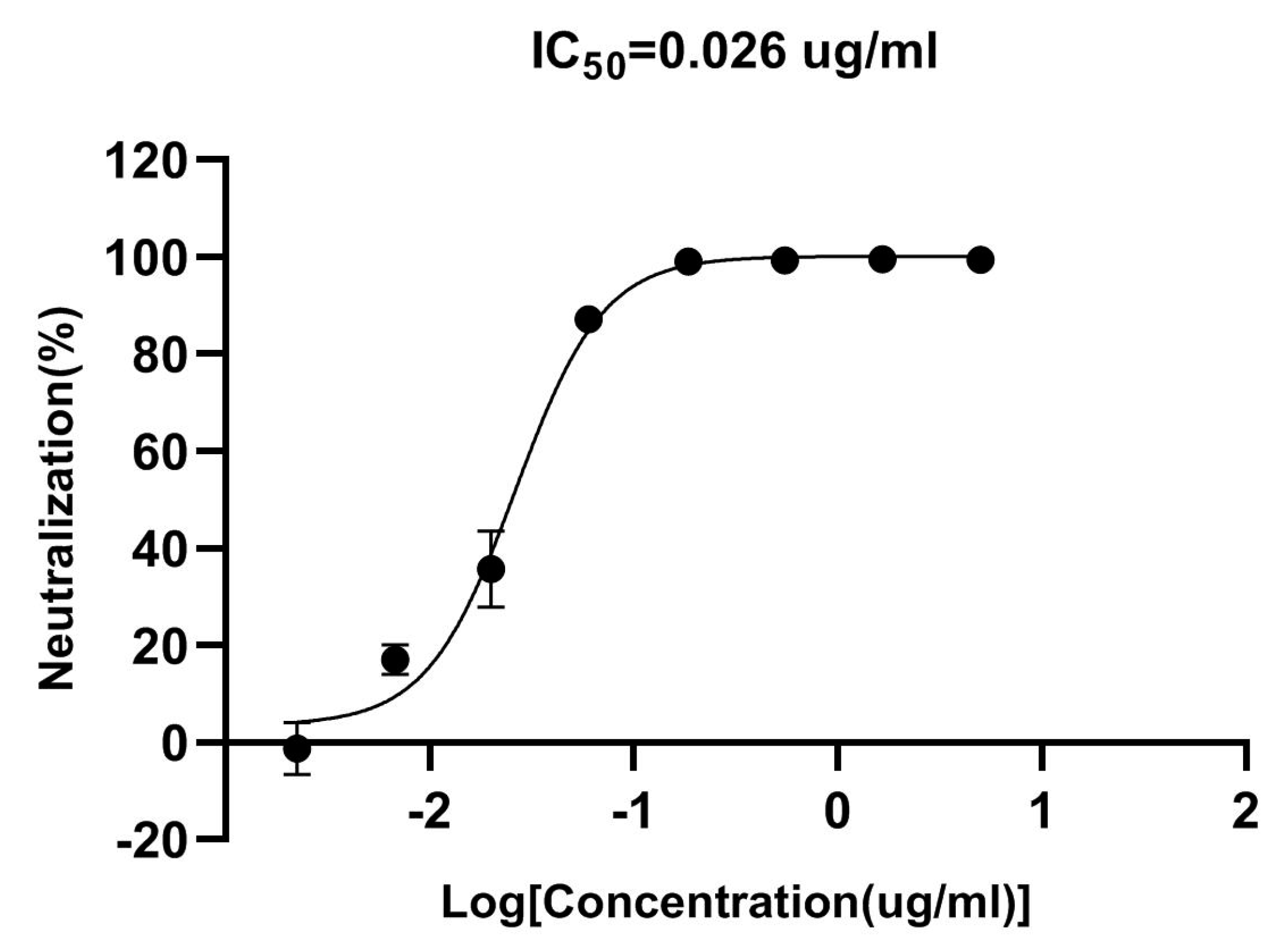
3.8. Pseudovirus Neutralization Ability of VHH5-05
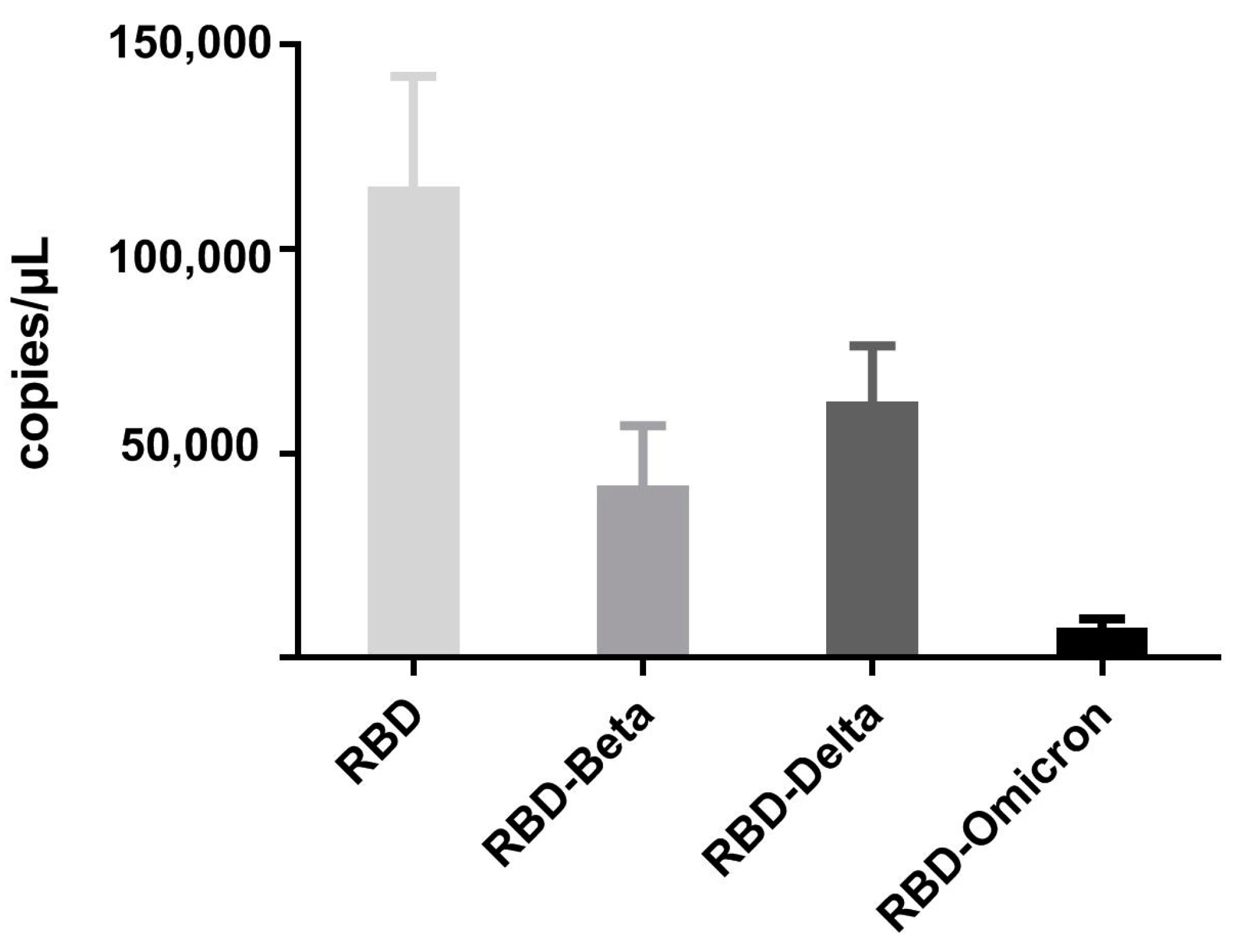
3.9. Binding of VHH5-05 to Recombinant Phages Displaying the RBD of SARS-CoV-2 Mutants
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Q.; Guan, X.H.; Wu, P.; Wang, X.Y.; Zhou, L.; Tong, Y.Q.; Ren, R.Q.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.S.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Ju, B.; Zhang, Q.; Ge, J.W.; Wang, R.K.; Sun, J.; Ge, X.Y.; Yu, J.Z.; Zhou, B.; Song, S.; Tang, X.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119. [Google Scholar] [CrossRef]
- Rogers, T.F.; Zhao, F.Z.; Huang, D.L.; Beutler, N.; Burns, A.; He, W.T.; Limbo, O.; Smith, C.; Song, G.; Woehl, J.; et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 2020, 369, 956–963. [Google Scholar] [CrossRef]
- Shi, R.; Shan, C.; Duan, X.M.; Chen, Z.H.; Liu, P.P.; Song, J.W.; Song, T.; Bi, X.S.; Han, C.; Wu, L.A.; et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020, 584, 120–124. [Google Scholar] [CrossRef]
- Taylor, P.C.; Adams, A.C.; Hufford, M.M.; de la Torre, I.; Winthrop, K.; Gottlieb, R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021, 21, 382–393. [Google Scholar] [CrossRef]
- Sui, J.H.; Li, W.H.; Murakami, A.; Tamin, A.; Matthews, L.J.; Wong, S.K.; Moore, M.J.; Tallarico, A.S.C.; Olurinde, M.; Choe, H.; et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. USA 2004, 101, 536–2541. [Google Scholar]
- Ter Meulen, J.; van den Brink, E.N.; Poon, L.L.M.; Marissen, W.E.; Leung, C.S.W.; Cox, F.; Cheung, C.Y.; Bakker, A.Q.; Bogaards, J.A.; van Deventer, E.; et al. Human monoclonal antibody combination against SARS coronavirus: Synergy and coverage of escape mutants. PLoS Med. 2006, 3, e237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.Y.; Chakraborti, S.; He, Y.; Roberts, A.; Sheahan, T.; Xiao, X.D.; Hensley, L.E.; Prabakaran, P.; Rockx, B.; Sidorov, I.A.; et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc. Natl. Acad. Sci. USA 2007, 104, 12123–12128. [Google Scholar] [CrossRef]
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hammers, C.; Bajyana Songa, E.; Bendahman, N.; Hammers, R. Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448. [Google Scholar] [CrossRef]
- Muyldermans, S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef]
- Wu, Y.; Jiang, S.; Ying, T. Single-Domain Antibodies As Therapeutics against Human Viral Diseases. Front. Immunol. 2017, 8, 1802. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef]
- Smith, G.P. Phage display: Simple evolution in a petri dish (Nobel Lecture). Angew. Chem. Int. Ed. 2019, 58, 14428–14437. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, A.D.; Duncan, A.R. Strategies for selection of antibodies by phage display. Curr. Opin. Biotechnol. 1998, 9, 102–108. [Google Scholar] [CrossRef]
- Coomber, D.W. Panning of antibody phage-display libraries. Standard protocols. Methods Mol. Biol. 2002, 178, 133–145. [Google Scholar]
- Smith, G.P.; Petrenko, V.A. Phage display. Chem. Rev. 1997, 97, 391–410. [Google Scholar] [CrossRef] [PubMed]
- Watkins, J.D.; Beuerlein, G.; Pecht, G.; McFadden, P.R.; Glaser, S.M.; Huse, W.D. Determination of the relative affinities of antibody fragments expressed in Escherichia coli by enzyme-linked immunosorbent assay. Anal. Biochem. 1997, 253, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.H.; Zhang, Y.F.; Wu, L.L.; Niu, S.; Song, C.L.; Zhang, Z.Y.; Lu, G.W.; Qiao, C.P.; Hu, Y.; Yuen, K.Y.; et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Ou, X.Y.; Liu, Y.; Lei, X.B.; Li, P.; Mi, D.; Ren, L.L.; Guo, L.; Guo, R.X.; Chen, T.; Hu, J.X.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef]
- Chen, H.Y.; Li, S.; Wang, J.L.; He, S.Q.; Wang, D.; Qian, Z.H.; Hu, D.D.; Qi, F.F.; Hu, K.P.; Luo, C.Y.; et al. Simultaneous measurement of the antibody responses against SARS-CoV-2 and its multiple variants by a phage display mediated immuno-multiplex quantitative PCR-based assay. Front. Microbiol. 2022, 13, 968036. [Google Scholar] [CrossRef]
- Harmsen, M.M.; Van Solt, C.B.; Fijten, H.P.D. Enhancement of toxin- and virus-neutralizing capacity of single-domain antibody fragments by N-glycosylation. Appl. Microbiol. Biotechnol 2009, 84, 1087–1094. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.T.; Man, Z.T.; Peng, C.L.; Wang, G.Z.; Sun, S. A specific affinity cyclic peptide enhances the adhesion, expansion and proliferation of rat bone mesenchymal stem cells on β-tricalcium phosphate scaffolds. Mol. Med. Rep. 2019, 20, 1157–1166. [Google Scholar] [CrossRef]
- Butler, J.E.; Ni, L.; Nessler, R.; Joshi, K.S.; Suter, M.; Rosenberg, B.; Chang, J.; Brown, W.R.; Cantarero, L.A. The physical and functional behavior of capture antibodies adsorbed on polystyrene. Immunol. Methods 1992, 150, 77–90. [Google Scholar] [CrossRef] [PubMed]
- He, F.L. Laemmli-sds-page. Bio-Protocol 2011, 101, e80. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef] [PubMed]
- Luan, J.; Lu, Y.; Jin, X.; Zhang, L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem. Biophys. Res. Commun. 2020, 526, 165–169. [Google Scholar] [CrossRef]
- Cao, Y.L.; Wang, J.; Jian, F.C.; Xiao, T.H.; Song, W.L.; Yisimayi, A.; Huang, W.J.; Li, Q.Q.; Wang, P.; An, R.; et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2021, 602, 657–663. [Google Scholar] [CrossRef]
- Planas, D.; Saunders, N.; Maes, P.; Guivel-Benhassine, F.; Planchais, C.; Buchrieser, J.; Bolland, W.H.; Porrot, F.; Staropoli, I.; Lemoine, F.; et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2021, 602, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.H.; Iketani, S.; Guo, Y.C.; Chan, J.F.W.; Wang, M.; Liu, L.Y.; Luo, Y.; Chu, H.; Huang, Y.M.; Nair, M.S.; et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 2021, 602, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Rothe, C.; Schunk, M.; Sothmann, P.; Bretzel, G.; Froeschl, G.; Wallrauch, C.; Zimmer, T.; Thiel, V.; Janke, C.; Guggemos, W.; et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020, 382, 970–971. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.Y.; He, L.; Sun, S.H.; Qiu, H.J.; Tai, W.B.; Chen, J.W.; Li, J.F.; Chen, Y.H.; Guo, Y.; Wang, Y.F.; et al. A novel nanobody targeting Middle East respiratory syndrome coronavirus (MERS-CoV) receptor-binding domain has potent cross-neutralizing activity and protective efficacy against MERS-CoV. J. Virol. 2018, 92, e00837-18. [Google Scholar] [CrossRef]
- Du, S.; Cao, Y.L.; Zhu, Q.Y.; Yu, P.; Qi, F.F.; Wang, G.P.; Du, X.X.; Bao, L.L.; Deng, W.; Zhu, H.; et al. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell 2020, 183, 1013–1023. [Google Scholar] [CrossRef]
- Van Heeke, G.; Allosery, K.; De Brabandere, V.; De Smedt, T.; Detalle, L.; de Fougerolles, A. Nanobodies as inhaled biotherapeutics for lung diseases. Pharmacol. Ther. 2017, 169, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhan, W.Q.; Yang, Z.L.; Tu, C.; Hu, G.W.; Zhang, X.; Song, W.P.; Du, S.J.; Zhu, Y.F.; Huang, K.K.; et al. Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody. Cell 2022, 185, 1389–1401.e18. [Google Scholar] [CrossRef]
- Cull, M.G.; Miller, J.F.; Schatz, P.J. Screening for receptor ligands using large libraries of peptides linked to the C terminus of the lac repressor. Proc. Natl. Acad. Sci. USA 1992, 89, 1865. [Google Scholar] [CrossRef]
- Ledsgaard, L.; Ljungars, A.; Rimbault, C.; Sorensen, C.V.; Tulika, T.; Wade, J.; Wouters, Y.; McCafferty, J.; Laustsen, A.H. Advances in antibody phage display technology. Drug Discov. Today 2022, 27, 2151–2169. [Google Scholar] [CrossRef]
- Wang, K.; Jia, Z.J.; Bao, L.L.; Wang, L.; Cao, L.; Chi, H.; Hu, Y.L.; Li, Q.Q.; Zhou, Y.J.; Jiang, Y.A.; et al. Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants. Nature 2022, 603, 919–925. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, P.; Wang, N.; Wang, L.; Fan, K.Y.; Zhu, Q.H.; Wang, K.; Chen, R.H.; Feng, R.; Jia, Z.J.; et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell 2022, 185, 860–871.e13. [Google Scholar] [CrossRef] [PubMed]
- Mannar, D.; Saville, J.W.; Zhu, X.; Srivastava, S.S.; Berezuk, A.M.; Tuttle, K.S.; Marquez, A.C.; Sekirov, I.; Subramaniam, S. SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein–ACE2 complex. Science 2022, 375, 760–764. [Google Scholar] [CrossRef]
- Iketani, S.; Liu, L.H.; Guo, Y.C.; Liu, L.Y.; Chan, J.F.W.; Huang, Y.M.; Wang, M.; Luo, Y.; Yu, J.; Chu, H.; et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature 2022, 604, 553–556. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.Q.; Wang, Y.F.; Wang, Y.X.; Zhang, C.; Hong, Q.; Gu, C.J.; Xu, R.; Wang, T.F.; Yang, Y.; Zang, J.K.; et al. Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein. Emerg. Microbes Infect. 2022, 11, 351–367. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Tian, X.L.; Jia, X.D.; Wan, J.K.; Lu, L.; Jiang, S.B.; Lan, F.; Lu, Y.Y.; Wu, Y.L.; Ying, T.L. The impact of receptor-binding domain natural mutations on antibody recognition of SARS-CoV-2. Signal Transduct. Target. Ther. 2021, 6, 132. [Google Scholar] [CrossRef]
- Tian, X.L.; Li, C.; Huang, A.L.; Xia, S.; Lu, S.C.; Shi, Z.L.; Lu, L.; Jiang, S.B.; Yang, Z.L.; Wu, Y.L.; et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef]
- Yuan, M.; Wu, N.C.; Zhu, X.; Lee, C.C.D.; So, R.T.Y.; Lv, H.B.; Mok, C.K.P.; Wilson, I.A. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science 2020, 368, 630–633. [Google Scholar] [CrossRef] [Green Version]
| RBD Concentration (µg/well) | ACE2 Concentration (µg/well) | Proportion of Tween-20 in PBST | Wash Times in Final Step | Input Titer (PFU) | Output Titer (PFU) | Recovery Efficiency | Fold Increase | |
|---|---|---|---|---|---|---|---|---|
| Round 1 | 0.5 | — | 0.05% | 10 | 1.4 × 1011 | 3.66 × 105 | 2.6 × 10−6 | — |
| Round 2 | 0.1 | 0.2 | 0.1% | 20 | 3.31 × 1011 | 5.35 × 107 | 1.6 × 10−4 | 62 |
| Round 3 | 0.05 | 0.1 | 0.2% | 30 | 2.93 × 1011 | 3.7 × 108 | 1.3 × 10−3 | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, S.; Wang, J.; Chen, H.; Qian, Z.; Hu, K.; Shi, B.; Wang, J. A Competitive Panning Method Reveals an Anti-SARS-CoV-2 Nanobody Specific for an RBD-ACE2 Binding Site. Vaccines 2023, 11, 371. https://doi.org/10.3390/vaccines11020371
He S, Wang J, Chen H, Qian Z, Hu K, Shi B, Wang J. A Competitive Panning Method Reveals an Anti-SARS-CoV-2 Nanobody Specific for an RBD-ACE2 Binding Site. Vaccines. 2023; 11(2):371. https://doi.org/10.3390/vaccines11020371
Chicago/Turabian StyleHe, Siqi, Jiali Wang, Hanyi Chen, Zhaohui Qian, Keping Hu, Bingjie Shi, and Jianxun Wang. 2023. "A Competitive Panning Method Reveals an Anti-SARS-CoV-2 Nanobody Specific for an RBD-ACE2 Binding Site" Vaccines 11, no. 2: 371. https://doi.org/10.3390/vaccines11020371
APA StyleHe, S., Wang, J., Chen, H., Qian, Z., Hu, K., Shi, B., & Wang, J. (2023). A Competitive Panning Method Reveals an Anti-SARS-CoV-2 Nanobody Specific for an RBD-ACE2 Binding Site. Vaccines, 11(2), 371. https://doi.org/10.3390/vaccines11020371





