HLA-I and HLA-II Peptidomes of SARS-CoV-2: A Review
Abstract
1. Introduction
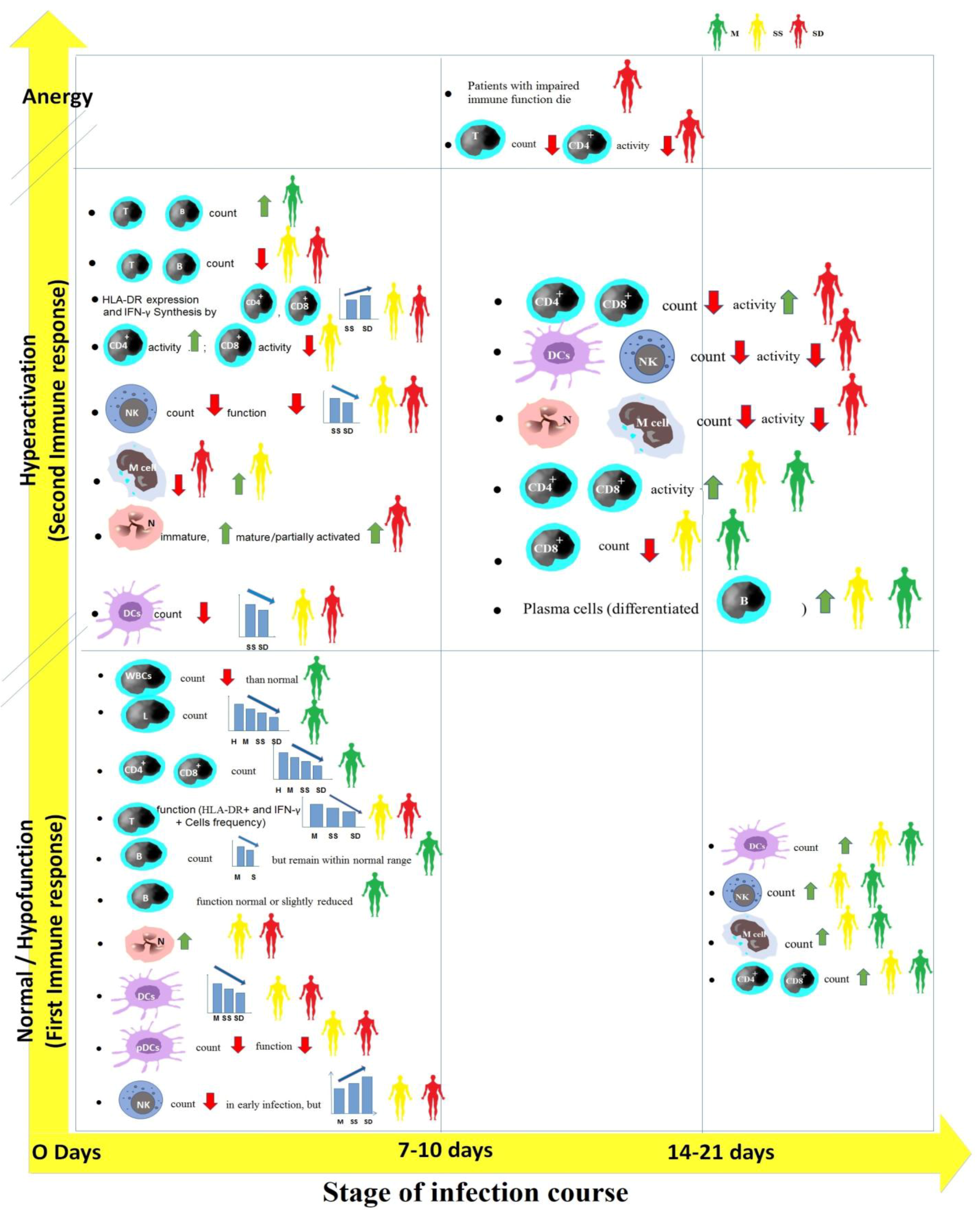
2. Cellular Immune Responses against SARS-CoV-2
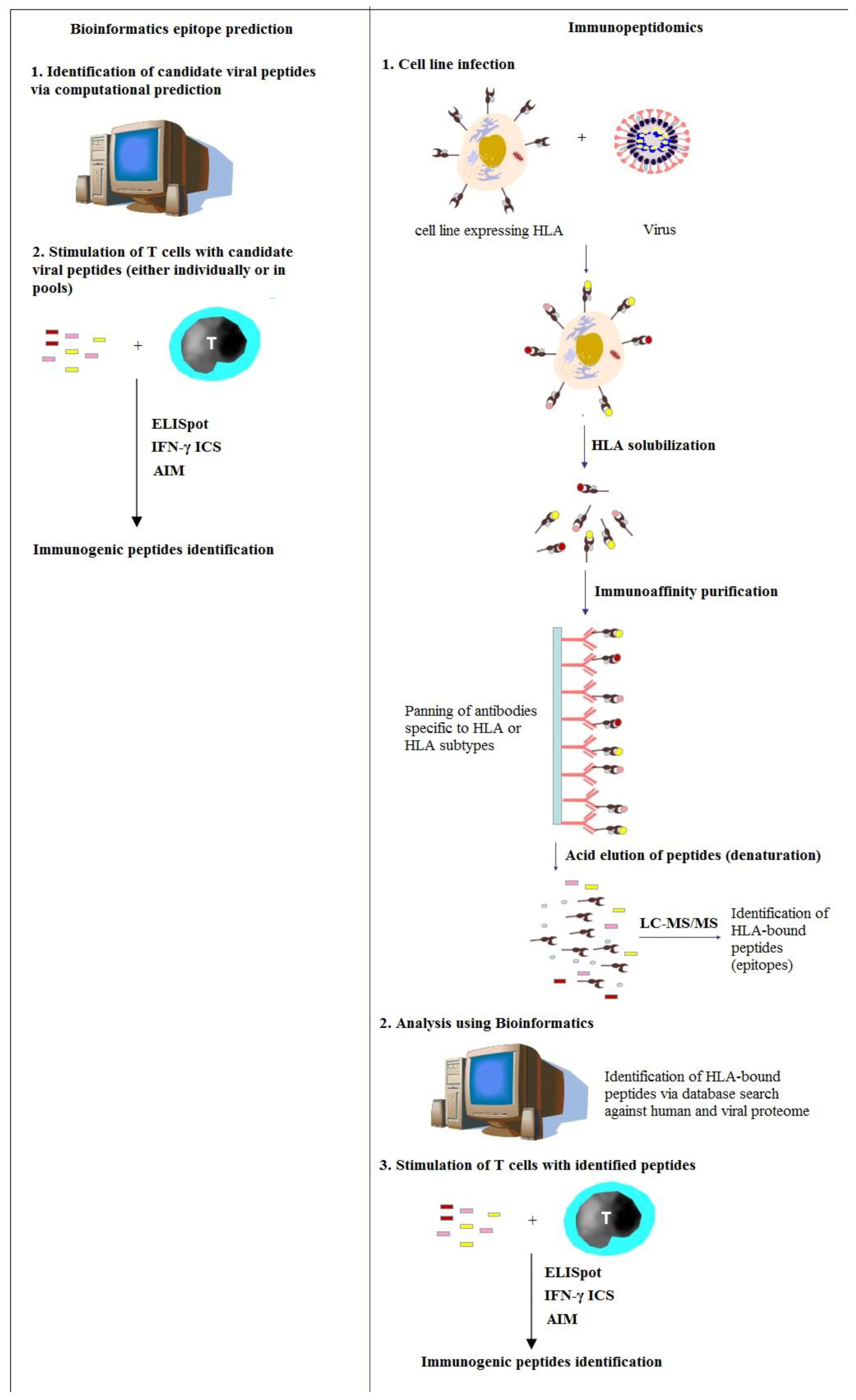
3. Immunopeptidomics to Study and Fight SARS-CoV-2
4. Immunopeptidomic Screenings for SARS-CoV-2 Using Bioinformatics
5. Immunopeptidomic Screenings for SARS-CoV-2 Using Mass Spectrometry
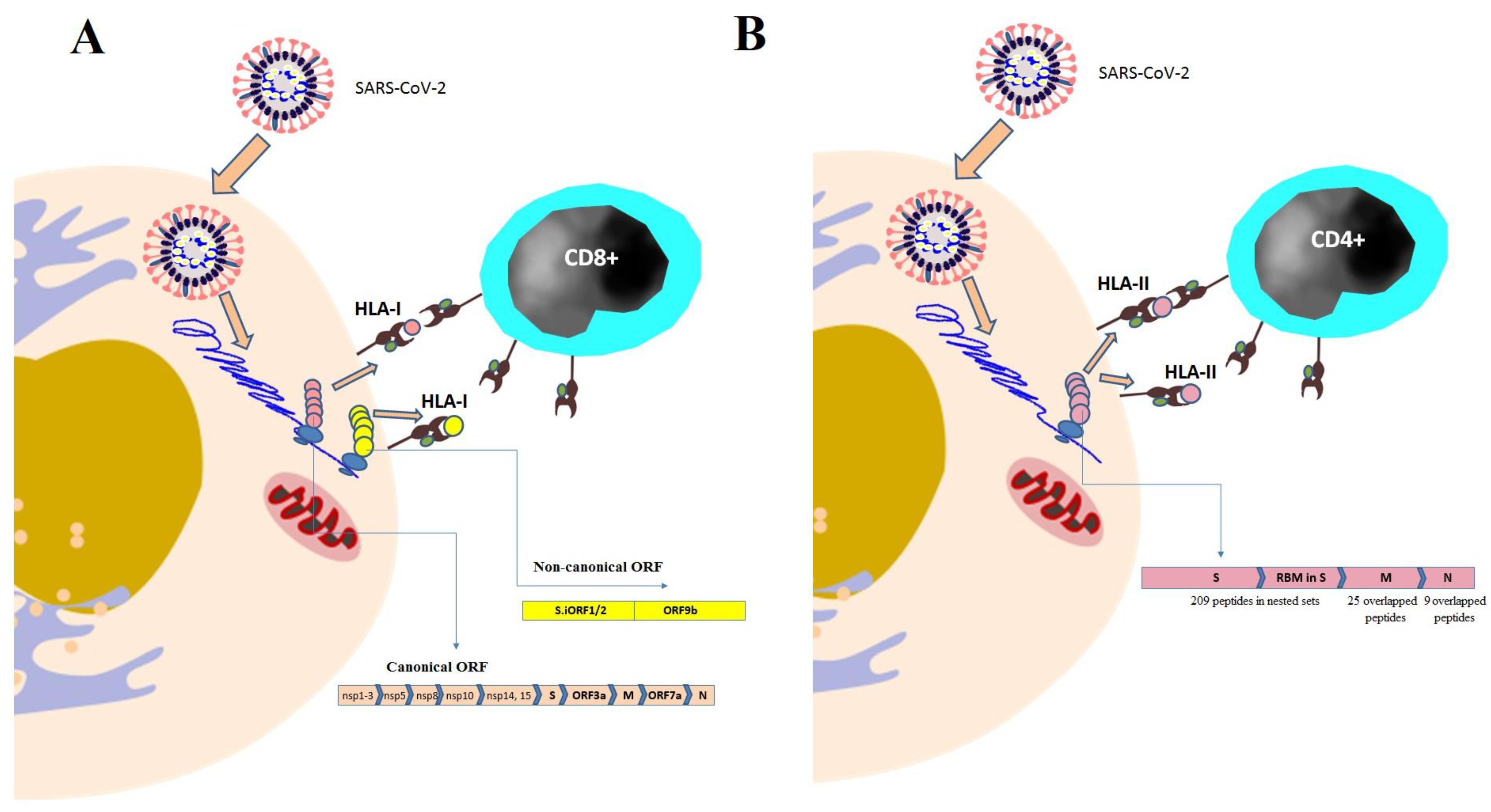
5.1. Profiling HLA-I Peptidome of SARS-CoV-2
5.2. Profiling HLA-II Peptidome of SARS-CoV-2
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. WHO Coronavirus (COVID-19) Dashboard. 2022. Available online: https://covid19.who.int/ (accessed on 28 December 2022).
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. mRNA-1273 Study Group. An mRNA vaccine against SARSCoV-2—Preliminary report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.E.; Frenck, R.W.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and immunogenicity of two RNA-based COVID-19 vaccine candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A.; et al. Publisher Correction: Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 2021, 590, E26. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Yuan, Y.; Zhou, Y.; Deng, Z.; Zhao, J.; Feng, F.; Zou, H.; Sun, C. Safety of SARS-CoV-2 vaccines: A systematic review and meta-analysis of randomized controlled trials. Infect. Dis. Poverty 2021, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Muller, L.; Andrée, M.; Moskorz, W.; Drexler, I.; Walotka, L.; Grothmann, R.; Ptok, J.; Hillebrandt, J.; Ritchie, A.; Rabl, D.; et al. Age-Dependent immune response to the Biontech/Pfizer BNT162b2 coronavirus disease 2019 vaccination. Clin. Infect. Dis. 2021, 73, 2065–2072. [Google Scholar] [CrossRef]
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Yu, E.D.; Faliti, C.E.; Grifoni, A.; Ramirez, S.I.; Haupt, S.; Frazier, A.; et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021, 371, eabf4063. [Google Scholar] [CrossRef]
- Rodda, L.B.; Netland, J.; Shehata, L.; Pruner, K.B.; Morawski, P.A.; Thouvenel, C.D.; Takehara, K.K.; Eggenberger, J.; Hemann, E.A.; Waterman, H.R.; et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell 2021, 184, 169–183.e17. [Google Scholar] [CrossRef]
- Bilich, T.; Nelde, A.; Heitmann, J.S.; Maringer, Y.; Roerden, M.; Bauer, J.; Rieth, J.; Wacker, M.; Peter, A.; Hörber, S.; et al. T cell and antibody kinetics delineate SARS-CoV-2 peptides mediating long-term immune responses in COVID-19 convalescent individuals. Sci. Transl. Med. 2021, 13, eabf7517. [Google Scholar] [CrossRef]
- Koutsakos, M.; Rowntree, L.C.; Hensen, L.; Chua, B.Y.; van de Sandt, C.E.; Habel, J.R.; Zhang, W.; Jia, X.; Kedzierski, L.; Kedzierski, T.M.; et al. Integrated immune dynamics define correlates of COVID-19 severity and antibody responses. Cell Rep. Med. 2021, 2, 100208. [Google Scholar] [CrossRef]
- Rydyznski Moderbacher, C.; Ramirez, S.I.; Dan, J.M.; Grifoni, A.; Hastie, K.M.; Weiskopf, D.; Belanger, S.; Abbott, R.K.; Kim, C.; Choi, J.; et al. Antigen-Specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 2020, 183, 996–1012.e19. [Google Scholar] [CrossRef]
- Yewdell, J.W. Antigenic drift: Understanding COVID-19. Immunity 2021, 54, 2681–2687. [Google Scholar] [CrossRef] [PubMed]
- Dejnirattisai, W.; Zhou, D.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.E.; Tuekprakhon, A.; Nutalai, R.; et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell 2021, 184, 2939–2954.e9. [Google Scholar] [CrossRef] [PubMed]
- Kundu, R.; Narean, J.S.; Wang, L.; Fenn, J.; Pillay, T.; Fernandez, N.D.; Conibear, E.; Koycheva, A.; Davies, M.; Tolosa-Wright, M.; et al. Cross-Reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat. Commun. 2022, 13, 80. [Google Scholar] [CrossRef] [PubMed]
- Croft, N.P.; Smith, S.A.; Pickering, J.; Sidney, J.; Peters, B.; Faridi, P.; Witney, M.J.; Sebastian, P.; Flesch, I.E.A.; Heading, S.L.; et al. Most viral peptides displayed by class I MHC on infected cells are immunogenic. Proc. Natl. Acad. Sci. USA 2019, 116, 3112–3117. [Google Scholar] [CrossRef]
- Yerly, D.; Heckerman, D.; Allen, T.M.; Chisholm, J.V., III; Faircloth, K.; Linde, C.H.; Frahm, N.; Timm, J.; Pichler, W.J.; Cerny, A.; et al. Increased cytotoxic T-lymphocyte epitope variant cross-recognition and functional avidity are associated with hepatitis C virus clearance. J. Virol. 2008, 82, 3147–3153. [Google Scholar] [CrossRef]
- Hensen, L.; Illing, P.T.; Rowntree, L.C.; Davies, J.; Miller, A.; Tong, S.Y.C.; Habel, J.R.; van de Sandt, C.E.; Flanagan, K.L.; Purcell, A.W.; et al. T cell epitope discovery in the context of distinct and unique indigenous HLA profiles. Front. Immunol. 2022, 13, 812393. [Google Scholar] [CrossRef]
- Weingarten-Gabbay, S.; Klaeger, S.; Sarkizova, S.; Pearlman, L.R.; Chen, D.Y.; Gallagher, K.M.E.; Bauer, M.R.; Taylor, H.B.; Dunn, W.A.; Tarr, C.; et al. Profiling SARS-CoV-2 HLA-I peptidome reveals T cell epitopes from out-of-frame ORFs. Cell 2021, 184, 3962–3980.e17. [Google Scholar] [CrossRef]
- Campbell, K.M.; Steiner, G.; Wells, D.K.; Ribas, A.; Kalbasi, A. Prioritization of SARS-CoV-2 epitopes using a pan-HLA and global population inference approach. bioRxiv 2020. Preprint. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef]
- Grifoni, A.; Sidney, J.; Zhang, Y.; Scheuermann, R.H.; Peters, B.; Sette, A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe 2020, 27, 671–680.e2. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.; Joshi, B.C.; Mannan, M.A.; Kaushik, V. Epitope based vaccine prediction for SARS-CoV-2 by deploying immuno-informatics approach. Inform. Med. Unlocked 2020, 19, 100338. [Google Scholar] [CrossRef]
- Kiyotani, K.; Toyoshima, Y.; Nemoto, K.; Nakamura, Y. Bioinformatic prediction of potential T cell epitopes for SARS-CoV-2. J. Hum. Genet. 2020, 65, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Ting, S.; Yufei, H.; Wendong, L.; Yubo, F.; Jing, Z. Epitope based peptide vaccines predicted against novel coronavirus disease caused by SARS-CoV-2. Virus Res. 2020, 288, 198082. [Google Scholar] [CrossRef]
- Nelde, A.; Bilich, T.; Heitmann, J.S.; Maringer, Y.; Salih, H.R.; Roerden, M.; Lubke, M.; Bauer, J.; Rieth, J.; Wacker, M.; et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021, 22, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Poran, A.; Harjanto, D.; Malloy, M.; Arieta, C.M.; Rothenberg, D.A.; Lenkala, D.; van Buuren, M.M.; Addona, T.A.; Rooney, M.S.; Srinivasan, L.; et al. Sequence-Based prediction of SARS-CoV-2 vaccine targets using a mass spectrometry-based bioinformatics predictor identifies immunogenic T cell epitopes. Genome Med. 2020, 12, 70. [Google Scholar] [CrossRef]
- Sekine, T.; Perez-Potti, A.; Rivera-Ballesteros, O.; Stralin, K.; Gorin, J.B.; Olsson, A.; Llewellyn-Lacey, S.; Kamal, H.; Bogdanovic, G.; Muschiol, S.; et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 2020, 183, 158–168.e14. [Google Scholar] [CrossRef] [PubMed]
- Woldemeskel, B.A.; Kwaa, A.K.; Garliss, C.C.; Laeyendecker, O.; Ray, S.C.; Blankson, J.N. Healthy donor T-cell responses to common cold coronaviruses and SARS-CoV-2. J. Clin. Investig. 2020, 130, 6631–6638. [Google Scholar] [CrossRef]
- Zhang, F.; Gan, R.; Zhen, Z.; Hu, X.; Li, X.; Zhou, F.; Liu, Y.; Chen, C.; Xie, S.; Zhang, B.; et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal. Transduct. Target. Ther. 2020, 5, 156. [Google Scholar] [CrossRef]
- Neefjes, J.; Jongsma, M.L.M.; Paul, P.; Bakke, O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011, 11, 823–836. [Google Scholar] [CrossRef]
- Sarkizova, S.; Klaeger, S.; Le, P.M.; Li, L.W.; Oliveira, G.; Keshishian, H.; Hartigan, C.R.; Zhang, W.; Braun, D.A.; Ligon, K.L.; et al. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat. Biotechnol. 2020, 38, 199–209. [Google Scholar] [CrossRef]
- Hansen, T.H.; Bouvier, M. MHC class I antigen presentation: Learning from viral evasion strategies. Nat. Rev. Immunol. 2009, 9, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Croft, N.P.; Smith, S.A.; Wong, Y.C.; Tan, C.T.; Dudek, N.L.; Flesch, I.E.A.; Lin, L.C.W.; Tscharke, D.C.; Purcell, A.W. Kinetics of antigen expression and epitope presentation during virus infection. PLoS Pathog. 2013, 9, e1003129. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Guan, J.; Handel, A.; Tscharke, D.C.; Sidney, J.; Sette, A.; Wakim, L.M.; Sng, X.Y.X.; Thomas, P.G.; Croft, N.P.; et al. Quantification of epitope abundance reveals the effect of direct and cross-presentation on influenza CTL responses. Nat. Commun. 2019, 10, 2846. [Google Scholar] [CrossRef]
- Nagler, A.; Kalaora, S.; Barbolin, C.; Gangaev, A.; Ketelaars, S.L.C.; Alon, M.; Pai, J.; Benedek, G.; Yahalom-Ronen, Y.; Erez, N.; et al. Identification of presented SARS-CoV-2 HLA class I and HLA class II peptides using HLA peptidomics. Cell Rep. 2021, 35, 109305. [Google Scholar] [CrossRef]
- Parker, R.; Partridge, T.; Wormald, C.; Kawahara, R.; Stalls, V.; Aggelakopoulou, M.; Parker, J.; Doherty, R.P.; Morejon, Y.A.; Lee, E.; et al. Mapping the SARS-CoV-2 spike glycoprotein-derived peptidome presented by HLA class II on dendritic cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Knierman, M.D.; Lannan, M.B.; Spindler, L.J.; McMillian, C.L.; Konrad, R.J.; Siegel, R.W. The human leukocyte antigen class II immunopeptidome of the SARS-CoV-2 spike glycoprotein. Cell Rep. 2020, 33, 108454. [Google Scholar] [CrossRef]
- Moss, P. The T cell immune response against SARS-CoV-2. Nat. Immunol. 2022, 23, 186–193. [Google Scholar] [CrossRef]
- Yan, L.-N.; Liu, P.-P.; Li, X.-G.; Zhou, S.-J.; Li, H.; Wang, Z.-Y.; Shen, F.; Lu, B.-C.; Long, Y.; Xiao, X.; et al. Neutralizing antibodies and cellular immune responses against SARS-CoV-2 sustained one and a half years after natural infection. Front. Microbiol. 2022, 12, 803031. [Google Scholar] [CrossRef]
- Dos Santos, W.G. Natural history of COVID-19 and current knowledge on treatment therapeutic options. BioMed. Pharmacother. 2020, 129, 110493. [Google Scholar] [CrossRef]
- Lin, L.; Lu, L.; Cao, W.; Li, T. Hypothesis for potential pathogenesis of SARSCoV-2 infection-a Review of immune changes in patients with viral Pneumonia. Emerg. Microbes. Infect. 2020, 9, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.H.; Li, Y.; Peng, M.W.; Kong, D.G.; Yang, X.B.; Wang, L.; Liu, M.-Q. SARS-CoV-2 detection in patients with influenza-like illness. Nat. Microbiol. 2020, 5, 675–678. [Google Scholar] [CrossRef]
- Chan, J.F.; Yuan, S.; Kok, K.H.; To, K.K.; Chu, H.; Yang, J.; Xing, F.; BNurs, J.L.; Yip, C.C.-Y.; Poon, R.W.-S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person to-person transmission: A study of a family cluster. Lancet. 2020, 395, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- National Health Commission of the People sRoC. COVID-19’s diagnosis and treatment plan (trial eighth edition). Infect. Dis. Immun. 2020, 1, E1. [Google Scholar] [CrossRef]
- Zhou, X.; Ye, Q. Cellular immune response to COVID-19 and potential immune modulators. Front. Immunol. 2021, 12, 646333. [Google Scholar] [CrossRef]
- Deng, Z.; Zhang, M.; Zhu, T.; Zhili, N.; Liu, Z.; Xiang, R.; Zhang, W.; Xu, Y. Dynamic changes in peripheral blood lymphocyte subsets in adult patients with COVID-19. Int. J. Infect. Dis. 2020, 98, 353–358. [Google Scholar] [CrossRef]
- Schulte-Schrepping, J.; Reusch, N.; Paclik, D.; Bassler, K.; Schlickeiser, S.; Zhang, B.; Krämer, B.; Krammer, T.; Brumhard, S.; Bonaguro, L.; et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 2020, 182, 1419–1440.e23. [Google Scholar] [CrossRef]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of immune response in patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020, 71, 762–768. [Google Scholar] [CrossRef]
- Lee, J.S.; Park, S.; Jeong, H.W.; Ahn, J.Y.; Choi, S.J.; Lee, H.; Choi, B.; Nam, S.K.; Sa, M.; Kwon, J.-S.; et al. Immunophenotyping of COVID-19 and Influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020, 5, eabd1554. [Google Scholar] [CrossRef]
- Kuri-Cervantes, L.; Pampena, M.B.; Meng, W.; Rosenfeld, A.M.; Ittner, C.A.G.; Weisman, A.R.; Agyekum, R.S.; Mathew, D.; Baxter, A.E.; Vella, L.A.; et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020, 5, eabd7114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.D.; Hodeib, S.; Korol, C.; et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020, 370, eabd4570. [Google Scholar] [CrossRef] [PubMed]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Front. Immunol. 2020, 11, 827. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Ye, F.; Cheng, M.L.; Feng, Y.; Deng, Y.Q.; Zhao, H.; Wei, P.; Ge, J.; Gou, M.; Li, X.; et al. Detection of SARSCoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity 2020, 52, 971–977.e3. [Google Scholar] [CrossRef]
- Li, C.K.; Wu, H.; Yan, H.; Ma, S.; Wang, L.; Zhang, M.; Tang, X.; Temperton, N.J.; Weiss, R.A.; Brenchley, J.M.; et al. T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008, 181, 5490–5500. [Google Scholar] [CrossRef]
- Wilk, A.J.; Rustagi, A.; Zhao, N.Q.; Roque, J.; Martinez-Colon, G.J.; McKechnie, J.L.; Ivison, G.T.; Ranganath, T.; Vergara, R.; Hollis, T.; et al. A single-cell Atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020, 26, 1070–1076. [Google Scholar] [CrossRef]
- Mitchison, N.A. T-cell–B-cell cooperation. Nat. Rev. Immunol. 2004, 4, 308–312. [Google Scholar] [CrossRef]
- Ferretti, A.P.; Kula, T.; Wang, Y.; Nguyen, D.M.V.; Weinheimer, A.; Dunlap, G.S.; Xu, Q.; Nabilsi, N.; Perullo, C.R.; Cristofaro, A.W.; et al. Unbiased screens show CD8+ T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike protein. Immunity 2020, 53, 1095–1107.e3. [Google Scholar] [CrossRef]
- Vabret, N.; Britton, G.J.; Gruber, C.; Hegde, S.; Kim, J.; Kuksin, M.; Levantovsky, R.; Malle, L.; Moreira, A.; Park, M.D.; et al. Immunology of COVID-19: Current state of the science. Immunity 2020, 52, 910–941. [Google Scholar] [CrossRef]
- Peng, H.; Yang, L.T.; Wang, L.Y.; Li, J.; Huang, J.; Lu, Z.Q.; Koup, R.A.; Bailer, R.T.; Wu, C.Y. Long-lived memory T lymphocyte responses against SARS coronavirus nucleocapsid protein in SARS-recovered patients. Virology 2006, 351, 466–475. [Google Scholar] [CrossRef]
- Tang, F.; Quan, Y.; Xin, Z.T.; Wrammert, J.; Ma, M.J.; Lv, H.; Wang, T.B.; Yang, H.; Richardus, J.H.; Liu, W.; et al. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: A six-year follow-up study. J. Immunol. 2011, 186, 7264–7268. [Google Scholar] [CrossRef] [PubMed]
- Seow, J.; Graham, C.; Merrick, B.; Acors, S.; Pickering, S.; Steel, K.J.A.; Hemmings, O.; O’Byrne, A.; Kouphou, N.; Galao, R.P.; et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020, 5, 1598–1607. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhao, J.; Perlman, S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J. Virol. 2010, 84, 9318–9325. [Google Scholar] [CrossRef] [PubMed]
- Channappanavar, R.; Fett, C.; Zhao, J.; Meyerholz, D.K.; Perlman, S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J. Virol. 2014, 88, 11034–11044. [Google Scholar] [CrossRef] [PubMed]
- Barker, D.J.; Maccari, G.; Georgiou, X.; Cooper, M.A.; Flicek, P.; Robinson, J.; Marsh, S.G.E. The IPD-IMGT/HLA Database. Nucleic Acids Res. 2023, 51, D1053–D1060. [Google Scholar] [CrossRef] [PubMed]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef]
- Li, C.; Revote, J.; Ramarathinam, S.H.; Chung, S.Z.; Croft, N.P.; Scull, K.E.; Huang, Z.; Ayala, R.; Braun, A.; Mifsud, N.A.; et al. Resourcing, annotating, and analysing synthetic peptides of SARS-CoV-2 for immunopeptidomics and other immunological studies. Proteomics 2021, 21, e2100036. [Google Scholar] [CrossRef]
- Chen, R.; Fulton, K.M.; Tran, A.; Duque, D.; Kovalchik, K.; Caron, E.; Twine, S.M.; Li, J. Integrated immunopeptidomics and proteomics study reveals imbalanced innate and adaptive immune responses to SARS-CoV-2 infection. bioRxiv 2022. [Google Scholar] [CrossRef]
- Nguyen, A.; David, J.K.; Maden, S.K.; Wood, M.A.; Weeder, B.R.; Nellore, A.; Thompson, R.F. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J. Virol. 2020, 94, e00510-20. [Google Scholar] [CrossRef]
- Saini, S.K.; Hersby, D.S.; Tamhane, T.; Povlsen, H.R.; Amaya Hernandez, S.P.; Nielsen, M.; Gang, A.O.; Hadrup, S.R. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8+ T cell activation in COVID-19 patients. Sci. Immunol. 2021, 6, eabf7550. [Google Scholar] [CrossRef]
- Tarke, A.; Sidney, J.; Kidd, C.K.; Dan, J.M.; Ramirez, S.I.; Yu, E.D.; Mateus, J.; da Silva Antunes, R.; Moore, E.; Rubiro, P.; et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021, 2, 100204. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.; Lundegaard, C.; Worning, P.; Lauemøller, S.L.; Lamberth, K.; Buus, S.; Brunak, S.; Lund, O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003, 12, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Andreatta, M.; Nielsen, M. Gapped sequence alignment using artificial neural networks: Application to the MHC class I system. Bioinformatics 2016, 32, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Becerra-Artiles, A.; Cruz, J.; Leszyk, J.D.; Sidney, J.; Sette, A.; Shaffer, S.A.; Stern, L.J. Naturally processed HLA-DR3-restricted HHV-6B peptides are recognized broadly with polyfunctional and cytotoxic CD4 T-cell responses. Eur. J. Immunol. 2019, 49, 1167–1185. [Google Scholar] [CrossRef] [PubMed]
- Bettencourt, P.; Müller, J.; Nicastri, A.; Cantillon, D.; Madhavan, M.; Charles, P.D.; Fotso, C.B.; Wittenberg, R.; Bull, N.; Pinpathomrat, N.; et al. Identification of antigens presented by MHC for vaccines against tuberculosis. NPJ Vaccines 2020, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Bourgonje, A.R.; Abdulle, A.E.; Timens, W.; Hillebrands, J.L.; Navis, G.J.; Gordijn, S.J.; Bolling, M.C.; Dijkstra, G.; Voors, A.A.; Osterhaus, A.D.; et al. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19). J. Pathol. 2020, 251, 228–248. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, W.; Chen, Z.; Lu, S.; Yang, F.; Bi, Z.; Bao, L.; Mo, F.; Li, X.; Huang, Y.; et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 2020, 586, 572–577. [Google Scholar] [CrossRef]
- Kim, D.; Lee, J.Y.; Yang, J.S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e10. [Google Scholar] [CrossRef]
- Finkel, Y.; Mizrahi, O.; Nachshon, A.; Weingarten-Gabbay, S.; Morgenstern, D.; Yahalom-Ronen, Y.; Tamir, H.; Achdout, H.; Stein, D.; Israeli, O.; et al. The coding capacity of SARS-CoV-2. Nature 2021, 589, 125–130. [Google Scholar] [CrossRef]
- Su, Y.; Chen, D.; Yuan, D.; Lausted, C.; Choi, J.; Dai, C.L.; Voillet, V.; Duvvuri, V.R.; Scherler, K.; Troisch, P.; et al. ISB-Swedish COVID-19 Biobanking Unit. Multi-Omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell 2020, 183, 1479–1495.e20. [Google Scholar] [CrossRef]
- Hong, S.; Zhang, Z.; Liu, H.; Tian, M.; Zhu, X.; Zhang, Z.; Wang, W.; Zhou, X.; Zhang, F.; Ge, Q.; et al. B cells are the dominant antigen-presenting cells that activate naive CD4+ T cells upon immunization with a virus-derived nanoparticle antigen. Immunity 2018, 49, 695–708.e4. [Google Scholar] [CrossRef] [PubMed]
- Pontelli, M.C.; Castro, I.A.; Martins, R.B.; Veras, F.P.; Serra, L.; Nascimento, D.C.; Cardoso, R.S.; Rosales, R.; Lima, T.M.; Souza, J.P.; et al. Infection of human lymphomononuclear cells by SARS-CoV-2. bioRxiv 2020. Preprint Update in J. Mol. Cell. Biol. 2022, 14. [Google Scholar] [CrossRef]
- Saveanu, L.; Carroll, O.; Hassainya, Y.; van Endert, P. Complexity, contradictions, and conundrums: Studying post-proteasomal proteolysis in HLA class I antigen presentation. Immunol. Rev. 2005, 207, 42–59. [Google Scholar] [CrossRef] [PubMed]
- Saulle, I.; Vicentini, C.; Clerici, M.; Biasin, M. An overview on ERAP roles in infectious diseases. Cells 2020, 9, 720. [Google Scholar] [CrossRef] [PubMed]
- Saulle, I.; Vicentini, C.; Clerici, M.; Biasin, M. Antigen presentation in SARS-CoV-2 infection: The role of class I HLA and ERAP polymorphisms. Hum. Immunol. 2021, 82, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, G.; Samiotaki, M.; Temponeras, I.; Panayotou, G.; Stratikos, E. Allotypic variation in antigen processing controls antigenic peptide generation from SARS-CoV-2 S1 spike glycoprotein. J. Biol. Chem. 2021, 297, 101329. [Google Scholar] [CrossRef]
- D’Amico, S.; Tempora, P.; Lucarini, V.; Melaiu, O.; Gaspari, S.; Algeri, M.; Fruci, D. ERAP1 and ERAP2 Enzymes: A Protective Shield for RAS against COVID-19? Int. J. Mol. Sci. 2021, 22, 1705. [Google Scholar] [CrossRef]
- Becerra-Artiles, A.; Nanaware, P.P.; Muneeruddin, K.; Weaver, G.C.; Shaffer, S.A.; Calvo-Calle, J.M.; Stern, L.J. Immunopeptidome profiling of human coronavirus OC43-infected cells identifies CD4 T cell epitopes specific to seasonal coronaviruses or cross-reactive with SARS-CoV-2. bioRxiv 2022. Preprint. [Google Scholar] [CrossRef]
- Liu, H.; Wilson, I.A. Protective neutralizing epitopes in SARS-CoV-2. Immunol. Rev. 2022, 310, 76–92. [Google Scholar] [CrossRef]
- Shapiro, I.E.; Bassani-Sternberg, M. The impact of immunopeptidomics: From basic research to clinical implementation. Semin. Immunol. 2023, 66, 101727. [Google Scholar] [CrossRef]
- Schroeder, S.M.; Nelde, A.; Walz, J.S. Viral T-cell epitopes—Identification, characterization and clinical application. Semin Immunol. 2023, 66, 101725. [Google Scholar] [CrossRef] [PubMed]
| HLA Allele | Peptide of SARS-CoV-2 | Parent Protein | Ref. |
|---|---|---|---|
| HLA-B15:03 | PRWYFYYLGTGP WSFNPETN QPPGTGKSH and VYTACSHAAVDALCEKA | Nucleocapsid Membrane ORF1ab | [70] |
| A01:01 B07:02 | TTDPSFLGRY SPRWYFYYL | ORF1 Nucleaocapside | [71] |
| B1*01:01, B1*03:01 B1*04:01, B1*04:05 B1*07:01, B1*08:02 B1*09:01, B1*11:01 B1*12:01, B1*13:02 B1*15:01, B3*01:01 B3*02:02, B4*01:01 B5*01:01, A1*01:03 A1*02:01, A1*03:01 A1*01:01, A1*01:02 A1*04:01, A1*05:01 | 1925 peptides | Non-structural Proteins (nsp1- nsp16) Spike ORF3a Envelope Membrane ORF6 ORF7a ORF8 Nucleaocapside ORF10 | [72] |
| A01:01, A02:01 A03:01, A11:01 A24:02, A26:01 A29:02, A30:01 A32:01, A68:01 B07:02, B08:01 B15:01, B35:01 B40:01, B44:02 B44:03, B51:01 | 5600 predicted binding epitopes | Non-structural Proteins (nsp1- nsp16) Spike ORF3a Envelope Membrane ORF6 ORF7a ORF8 Nucleocapsid ORF10 | [72] |
| 1022 HLA-A, -B, and -C | 1103 peptides | ORF1ab Spike Nucleocapsid ORF3a Membrane ORF8 ORF7a Envelope ORF6 ORF10 ORF7b | [19] |
| B1*04:01 | IRGWIFGTTLDSKTQSLL CTFEYVSQPFLMD QPFLMDLEGKQGN TRFQTLLALHRSYLTPGD SSSGW KSFTVEKGIYQTSNFRVQ YLYRLFRKSNLKPFERDI KPFERDISTEIYQ QSIIAYTMSLGAENSVAY VKQIYKTPPIKDFGGFNF DSLSSTASALGKLQDVV QLIRAAEIRASANLAATK HWFVTQRNFYEPQII | Spike Spike Spike Spike Spike Spike Spike Spike Spike Spike Spike Spike | [21] |
| B1*07:01 | KSFTVEKGIYQTSNFRVQ SASFSTFKCYGVSPTKL QSIIAYTMSLGAENSVAY | Spike Spike Spike | [21] |
| A*02:01 | KLPDDFTGCV SIIAYTMSL ALNTLVKQL VLNDILSRL LITGRLQSL RLNEVAKNL NLNESLIDL FIAGLIAIV TLACFVLAAV GLMWLSYFI ALNTPKDHI LQLPQGTTL LALLLLDRL LLLDRLNQL RLNQLESKM GMSRIGMEV CLEASFNYL WLMWLIINL ILLLDQALV SACVLAAEC SLPGVFCGV TLMNVLTLV SMWALIISV | Spike Spike Spike Spike Spike Spike Spike Spike Membrane Membrane Nucleocapsid Nucleocapsid Nucleocapsid Nucleocapsid Nucleocapsid Nucleocapsid ORF1ab ORF1ab ORF1ab ORF1ab ORF1ab ORF1ab ORF1ab | [21] |
| HLA-I | HLRIAGHHL TKAYNVTQAF | Membrane Nucleocapsid | [21] |
| HLA-II | NLDSKVGGNYNYLYRLFR | Spike | [21] |
| B58:01 | RIFTIGTVTLKQGEI TVTLKQGEI | ORF3a ORF3a | [21] |
| B40:01 | GDAALALLLL MEVTPSGTWL | Nucleocapsid Nucleocapsid | [21] |
| A24:02 | NFKDQVILL | Nucleocapsid | [21] |
| HLA-A02:01 | KLDDKDPNF FGDDTVIEV FLAFVVFL KLNDLCFTNV FLFLTWICL | Nucleocapsid ORF1ab Envelope Spike Membrane | [26] |
| A02:01 | KLWAQCVQL YLFDESGEFKL LLYDANYFL ALWEIQQVV YLQPRTFLL LLLDRLNQL | ORF1ab ORF1ab ORF3a ORF1ab Spike Nucleocapsid | [59] |
| A01:01 | GTDLEGNFY NTCDGTTFTY CTDDNALAYY TTDPSFLGRY PTDNYITTY FTSDYYQLY ATSRTLSYY DTDFVNEFY | ORF1ab ORF1ab ORF1ab ORF1ab ORF1ab ORF3a Membrane ORF1ab | [59] |
| A03:01 | KTIQPRVEK VTNNTFTLK KCYGVSPTK KTFPPTEPK | ORF1ab ORF1ab Spike Nucleocapsid | [59] |
| A11:01 | VTDTPKGPK ATSRTLSYYK ASAFFGMSR ATEGALNTPK KTF KTFPPTEPK | ORF1ab Membrane Nucleocapsid Nucleocapsid Nucleocapsid | [59] |
| A24:02 | VYIGDPAQL VYFLQSINF QYIKWPWYI | ORF1ab ORF3a Spike | [59] |
| B07:02 | IPRRNVATL RPDTRYVL SPRWYFYYL | ORF1ab ORF1ab Nucleocapsid | [59] |
| HLA-I Allele | Peptide of SARS-CoV-2 | Parent Protein | Ref. |
|---|---|---|---|
| A02:01 | GPMVLRGLIT GLITLSYHL MLLGSMLYM LEDKAFQL DEFVVVTV SLEDKAFQL KAFQLTPIAV ELPDEFVVV ELPDEFVVVTV | Out-of-frame S.iORF1/2 (also known as ORF2b) Out-of-frame ORF9b | [18] |
| A02:01 | YLNSTNVTI STSAFVETV FGDDTVIEV FASEAARVV | nsp3 nsp2 nsp3 nsp2 | [18] |
| A24:02, A02:05, A68:01 | APHGHVMVEL EIKESVQTF LATNNLVVM EEFEPSTQYEY SEFSSLPSY FAVDAAKAY KRVDWTIEY VATSRTLSY IRQEEVQEL APRITFGGP EILDITPCSF EILDITPCSFG HADQLTPTW KNIDGYFKIY NATNVVIKV QLTPTWRVY VGYLQPRTF | nsp1 nsp2 nsp2 nsp3 nsp8 nsp10 nsp14 Membrane ORF7a Nucleocapside Spike Spike Spike Spike Spike Spike Spike | [18] |
| B07:02 A02:01 and C07:02 | GPMVLRGLIT SLEDKAFQL | ORF S.iORF1/2 ORF9b | [36] |
| B40:01 A68:01 A68:01 A24:02 A68:01 C15:02 B07:02 A03:01 A68:01 A68:01 | NEVAKNLNESL TGSNVFQTR STTTNIVTR YYTSNPTTF FTIGTVTLK HSSGVTREL APRITFGGP RITFGGPSD NAPRITFGGP ITFGGPSDSTGSNQNGER | Spike Spike nsp3 nsp3 ORF3a nsp1 Nucleocapside Nucleocapside Nucleocapside Nucleocapside | [36] |
| HLA-II Allele | Peptide of SARS-CoV-2 | Parent Protein | Ref. |
|---|---|---|---|
| A11:01 | KDGIIWVATEGALN | Nucleocapsid | [36] |
| DRB1*01:02 | VYSRVKNLNSSRVPD | Envelope | [36] |
| DRB1*01:02 | YYKLGASQRVAGDS SYYKLGASQRVAGDSG SYYKLGASQRVAGDS SYYKLGASQRVAG SYYKLGASQRVA LSYYKLGASQRVAGDSG LSYYKLGASQRVAGDS LSYYKLGASQRVAGD LSYYKLGASQRVAG LSYYKLGASQRVA TLSYYKLGASQRVAGDSG TLSYYKLGASQRVAGDS TLSYYKLGASQRVAGD TLSYYKLGASQRVAG TLSYYKLGASQRVA RTLSYYKLGASQRVAGDSG RTLSYYKLGASQRVAGDS RTLSYYKLGASQRVAGD RTLSYYKLGASQRVAG RTLSYYKLGASQRVA | Membrane | [36] |
| DRB1*01:02 | TKAYNVTQAFGRRGPE TKAYNVTQAFGRRGP ATKAYNVTQAFGRRGPE ATKAYNVTQAFGRRGP TATKAYNVTQAFGRRGPEQ TATKAYNVTQAFGRRGPE TATKAYNVTQAFGRRGP KPRQKRTATKAYNVTQAF KPRQKRTATKAYNVTQA | Nucleocapsid | [36] |
| DRB1*01:02 | RDISTEIYQAGSTPCNGVEG DISTEIYQAGSTPCNG DISTEIYQAGSTPC ISTEIYQAGSTPCNG | RBM located within the RBD of spike | [37] |
| B1*03:01 B1*04:01 B1*07:01 B1*07:01 B3*02:02 B4*01:03 B1*04:01 B1*15:01 B3*02:02 B1*04:01 B1*07:01 B1*04:01 B3*01:01 | FTRGVYYPDKVFRS SFTRGVYYPDKVFRSSVLHS PPAYTNSFTRGVYYPD SSVLHSTQDLFLPF TRGVYYPDKVFRSSVLH LLPLVSSQCVNLTT LHSTQDLFLPFFSN LHSTQDLFLPFFSNVT TTLDSKTQSLLIVNNATNVVIKV LGVYYHKNNKSW NIDGYFKIYSKHTPINLVRD SETKCTLKSFTVEKGIYQTS TGTGVLTESNKKFLPFQQFGRDIA | Spike | [37] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abd El-Baky, N.; Amara, A.A.; Redwan, E.M. HLA-I and HLA-II Peptidomes of SARS-CoV-2: A Review. Vaccines 2023, 11, 548. https://doi.org/10.3390/vaccines11030548
Abd El-Baky N, Amara AA, Redwan EM. HLA-I and HLA-II Peptidomes of SARS-CoV-2: A Review. Vaccines. 2023; 11(3):548. https://doi.org/10.3390/vaccines11030548
Chicago/Turabian StyleAbd El-Baky, Nawal, Amro A. Amara, and Elrashdy M. Redwan. 2023. "HLA-I and HLA-II Peptidomes of SARS-CoV-2: A Review" Vaccines 11, no. 3: 548. https://doi.org/10.3390/vaccines11030548
APA StyleAbd El-Baky, N., Amara, A. A., & Redwan, E. M. (2023). HLA-I and HLA-II Peptidomes of SARS-CoV-2: A Review. Vaccines, 11(3), 548. https://doi.org/10.3390/vaccines11030548






