Cell Surface Fibroblast Activation Protein-2 (Fap2) of Fusobacterium nucleatum as a Vaccine Candidate for Therapeutic Intervention of Human Colorectal Cancer: An Immunoinformatics Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Retrieval of Protein Sequences and Prediction of Linear B-Cell Epitopes
2.2. Prediction of MHC-I and MHC-II Epitopes and Determination of Their Antigenicity
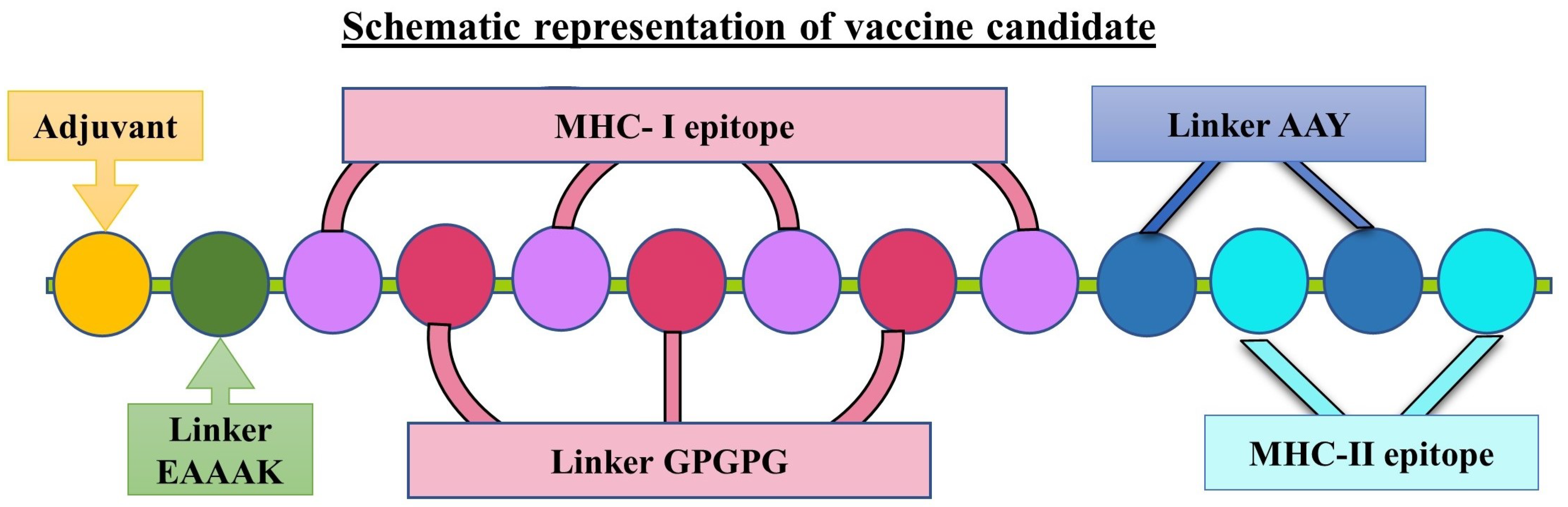
2.3. Designing of Vaccine Candidates and Prediction of Secondary Structures
2.4. Analyses of the Antigenicity, Allergenicity, and Physicochemical Properties of the Designed Vaccine Candidates
2.5. Modelling of Tertiary Structure, Refinement, and Validation
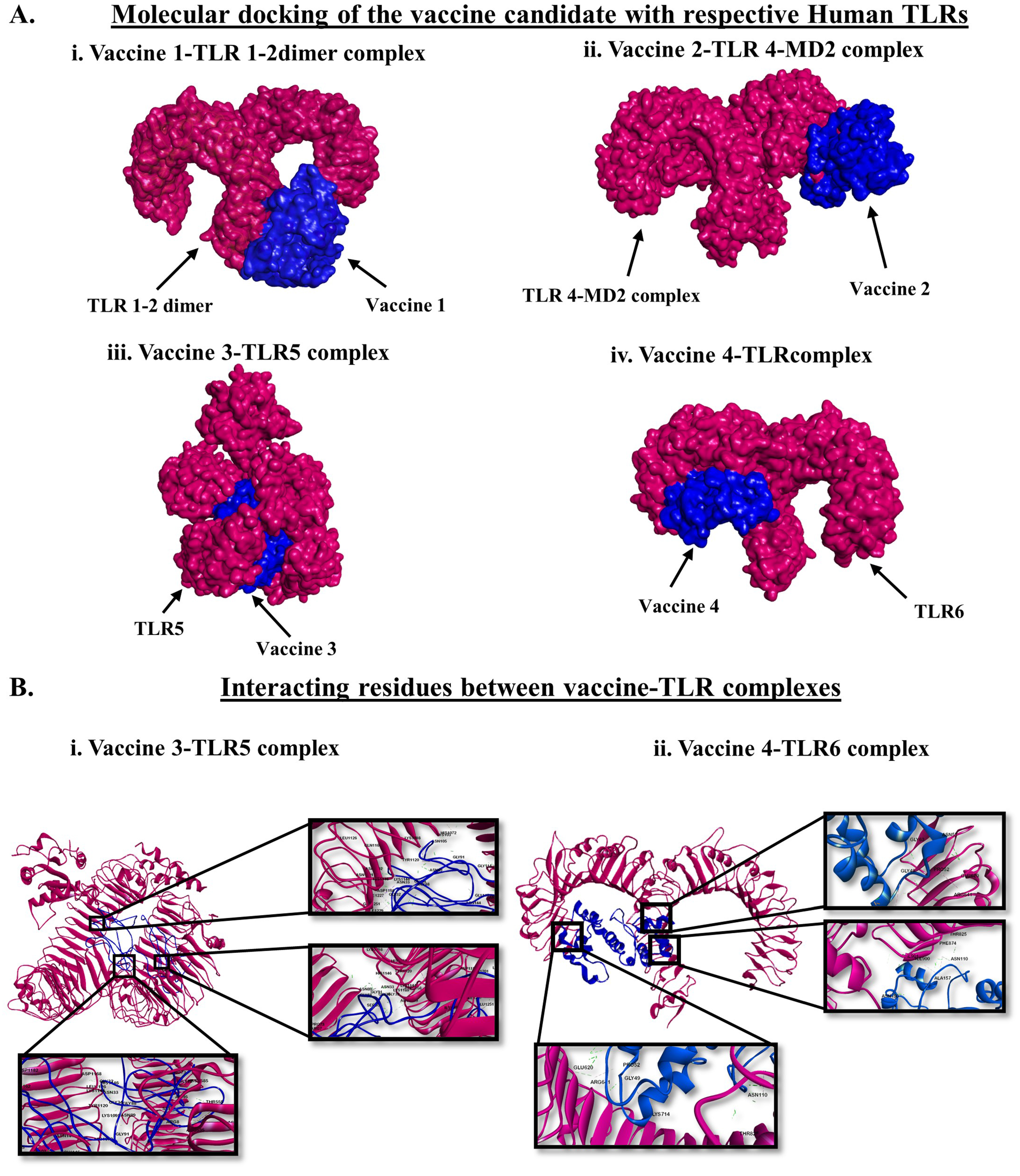
2.6. Molecular Docking and Determination of Biophysical Interactions
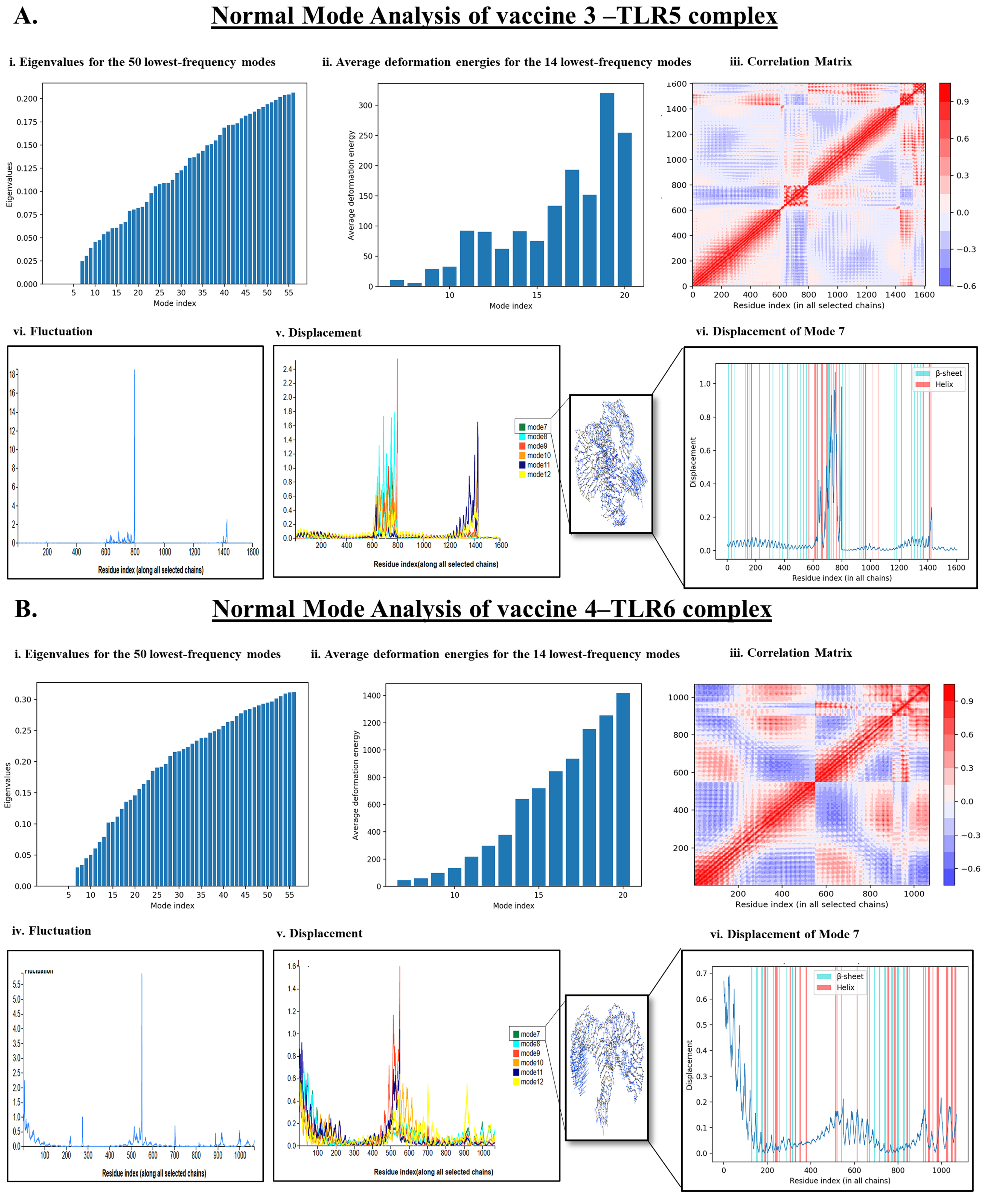
2.7. Normal Mode Analysis
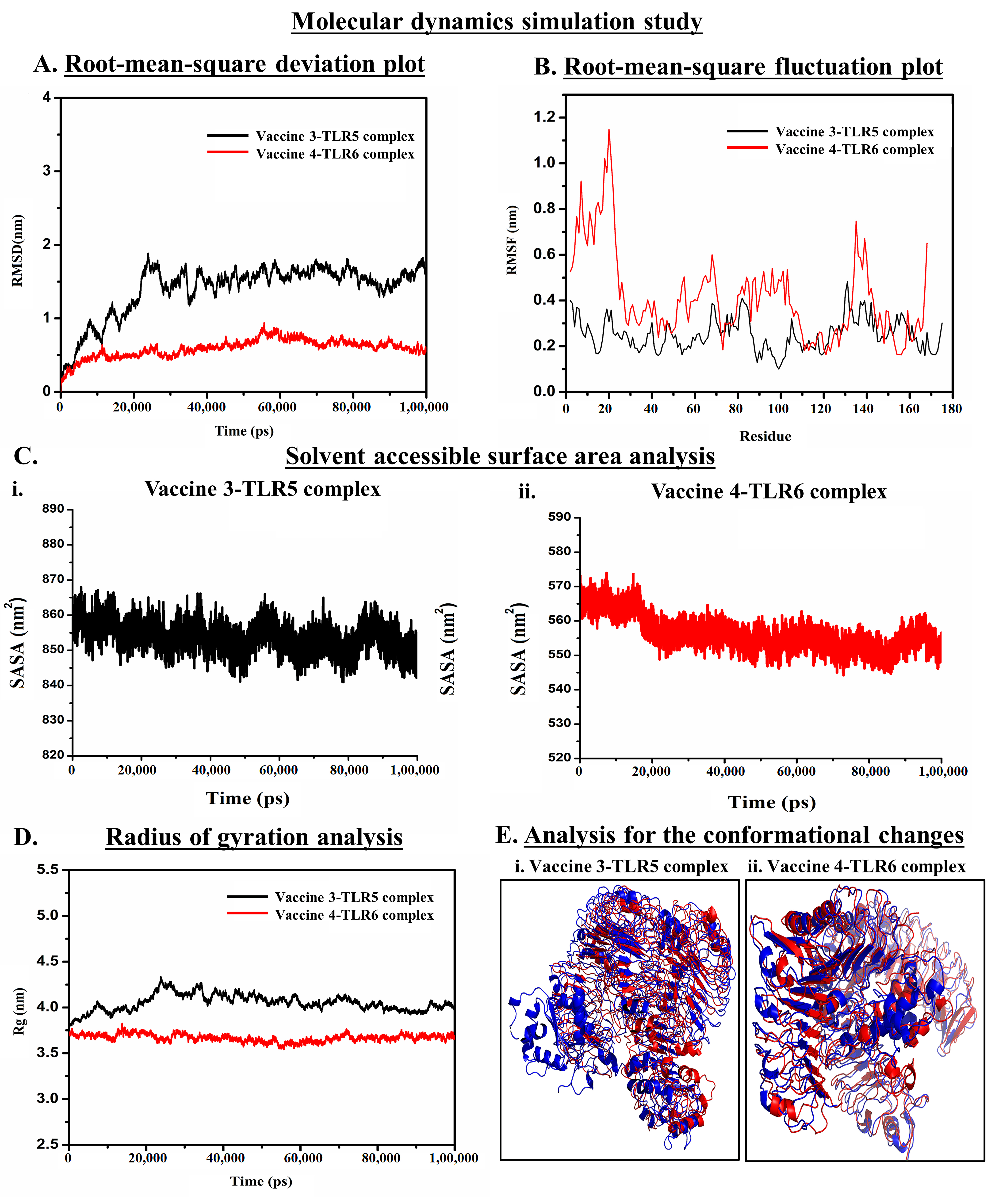
2.8. Molecular Dynamic Simulation
2.9. Codon Optimization and In Silico Cloning Preparation
2.10. Determination of the Structure of the mRNA Encoding the Vaccine Peptide
2.11. Immune Simulation
3. Results
3.1. Presence of B-Cell, MHC-I, and MHC-II Epitopes in Fap2 Protein and Their Antigenicity
3.2. Designing the Vaccine Candidates and Prediction of the Secondary and Tertiary Structures
3.3. Molecular Docking of the Vaccines with Human TLRs and Exploration of the Biophysical Interactions
3.4. Normal Mode Analysis (NMA)
3.5. Molecular Dynamics Simulation (MDS) Trajectory Analysis
3.5.1. Analysis of the Stability of Vaccine–TLR Complexes Using RMSD
3.5.2. Analysis of Residual Flexibility
3.5.3. Analysis of Solvent-Accessible Surface Area
3.5.4. Analysis of Compactness
3.5.5. Determining the Conformational Changes
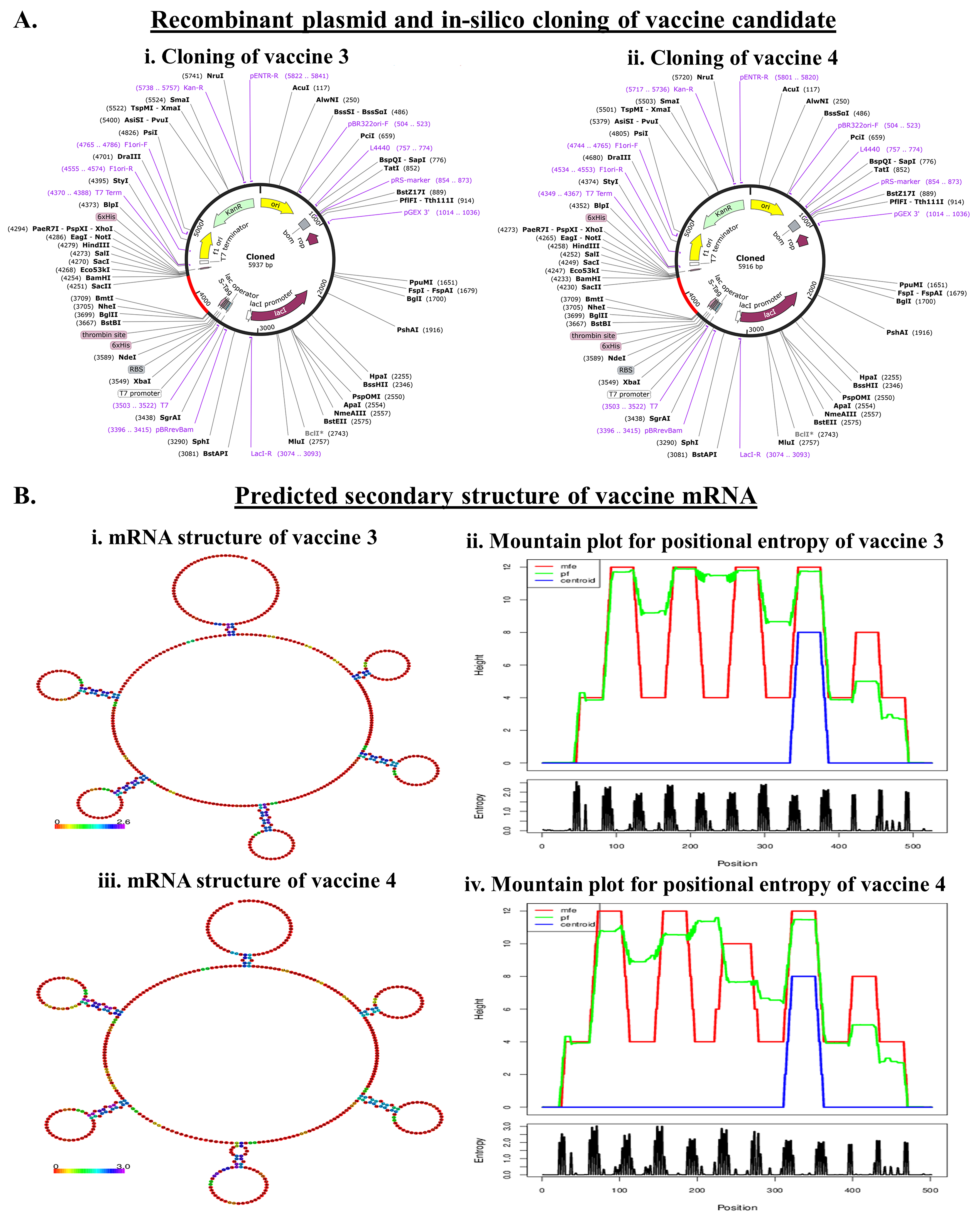
3.6. Codon Optimization and In Silico Cloning for the Production of Recombinant Protein
3.7. mRNA Structure Prediction and Durability Analysis of the Designed Vaccine
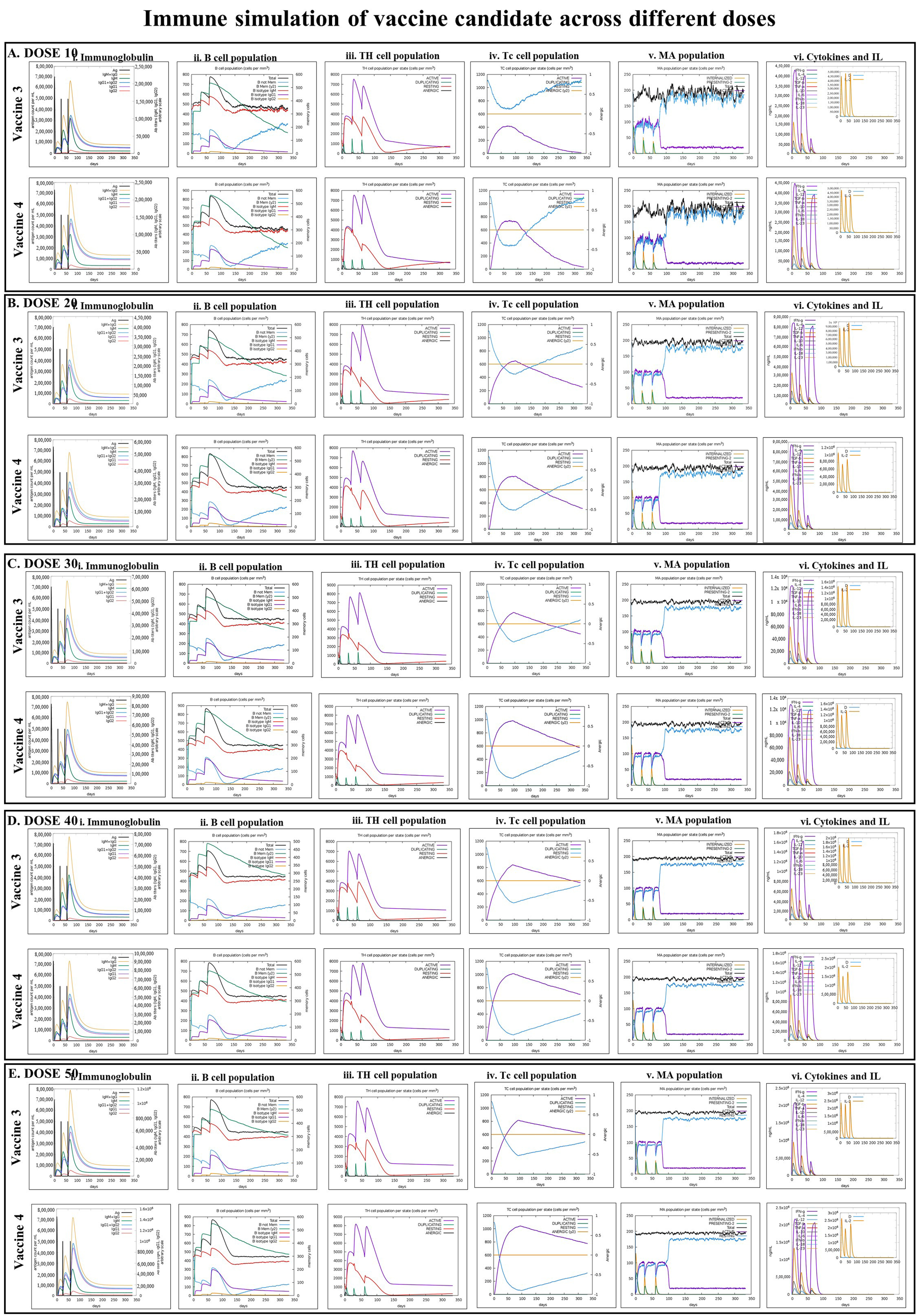
3.8. Immune Simulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kuipers, E.J.; Grady, W.M.; Lieberman, D.; Seufferlein, T.; Sung, J.J.; Boelens, P.G.; van de Velde, C.J.H.; Watanabe, T. Colorectal cancer. Nat. Rev. Dis. Prim. 2015, 1, 15065. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed]
- Patil, P.S.; Saklani, A.; Gambhire, P.; Mehta, S.; Engineer, R.; De’Souza, A.; Chopra, S.; Bal, M. Colorectal Cancer in India: An Audit from a Tertiary Center in a Low Prevalence Area. Indian J. Surg. Oncol. 2017, 8, 484–490. [Google Scholar] [CrossRef] [PubMed]
- Patra, R.; Mukherjee, S.; Das, N.C. Exploring the differential expression and prognostic significance of the COL11A1 gene in human colorectal carcinoma: An integrated bioinformatics approach. Front. Genet. 2021, 12, 88. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Joardar, N.; Sengupta, S.; Babu, S.P.S. Gut microbes as future therapeutics in treating inflammatory and infectious diseases: Lessons from recent findings. J. Nutr. Biochem. 2018, 61, 111–128. [Google Scholar] [CrossRef]
- Wong, S.H.; Yu, J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 690–704. [Google Scholar] [CrossRef]
- Kim, J.; Lee, H.K. Potential Role of the Gut Microbiome in Colorectal Cancer Progression. Front. Immunol. 2021, 12, 807648. [Google Scholar] [CrossRef]
- Vieira, A.; Teixeira, M.; Martins, F. The Role of Probiotics and Prebiotics in Inducing Gut Immunity. Front. Immunol. 2013, 4, 445. [Google Scholar] [CrossRef]
- Das, N.C.; Patra, R.; Dey, A.; Mukherjee, S. Probiotics as Efficacious Therapeutic Option for Treating Gut-Related Diseases: Molecular and Immunobiological Perspectives BT—Prebiotics, Probiotics and Nutraceuticals; Behera, K.K., Bist, R., Mohanty, S., Bhattacharya, M., Eds.; Springer Nature Singapore: Singapore, 2022; pp. 69–93. ISBN 978-981-16-8990-1. [Google Scholar]
- Abed, J.; Emgård, J.E.M.; Zamir, G.; Faroja, M.; Almogy, G.; Grenov, A.; Sol, A.; Naor, R.; Pikarsky, E.; Atlan, K.A.; et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe 2016, 20, 215–225. [Google Scholar] [CrossRef]
- Ganesan, K.; Guo, S.; Fayyaz, S.; Zhang, G.; Xu, B. Targeting Programmed Fusobacterium nucleatum Fap2 for Colorectal Cancer Therapy. Cancers 2019, 11, 1592. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via Its FadA Adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef]
- Coppenhagen-Glazer, S.; Sol, A.; Abed, J.; Naor, R.; Zhang, X.; Han, Y.W.; Bachrach, G. Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect. Immun. 2015, 83, 1104–1113. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, L.; Zheng, S.; Li, M.; Xu, C.; Jia, D.; Qi, Y.; Hou, T.; Wang, L.; Wang, B.; et al. Fusobacterium nucleatum promotes colorectal cancer cells adhesion to endothelial cells and facilitates extravasation and metastasis by inducing ALPK1/NF-κB/ICAM1 axis. Gut Microbes 2022, 14, 2038852. [Google Scholar] [CrossRef]
- Alon-Maimon, T.; Mandelboim, O.; Bachrach, G. Fusobacterium nucleatum and cancer. Periodontol. 2000 2022, 89, 166–180. [Google Scholar] [CrossRef]
- Zhou, W.-Y.; Shi, Y.; Wu, C.; Zhang, W.-J.; Mao, X.-H.; Guo, G.; Li, H.-X.; Zou, Q.-M. Therapeutic efficacy of a multi-epitope vaccine against Helicobacter pylori infection in BALB/c mice model. Vaccine 2009, 27, 5013–5019. [Google Scholar] [CrossRef]
- Wu, J.; Li, Q.; Fu, X. Fusobacterium nucleatum Contributes to the Carcinogenesis of Colorectal Cancer by Inducing Inflammation and Suppressing Host Immunity. Transl. Oncol. 2019, 12, 846–851. [Google Scholar] [CrossRef]
- Wang, N.; Fang, J.-Y. Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. 2023, 31, 159–172. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Y.; Li, J.; Zhao, L.; Yan, W.; Lin, B.; Guo, X.; Wei, Y. Fusobacterium nucleatum Acts as a Pro-carcinogenic Bacterium in Colorectal Cancer: From Association to Causality. Front. Cell Dev. Biol. 2021, 9, 710165. [Google Scholar] [CrossRef]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S.; et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef]
- Das, N.C.; Gupta, P.S.S.; Panda, S.K.; Rana, M.K.; Mukherjee, S. Reverse vaccinology assisted design of a novel multi-epitope vaccine to target Wuchereria bancrofti cystatin: An immunoinformatics approach. Int. Immunopharmacol. 2023, 115, 109639. [Google Scholar] [CrossRef] [PubMed]
- Nourmohammadi, H.; Javanmardi, E.; Shams, M.; Shamsinia, S.; Nosrati, M.C.; Yousefi, A.; Nemati, T.; Fatollahzadeh, M.; Ghasemi, E.; Kordi, B.; et al. Multi-epitope vaccine against cystic echinococcosis using immunodominant epitopes from EgA31 and EgG1Y162 antigens. Inform. Med. Unlocked 2020, 21, 100464. [Google Scholar] [CrossRef]
- Ashfaq, U.A.; Saleem, S.; Masoud, M.S.; Ahmad, M.; Nahid, N.; Bhatti, R.; Almatroudi, A.; Khurshid, M. Rational design of multi epitope-based subunit vaccine by exploring MERS-COV proteome: Reverse vaccinology and molecular docking approach. PLoS ONE 2021, 16, e0245072. [Google Scholar] [CrossRef] [PubMed]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Duthie, M.S.; Windish, H.P.; Fox, C.B.; Reed, S.G. Use of defined TLR ligands as adjuvants within human vaccines. Immunol. Rev. 2011, 239, 178–196. [Google Scholar] [CrossRef]
- Luchner, M.; Reinke, S.; Milicic, A. TLR Agonists as Vaccine Adjuvants Targeting Cancer and Infectious Diseases. Pharmaceutics 2021, 13, 142. [Google Scholar] [CrossRef]
- Mukherjee, S.; Karmakar, S.; Babu, S.P.S. TLR2 and TLR4 mediated host immune responses in major infectious diseases: A review. Braz. J. Infect. Dis. 2016, 20, 193–204. [Google Scholar] [CrossRef]
- Das, N.C.; Patra, R.; Gupta, P.S.S.; Ghosh, P.; Bhattacharya, M.; Rana, M.K.; Mukherjee, S. Designing of a novel multi-epitope peptide based vaccine against Brugia malayi: An in silico approach. Infect. Genet. Evol. 2021, 87, 104633. [Google Scholar] [CrossRef]
- Magnan, C.N.; Zeller, M.; Kayala, M.A.; Vigil, A.; Randall, A.; Felgner, P.L.; Baldi, P. High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics 2010, 26, 2936–2943. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v.2—A server for in silico prediction of allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server BT—The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. ISBN 978-1-59259-890-8. [Google Scholar]
- Hebditch, M.; Carballo-Amador, M.A.; Charonis, S.; Curtis, R.; Warwicker, J. Protein-Sol: A web tool for predicting protein solubility from sequence. Bioinformatics 2017, 33, 3098–3100. [Google Scholar] [CrossRef]
- Lee, G.R.; Won, J.; Heo, L.; Seok, C. GalaxyRefine2: Simultaneous refinement of inaccurate local regions and overall protein structure. Nucleic Acids Res. 2019, 47, W451–W455. [Google Scholar] [CrossRef]
- Das, N.C.; Sen Gupta, P.S.; Biswal, S.; Patra, R.; Rana, M.K.; Mukherjee, S. In-silico evidences on filarial cystatin as a putative ligand of human TLR4. J. Biomol. Struct. Dyn. 2021, 40, 8808–8824. [Google Scholar] [CrossRef]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure 2020, 28, 1071–1081.e3. [Google Scholar] [CrossRef]
- Tiwari, S.P.; Fuglebakk, E.; Hollup, S.M.; Skjærven, L.; Cragnolini, T.; Grindhaug, S.H.; Tekle, K.M.; Reuter, N. WEBnm@ v2.0: Web server and services for comparing protein flexibility. BMC Bioinform. 2014, 15, 427. [Google Scholar] [CrossRef]
- Fu, H.; Liang, Y.; Zhong, X.; Pan, Z.; Huang, L.; Zhang, H.; Xu, Y.; Zhou, W.; Liu, Z. Codon optimization with deep learning to enhance protein expression. Sci. Rep. 2020, 10, 17617. [Google Scholar] [CrossRef]
- Grote, A.; Hiller, K.; Scheer, M.; Münch, R.; Nörtemann, B.; Hempel, D.C.; Jahn, D. JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005, 33, W526–W531. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- Sato, K.; Akiyama, M.; Sakakibara, Y. RNA secondary structure prediction using deep learning with thermodynamic integration. Nat. Commun. 2021, 12, 941. [Google Scholar] [CrossRef]
- Rapin, N.; Lund, O.; Bernaschi, M.; Castiglione, F. Computational immunology meets bioinformatics: The use of prediction tools for molecular binding in the simulation of the immune system. PLoS ONE 2010, 5, e9862. [Google Scholar] [CrossRef]
- Tahir Ul Qamar, M.; Rehman, A.; Tusleem, K.; Ashfaq, U.A.; Qasim, M.; Zhu, X.; Fatima, I.; Shahid, F.; Chen, L.-L. Designing of a next generation multiepitope based vaccine (MEV) against SARS-COV-2: Immunoinformatics and in silico approaches. PLoS ONE 2020, 15, e0244176. [Google Scholar] [CrossRef]
- Mukherjee, S.; Huda, S.; Sinha Babu, S.P. Toll-like receptor polymorphism in host immune response to infectious diseases: A review. Scand. J. Immunol. 2019, 90, e12771. [Google Scholar] [CrossRef] [PubMed]
- Das, C.N.; Labala, K.R.; Patra, R.; Chattoraj, A.; Mukherjee, S. In Silico Identification of New Anti-SARS-CoV-2 Agents from Bioactive Phytocompounds Targeting the Viral Spike Glycoprotein and Human TLR4. Lett. Drug Des. Discov. 2022, 19, 175–191. [Google Scholar] [CrossRef]
- Hamada, T.; Zhang, X.; Mima, K.; Bullman, S.; Sukawa, Y.; Nowak, J.A.; Kosumi, K.; Masugi, Y.; Twombly, T.S.; Cao, Y.; et al. Fusobacterium nucleatum in Colorectal Cancer Relates to Immune Response Differentially by Tumor Microsatellite Instability Status. Cancer Immunol. Res. 2018, 6, 1327–1336. [Google Scholar] [CrossRef] [PubMed]
- Luddy, K.A.; Robertson-Tessi, M.; Tafreshi, N.K.; Soliman, H.; Morse, D.L. The role of toll-like receptors in colorectal cancer progression: Evidence for epithelial to leucocytic transition. Front. Immunol. 2014, 5, 429. [Google Scholar] [CrossRef]
- Shey, R.A.; Ghogomu, S.M.; Shintouo, C.M.; Nkemngo, F.N.; Nebangwa, D.N.; Esoh, K.; Yaah, N.E.; Manka’aFri, M.; Nguve, J.E.; Ngwese, R.A.; et al. Computational Design and Preliminary Serological Analysis of a Novel Multi-Epitope Vaccine Candidate Against Onchocerciasis and Related Filarial Diseases. Pathogens 2021, 10, 99. [Google Scholar] [CrossRef]
- Masignani, V.; Pizza, M.; Moxon, E.R. The Development of a Vaccine against Meningococcus B Using Reverse Vaccinology. Front. Immunol. 2019, 10, 751. [Google Scholar] [CrossRef]
- Serruto, D.; Bottomley, M.J.; Ram, S.; Giuliani, M.M.; Rappuoli, R. The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: Immunological, functional and structural characterization of the antigens. Vaccine 2012, 30, B87–B97. [Google Scholar] [CrossRef]
- Choudhury, A.; Sen Gupta, P.S.; Panda, S.K.; Rana, M.K.; Mukherjee, S. Designing AbhiSCoVac—A single potential vaccine for all ‘corona culprits’: Immunoinformatics and immune simulation approaches. J. Mol. Liq. 2022, 351, 118633. [Google Scholar] [CrossRef]

| Sl. No. | B-Cell Epitopes | MHC-I Allele | MHC-I Epitopes | VaxiJen Score |
|---|---|---|---|---|
| 1. | NADGSNNTTMTNMVNK | HLA-A1 | NADGSNNTT | 2.3648 |
| 2. | NININGDSSIGVGLLQ | HLA-A1 | NGDSSIGVG | 1.7041 |
| 3. | NININGDSSIGVGLLQ | HLA-A-0205 | GDSSIGVGL | 1.6222 |
| 4. | ASKATNDSNGTITLDT | HLA-A1 | TNDSNGTIT | 1.6262 |
| HLA-A-0205 | KATNDSNGT | 2.2960 | ||
| 5. | SVGIFAKNNGTNDTAK | HLA-A1 | NNGTNDTAK | 1.6448 |
| HLA-A-0205 | KNNGTNDTA | 1.5250 | ||
| 6. | VGTITLKNSTVSNGSS | HLA-A1 | NSTVSNGSS | 1.7004 |
| 7. | PASPDPNKLEIETTSN | HLA-A1 | KLEIETTSN | 1.5014 |
| HLA-A-0205 |
| Sl. No. | B-Cell Epitopes Sequence | MHC-II Allele | MHC-II Epitopes | VaxiJen Score |
|---|---|---|---|---|
| 1. | NININGDSSIGVGLLQ | HLA-DRB1_0301 | INGDSSIGV | 1.3077 |
| 2. | SGTIIMKNQNSVGILG | HLA-DRB1_0101 HLA-DRB1_0102 | MKNQNSVGI | 1.1398 |
| 3. | VGTITLKNSTVSNGSS | HLA-DRB1_0301 | LKNSTVSNG | 1.0256 |
| Characteristics | VACCINE 1 | VACCINE 2 | VACCINE 3 | VACCINE 4 |
|---|---|---|---|---|
| VaxiJen Score | 1.308 | 0.9979 | 1.3648 | 1.4968 |
| Antigen-Pro Score | 0.81 | 0.87 | 0.76 | 0.75 |
| Solubility | 0.664 | 0.85 | 0.731 | 0.728 |
| No. of amino acids | 284 | 292 | 175 | 168 |
| Molecular weight (in Dalton) | 28,348.97 | 28,388.16 | 16,402.45 | 15,668.6 |
| Theoretical Isoelectric point (pI) | 5.58 | 4.54 | 4.62 | 6.37 |
| Formula | C1190H1906N358O425S10 | C1215H1963N341O434S3 | C673H1079N209O261S4 | C644H1032N202O250S2 |
| Total no. of atoms | 3889 | 3956 | 2226 | 2130 |
| Estimated half-life | 30 h (mammalian reticulocytes, in vitro). >20 h (yeast, in vivo). >10 h (Escherichia coli, in vivo) | 30 h (mammalian reticulocytes, in vitro) >20 h (yeast, in vivo). >10 h (Escherichia coli, in vivo) | 30 h (mammalian reticulocytes, in vitro). >20 h (yeast, in vivo). >10 h (Escherichia coli, in vivo) | 1.2 h (mammalian reticulocytes, in vitro) >20 h (yeast, in vivo) >10 h (Escherichia coli, in vivo) |
| Instability index | 25.11 | 10.71 | 5.87 | 3.51 |
| Aliphatic index | 57.01 | 69.97 | 46.34 | 41.31 |
| Grand Avg. of hydropathicity (GRAVY) | −0.621 | −0.339 | −0.655 | −0.812 |
| Name of Vaccine | ERRAT Value | Percentage of Protein Residue in the Favored Region (Ramachandran Plot) | Docking Score |
|---|---|---|---|
| VACCINE 1 | 0.81 | 91.8% | −693.1 |
| VACCINE 2 | 0.87 | 95.1% | −624.4 |
| VACCINE 3 | 0.76 | 95.8% | −1096.6 |
| VACCINE 4 | 0.75 | 97.3% | −853.8 |
| Vaccine Name | Targeted TLR | Hydrogen Bond | Hydrophobic Interaction | Electrostatic Bond |
|---|---|---|---|---|
| Vaccine 3 | TLR 5 | 57 | 9 | 8 |
| Vaccine 4 | TLR 6 | 25 | 4 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padma, S.; Patra, R.; Sen Gupta, P.S.; Panda, S.K.; Rana, M.K.; Mukherjee, S. Cell Surface Fibroblast Activation Protein-2 (Fap2) of Fusobacterium nucleatum as a Vaccine Candidate for Therapeutic Intervention of Human Colorectal Cancer: An Immunoinformatics Approach. Vaccines 2023, 11, 525. https://doi.org/10.3390/vaccines11030525
Padma S, Patra R, Sen Gupta PS, Panda SK, Rana MK, Mukherjee S. Cell Surface Fibroblast Activation Protein-2 (Fap2) of Fusobacterium nucleatum as a Vaccine Candidate for Therapeutic Intervention of Human Colorectal Cancer: An Immunoinformatics Approach. Vaccines. 2023; 11(3):525. https://doi.org/10.3390/vaccines11030525
Chicago/Turabian StylePadma, Somrita, Ritwik Patra, Parth Sarthi Sen Gupta, Saroj Kumar Panda, Malay Kumar Rana, and Suprabhat Mukherjee. 2023. "Cell Surface Fibroblast Activation Protein-2 (Fap2) of Fusobacterium nucleatum as a Vaccine Candidate for Therapeutic Intervention of Human Colorectal Cancer: An Immunoinformatics Approach" Vaccines 11, no. 3: 525. https://doi.org/10.3390/vaccines11030525
APA StylePadma, S., Patra, R., Sen Gupta, P. S., Panda, S. K., Rana, M. K., & Mukherjee, S. (2023). Cell Surface Fibroblast Activation Protein-2 (Fap2) of Fusobacterium nucleatum as a Vaccine Candidate for Therapeutic Intervention of Human Colorectal Cancer: An Immunoinformatics Approach. Vaccines, 11(3), 525. https://doi.org/10.3390/vaccines11030525






