Immune Control of Avian Influenza Virus Infection and Its Vaccine Development
Abstract
:1. Introduction
2. Epidemiology of Avian Influenza Viruses
| Influenza Strain | Year | Location | Clinical Symptoms | References |
|---|---|---|---|---|
| H5N1 | 1997 2003 2008 2003–2017 2015–2019 | China China India Thailand, Indonesia, China, Vietnam, Azerbaijan, Cambodia, Iraq, Egypt, Turkey, Laos, Nigeria, Myanmar, Canada, Bangladesh, Djibouti, Laos Egypt | Flu-like symptoms Pneumonia Influenza-like illness Pneumonia Influenza-like illness | [2,29] [52,53] [54] [53] [55] |
| H5N2 | 2015 | United States | Neurological signs, lethargy | [56] |
| H5N8 | 2014 2021 2020 | China China Russia | Fever Headache, nasal stiffness, cough, fever Asymptomatic | [57] [58] [59,60] |
| H6N1 | 2013 | China | Cough, fever, muscle ache, headache | [35,61] |
| H7N2 | 2002 2007 2016–2017 | Virginia UK USA | Influenza-like illness Conjunctivitis, influenza-like illness Conjunctivitis, sore throat, muscle aches, cough | [45] [38] [62] |
| H7N3 | 2004 2006 2012 | Canada UK Mexico | Conjunctivitis Conjunctivitis Conjunctivitis | [38] |
| H7N7 | 1996 2013 2015 | UK Italy Netherlands | Conjunctivitis Conjunctivitis Conjunctivitis, mild influenza-like illness | [38] [38] [38,63] |
| H7N9 | 2013 | China | Pneumonia-like symptoms | [29,36] |
| H9N2 | 1998 1999 2003 2011 2021 | China China China Bangladesh Cambodia | Symptoms similar to flu Symptoms similar to flu Symptoms similar to flu Symptoms similar to flu Influenza-like illness | [64] [64] [64] [65] [66,67] |
| H10N7 | 2004 2010 | Egypt Australia | Cough and fever Conjunctivitis | [51] [2,68] |
| H10N8 | 2013 | China | Pneumonia | [2,51] |
3. Pathogenesis and Clinical Features of Severe Disease
3.1. Pathogenesis of AIV Illness in Gallinaceous Birds
3.2. Pathogenesis of AIV Illness in Non-Gallinaceous Birds
3.3. Pathogenesis of AIV (H5N1) Infection in People

3.4. Clinical Findings of H5N1 Infection
3.5. Clinical Findings in H7N9 Infection
3.6. Clinical Findings in H5N6 Infection
3.7. Clinical Findings in H7N2 Infection
3.8. Clinical Findings in H7N7 Infection
3.9. Clinical Findings in H7N3 Infection
3.10. Clinical Findings in H3N2 Infection
4. Diagnosis of Avian Influenza Virus
4.1. Serology: A Diagnosis of Influenza Infection can Be Made Retrospectively Using Serologic Assays
4.2. Virus Isolation Test
4.3. Rapid Influenza Diagnostic Tests
4.4. Molecular Influenza Diagnostic Tests
5. Immune Response to Avian Influenza Viruses
5.1. Innate Immunity
5.1.1. Interferons
5.1.2. Antigen-Presenting Cells
5.1.3. Natural Killer Cells
5.1.4. Inducible Antimicrobial Components
5.1.5. Polymorphonuclear Leukocytes
5.1.6. Role of Cytokine Storm in Innate Immunity Response
5.1.7. Role of Toll-Like Receptors (TLRs) in Innate Immunity Response
5.2. Adaptive Immunity
5.2.1. T Cell Immunity against Avian Influenza Virus
| S. No. | Subtype of Avian Influenza Virus | T Cells | Important Remarks | Reference |
|---|---|---|---|---|
| 1 | H5N1 | CD8+ T cells | The importance of the duck cytotoxic T cell response in eradicating H5N1 infection in vivo was suggested through the detection of significantly more CD8+ cells and increased stimulation of cytotoxicity-associated genes, such as granzyme A and IFN, in PBMCs from 5 to 9 days post-infection in mallard duck infection experiments. | [255] |
| 2 | H5N1 | CD4+ T cells, CD8+ T cells | The impact of exacerbated innate immune responses on the impairment of subsequent T cell adaptive immune responses in the lungs was reported. | [257] |
| 3 | H7N9 | CD8+ T cells, γδ-T cells | Considerable increase in CD8+ T cells and γδ-T cells’ percentage and quantity and a considerable overexpression of CD25 expression was observed in the lungs. | [256] |
| 4 | H7N9 | CD38+HLA-DR+ CD8+ T cells | Those who recuperated from avian H7N9 IAV infection were reported to exhibit strong IFN-γ+CD8+ T cell responses, whereas those who died from infection had some or no IFN-γ-producing cells and showed protracted activation of exhausted PD-1-expressing CD38+HLA-DR+ CD8+ T cells. | [254] |
| 5 | H5N6 | CD8+ T cells, CD45RA+ CCR7- T cells | A thorough examination of the IFN-γ+ T cell response in the survivor revealed a preference for CD8+ T-cell-mediated immunity and an increase in virus-specific effector T cells (CD45RA+ CCR7-) between 10–18 days following the beginning of symptoms. | [31] |
| 6 | H7N9 | CD8+ T cells, CD4+ T cells | High numbers of CD8+ T cells and CD4+ T cells were corresponded with better therapeutic consequences in H7N9 patients. | [235] |
| 7 | H7N9 | CD8+ T cells | Early, strong CD8+ T cell responses that were specific for H7N9 were reported for patients who were released from the hospital within two to three weeks, whereas delayed enagagement of CD8+/CD4+ T cells and antibodies at the same time, which was further delayed by strong NK cell responses, were reported in patients who required lengthy hospital stays. | [241] |
| 8 | H7N9 | CTLs’ response to PBMCs obtained from healthy indigenous people from Australia | Mutations that prevent CTL recognition as well as conserved immunogenic peptides that can trigger potent CTL responses against any human IAV, including the H7N9 virus, were reported. | [236] |
| 9 | H7N9 | CD8+ T cells | It was determined that CD8+ T cells against seasonal influenza viruses have significant cross-reactivity with the new H7N9 virus, in addition to recognizing specific H7N9 variant epitopes. | [242] |
| 10 | H3N2, H1N1 | CD4+ T cells | A correlation between lesser viral shedding and milder disease and preexisting CD4+, but not CD8+, T lymphocytes reacting to influenza internal proteins was reported. | [259] |
5.2.2. Antibody Responses against Avian Influenza Virus
6. Vaccine Development
6.1. The Objective of a Pandemic Influenza Vaccine
6.2. Types of Vaccines
6.2.1. Inactivated Virus Vaccines
6.2.2. Live-Attenuated Influenza Viruses
6.2.3. Vector-Based Vaccines
6.2.4. Universal Influenza Virus Vaccines
6.3. Challenges in the Formulation of AIV Vaccines
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yoon, S.W.; Webby, R.J.; Webster, R.G. Evolution and ecology of influenza a viruses. Curr. Top. Microbiol. Immunol. 2014, 385, 359–375. [Google Scholar] [CrossRef]
- Wang, Z.; Loh, L.; Kedzierski, L.; Kedzierska, K. Avian influenza viruses, inflammation, and CD8+ T cell immunity. Front. Immunol. 2016, 7, 60. [Google Scholar] [CrossRef]
- Verhagen, J.H.; Fouchier, R.A.M.; Lewis, N. Highly pathogenic avian influenza viruses at the wild–domestic bird interface in europe: Future directions for research and surveillance. Viruses 2021, 13, 212. [Google Scholar] [CrossRef]
- Bao, P.; Liu, Y.; Zhang, X.; Fan, H.; Zhao, J.; Mu, M.; Li, H.; Wang, Y.; Ge, H.; Li, S.; et al. Human infection with a reassortment avian influenza A H3N8 virus: An epidemiological investigation study. Nat. Commun. 2022, 13, 6817. [Google Scholar] [CrossRef]
- Koutsakos, M.; Kedzierska, K.; Subbarao, K. Immune Responses to Avian Influenza Viruses. J. Immunol. 2019, 202, 382–391. [Google Scholar] [CrossRef]
- Peiris, J.S.M.; De Jong, M.D.; Guan, Y. Avian influenza virus (H5N1): A threat to human health. Clin. Microbiol. Rev. 2007, 20, 243–267. [Google Scholar] [CrossRef]
- Emergence of a Novel Swine-Origin Influenza A (H1N1) Virus in Humans. N. Engl. J. Med. 2009, 360, 2605–2615. [CrossRef]
- Centers for Disease Control and Prevention (CDC) Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. Morb. Mortal. Wkly. Rep. 2009, 58, 521–524.
- Ye, Y.; Ye, Z.; Yang, L.; Xiang, B.; Zheng, C. Unignorable public health risk of avian influenza virus during COVID-19 pandemic. J. Med. Virol. 2022, 94, 4058–4060. [Google Scholar] [CrossRef]
- de Vries, R.D.; Herfst, S.; Richard, M. Avian influenza A virus pandemic preparedness and vaccine development. Vaccines 2018, 6, 46. [Google Scholar] [CrossRef]
- Goraya, M.U.; Ali, L.; Younis, I. Innate Immune Responses Against Avian Respiratory Viruses. Hosts Viruses 2017, 4, 78–87. [Google Scholar] [CrossRef]
- Sun, Y.; Qin, K.; Wang, J.; Pu, J.; Tang, Q.; Hu, Y.; Bi, Y.; Zhao, X.; Yang, H.; Shu, Y.; et al. High genetic compatibility and increased pathogenicity of reassortants derived from avian H9N2 and pandemic H1N1/2009 influenza viruses. Proc. Natl. Acad. Sci. USA 2011, 108, 4164–4169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Q.; Kong, H.; Jiang, Y.; Gao, Y.; Deng, G.; Shi, J.; Tian, G.; Liu, L.; Liu, J.; et al. H5N1 hybrid viruses bearing 2009/H1N1 virus genes transmit in guinea pigs by respiratory droplet. Science 2013, 340, 1459–1463. [Google Scholar] [CrossRef] [PubMed]
- Lowen, A.C. Constraints, Drivers, and Implications of Influenza A Virus Reassortment. Annu. Rev. Virol. 2017, 4, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Shoham, D. The Modes of Evolutionary Emergence of Primal and Late Pandemic Influenza Virus Strains from Viral Reservoir in Animals: An Interdisciplinary Analysis. Influenza Res. Treat. 2011, 2011, 861792. [Google Scholar] [CrossRef]
- Taubenberger, J.K.; Morens, D.M. Influenza: The once and future pandemic. Public Health Rep. 2010, 125, 15–26. [Google Scholar] [CrossRef]
- Fauci, A.S. Emerging and re-emerging infectious diseases: Influenza as a prototype of the host-pathogen balancing act. Cell 2006, 124, 665–670. [Google Scholar] [CrossRef]
- Oldstone, M.B.A.; Teijaro, J.R.; Walsh, K.B.; Rosen, H. Dissecting influenza virus pathogenesis uncovers a novel chemical approach to combat the infection. Virology 2013, 435, 92–101. [Google Scholar] [CrossRef]
- Herold, S.; Becker, C.; Ridge, K.M.; Budinger, G.R.S. Influenza virus-induced lung injury: Pathogenesis and implications for treatment. Eur. Respir. J. 2015, 45, 1463–1478. [Google Scholar] [CrossRef]
- Wei, H.; Wang, S.; Chen, Q.; Chen, Y.; Chi, X.; Zhang, L.; Huang, S.; Gao, G.F.; Chen, J.L. Suppression of Interferon Lambda Signaling by SOCS-1 Results in Their Excessive Production during Influenza Virus Infection. PLoS Pathog. 2014, 10, e1003845. [Google Scholar] [CrossRef]
- Chen, X.; Liu, S.; Goraya, M.U.; Maarouf, M.; Huang, S.; Chen, J.L. Host immune response to influenza A virus infection. Front. Immunol. 2018, 9, 320. [Google Scholar] [CrossRef]
- Influenza. Available online: https://www.who.int/ (accessed on 25 November 2022).
- Bay, A.; Etlik, Ö.; Öner, A.F.; Ünal, Ö.; Arslan, H.; Bora, A.; Davran, R.; Yuca, S.A.; Dogan, M. Radiological and clinical course of pneumonia in patients with avian influenza H5N1. Eur. J. Radiol. 2007, 61, 245–250. [Google Scholar] [CrossRef]
- Yang, R.; Sun, H.; Gao, F.; Luo, K.; Huang, Z.; Tong, Q.; Song, H.; Han, Q.; Liu, J.; Lan, Y.; et al. Human infection of avian influenza A {H3N8} virus and the viral origins: A descriptive study. Lancet Microbe 2022, 3, e824–e834. [Google Scholar] [CrossRef]
- Trinh, T.T.T.; Duong, B.T.; Nguyen, A.T.V.; Tuong, H.T.; Hoang, V.T.; Than, D.D.; Nam, S.; Sung, H.W.; Yun, K.J.; Yeo, S.J.; et al. Emergence of novel reassortant h1n1 avian influenza viruses in Korean wild ducks in 2018 and 2019. Viruses 2021, 13, 30. [Google Scholar] [CrossRef]
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Prim. 2018, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Shi, J.; Kong, X.; Ma, S.; Zhang, Y.; Yin, X.; He, X.; Liu, L.; Suzuki, Y.; Li, C.; et al. H3N2 avian influenza viruses detected in live poultry markets in China bind to human-type receptors and transmit in guinea pigs and ferrets. Emerg. Microbes Infect. 2019, 8, 1280–1290. [Google Scholar] [CrossRef] [PubMed]
- Sutton, T.C. The pandemic threat of emerging H5 and H7 avian influenza viruses. Viruses 2018, 10, 461. [Google Scholar] [CrossRef]
- WHO. Avian and Other Zoonotic Influenza. 2018. Available online: https://www.who.int/news-room/fact-sheets/detail/influenza-(avian-and-other-zoonotic) (accessed on 25 November 2022).
- Lai, S.; Qin, Y.; Cowling, B.J.; Ren, X.; Wardrop, N.A.; Gilbert, M.; Tsang, T.K.; Wu, P.; Feng, L.; Jiang, H.; et al. Global epidemiology of avian influenza A H5N1 virus infection in humans, 1997-2015: A systematic review of individual case data. Lancet Infect. Dis. 2016, 16, e108–e118. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Tan, S.; Yang, Y.; Wong, G.; Zhao, M.; Zhang, Q.; Wang, Q.; Zhao, X.; Li, L.; Yuan, J.; et al. Clinical and Immunological Characteristics of Human Infections With H5N6 Avian Influenza Virus. Clin. Infect. Dis. 2019, 68, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Li, F.C.K.; Choi, B.C.K.; Sly, T.; Pak, A.W.P. Finding the real case-fatality rate of H5N1 avian influenza. J. Epidemiol. Community Health 2008, 62, 555–559. [Google Scholar] [CrossRef]
- Cui, P.; Zeng, X.; Li, X.; Li, Y.; Shi, J.; Zhao, C.; Qu, Z.; Wang, Y.; Guo, J.; Gu, W.; et al. Genetic and biological characteristics of the globally circulating H5N8 avian influenza viruses and the protective efficacy offered by the poultry vaccine currently used in China. Sci. China Life Sci. 2022, 65, 795–808. [Google Scholar] [CrossRef] [PubMed]
- Pyankova, O.G.; Susloparov, I.M.; Moiseeva, A.A.; Kolosova, N.P.; Onkhonova, G.S.; Danilenko, A.V.; Vakalova, E.V.; Shendo, G.L.; Nekeshina, N.N.; Noskova, L.N.; et al. Isolation of clade 2.3.4.4b A(H5N8), a highly pathogenic avian influenza virus, from a worker during an outbreak on a poultry farm, Russia, December 2020. Eurosurveillance 2021, 26, 2100439. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, F.; Liu, F.; Lu, R.; Peng, X.; Chen, B.; Yao, H.; Wu, N. Isolation and characterization of novel reassortant H6N1 avian influenza viruses from chickens in Eastern China. Virol. J. 2018, 15, 164. [Google Scholar] [CrossRef]
- Wu, X.; Xiao, L.; Li, L. Research progress on human infection with avian influenza H7N9. Front. Med. 2020, 14, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Tan, S.; Zhao, M.; Quan, C.; Bi, Y.; Wu, Y.; Zhang, S.; Zhang, H.; Xiao, H.; Qi, J.; et al. Cross-immunity against avian influenza a (H7N9) virus in the healthy population is affected by antigenicity-dependent substitutions. J. Infect. Dis. 2016, 214, 1937–1946. [Google Scholar] [CrossRef] [PubMed]
- Naguib, M.M.; Verhagen, J.H.; Mostafa, A.; Wille, M.; Li, R.; Graaf, A.; Järhult, J.D.; Ellström, P.; Zohari, S.; Lundkvist, Å.; et al. Global patterns of avian influenza A (H7): Virus evolution and zoonotic threats. FEMS Microbiol. Rev. 2019, 43, 608–621. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, L.; Zhou, M.; Chen, Z.; Li, F.; Wu, H.; Xiang, N.; Chen, E.; Tang, F.; Wang, D.; et al. Epidemiology of Human Infections with Avian Influenza A(H7N9) Virus in China. N. Engl. J. Med. 2014, 370, 520–532. [Google Scholar] [CrossRef]
- Taubenberger, J.K.; Morens, D.M. Influenza revisited. Emerg. Infect. Dis. 2006, 12, 1–2. [Google Scholar] [CrossRef]
- Taubenberger, J.K.; Morens, D.M. 1918 Influenza: The mother of all pandemics. Emerg. Infect. Dis. 2006, 12, 15–22. [Google Scholar] [CrossRef]
- Zhao, M.; Chen, J.; Tan, S.; Dong, T.; Jiang, H.; Zheng, J.; Quan, C.; Liao, Q.; Zhang, H.; Wang, X.; et al. Prolonged Evolution of Virus-Specific Memory T Cell Immunity after Severe Avian Influenza A (H7N9) Virus Infection. J. Virol. 2018, 92, e01024-18. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC) Notes from the field: Highly pathogenic avian influenza A (H7N3) virus infection in two poultry workers--Jalisco, Mexico, July 2012. Morb. Mortal. Wkly. Rep. 2012, 61, 726–727.
- Sekine, W.; Takenaka-Uema, A.; Kamiki, H.; Ishida, H.; Matsugo, H.; Murakami, S.; Horimoto, T. Adaptation of the H7N2 Feline Influenza Virus to Human Respiratory Cell Culture. Viruses 2022, 14, 1091. [Google Scholar] [CrossRef] [PubMed]
- Terebuh, P.; Adija, A.; Edwards, L.; Rowe, T.; Jenkins, S.; Kleene, J.; Fukuda, K.; Katz, J.M.; Bridges, C.B. Human infection with avian influenza A(H7N2) virus—Virginia, 2002. Influenza Other Respi. Viruses 2018, 12, 529–532. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.P.; James, J.; Sealy, J.E.; Iqbal, M. A global perspective on h9n2 avian influenza virus. Viruses 2019, 11, 620. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Lu, L.; Li, J.; Yin, Y.; Zhang, Y.; Gao, H.; Qin, Z.; Zeshan, B.; Liu, J.; Sun, L.; et al. Novel genetic reassortants in H9N2 influenza A viruses and their diverse pathogenicity to mice. Virol. J. 2011, 8, 505. [Google Scholar] [CrossRef]
- Pu, J.; Wang, S.; Yin, Y.; Zhang, G.; Carter, R.A.; Wang, J.; Xu, G.; Sun, H.; Wang, M.; Wen, C.; et al. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc. Natl. Acad. Sci. USA 2015, 112, 548–553. [Google Scholar] [CrossRef]
- Liu, M.; Li, X.; Yuan, H.; Zhou, J.; Wu, J.; Bo, H.; Xia, W.; Xiong, Y.; Yang, L.; Gao, R.; et al. Genetic diversity of avian influenza A (H10N8) virus in live poultry markets and its association with human infections in China. Sci. Rep. 2015, 5, 7632. [Google Scholar] [CrossRef]
- Ma, C.; Lam, T.T.-Y.; Chai, Y.; Wang, J.; Fan, X.; Hong, W.; Zhang, Y.; Li, L.; Liu, Y.; Smith, D.K.; et al. Emergence and Evolution of H10 Subtype Influenza Viruses in Poultry in China. J. Virol. 2015, 89, 3534–3541. [Google Scholar] [CrossRef]
- Guan, M.; Hall, J.S.; Zhang, X.; Dusek, R.J.; Olivier, A.K.; Liu, L.; Li, L.; Krauss, S.; Danner, A.; Li, T.; et al. Aerosol Transmission of Gull-Origin Iceland Subtype H10N7 Influenza A Virus in Ferrets. J. Virol. 2019, 93, e00282-19. [Google Scholar] [CrossRef]
- Horwood, P.F. Avian influenza H5N1: Still a pandemic threat? Microbiol. Aust. 2021, 42, 152–155. [Google Scholar] [CrossRef]
- Hamid, S.; Arima, Y.; Dueger, E.; Konings, F.; Bell, L.; Lee, C.-K.; Luo, D.; Otsu, S.; Olowokure, B.; Li, A.; et al. From H5N1 to HxNy: An epidemiologic overview of human infections with avian influenza in the Western Pacific Region, 2003–2017. West. Pacific Surveill. Response J. 2018, 9, 53. [Google Scholar] [CrossRef]
- Tosh, C.; Nagarajan, S.; Murugkar, H.V.; Jain, R.; Behera, P.; Katare, M.; Kulkarni, D.D.; Dubey, S.C. Phylogenetic evidence of multiple introduction of H5N1 virus in Malda district of West Bengal, India in 2008. Vet. Microbiol. 2011, 148, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, M.R.; El Rifay, A.S.; Zeid, D.A.; Elabd, M.A.; Elabd, E.; Kandeil, A.; Shama, N.M.A.; Kamel, M.N.; Marouf, M.A.; Barakat, A.; et al. Incidence and seroprevalence of avian influenza in a cohort of backyard poultry growers, Egypt, August 2015-March 2019. Emerg. Infect. Dis. 2020, 26, 2129–2136. [Google Scholar] [CrossRef]
- Spackman, E.; Pantin-Jackwood, M.J.; Kapczynski, D.R.; Swayne, D.E.; Suarez, D.L. H5N2 Highly Pathogenic Avian Influenza Viruses from the US 2014-2015 outbreak have an unusually long pre-clinical period in turkeys. BMC Vet. Res. 2016, 12, 260. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhao, X.; Li, X.; Bo, H.; Li, D.; Liu, J.; Wang, D. Case report for human infection with a highly pathogenic avian influenza A(H5N6) virus in Beijing, China 2019. Biosaf. Health 2020, 2, 49–52. [Google Scholar] [CrossRef]
- Xiao, C.; Xu, J.; Lan, Y.; Huang, Z.; Zhou, L.; Guo, Y.; Li, X.; Yang, L.; Gao, G.F.; Wang, D.; et al. Five Independent Cases of Human Infection with Avian Influenza H5N6—Sichuan Province, China, 2021. China CDC Wkly. 2021, 3, 751–756. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Gao, G.F. Emerging H5N8 avian influenza viruses. Science 2021, 372, 784–786. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Threat Assessment Brief: First Identification of Human Cases of Avian Influenza {A(H5N8}) Infection; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2021. [Google Scholar]
- Attaran, H.; Jin, W.; Luo, J.; Wang, C.; He, H.; He, H.; Attaran, H. Isolation and characterizations of a novel H6N1 avian influenza virus from Von Schrenck’s Bittern (Ixobrychus eurhythmus) in Jiangxi province, China. bioRxiv Mol. Biol. 2021, 2021-06. [Google Scholar] [CrossRef]
- Poirot, E.; Levine, M.Z.; Russell, K.; Stewart, R.J.; Pompey, J.M.; Chiu, S.; Fry, A.M.; Gross, L.; Havers, F.P.; Li, Z.N.; et al. Detection of avian influenza A(H7N2) virus infection among animal shelter workers using a novel serological approach-New York City, 2016–2017. J. Infect. Dis. 2019, 219, 1688–1696. [Google Scholar] [CrossRef]
- Uyeki, T.M.; Peiris, M. Novel Avian Influenza A Virus Infections of Humans. Infect. Dis. Clin. N. Am. 2019, 33, 907–932. [Google Scholar] [CrossRef]
- Gu, M.; Xu, L.; Wang, X.; Liu, X. Current situation of H9N2 subtype avian influenza in China. Vet. Res. 2017, 48, 1–10. [Google Scholar] [CrossRef]
- Pusch, E.A.; Suarez, D.L. The multifaceted zoonotic risk of H9N2 avian influenza. Vet. Sci. 2018, 5, 82. [Google Scholar] [CrossRef]
- Schwartz, I.S.; Govender, N.P.; Sigler, L.; Jiang, Y.; Maphanga, T.G.; Toplis, B.; Botha, A.; Dukik, K.; Claire Hoving, J.; Muñoz, J.F.; et al. Emergomyces: The global rise of new dimorphic fungal pathogens. PLoS Pathog. 2019, 15, e1007977. [Google Scholar] [CrossRef]
- Carnaccini, S.; Perez, D.R. H9 influenza viruses: An emerging challenge. Cold Spring Harb. Perspect. Med. 2020, 10, a038588. [Google Scholar] [CrossRef] [PubMed]
- Herfst, S.; Zhang, J.; Richard, M.; McBride, R.; Lexmond, P.; Bestebroer, T.M.; Spronken, M.I.J.; de Meulder, D.; van den Brand, J.M.; Rosu, M.E.; et al. Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals. Cell Host Microbe 2020, 28, 602–613. [Google Scholar] [CrossRef]
- Juno, J.; Fowke, K.R.; Keynan, Y. Immunogenetic factors associated with severe respiratory illness caused by zoonotic H1N1 and H5N1 influenza viruses. Clin. Dev. Immunol. 2012, 2012, 797180. [Google Scholar] [CrossRef]
- Lin, T.Y.; Brass, A.L. Host genetic determinants of influenza pathogenicity. Curr. Opin. Virol. 2013, 3, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Hale, B.G.; Albrecht, R.A.; García-Sastre, A. Innate immune evasion strategies of influenza viruses. Future Microbiol. 2010, 5, 23–41. [Google Scholar] [CrossRef]
- Bosch, F.X.; Orlich, M.; Klenk, H.D.; Rott, R. The structure of the hemagglutinin, a determinant for the pathogenicity of influenza viruses. Virology 1979, 95, 197–207. [Google Scholar] [CrossRef]
- Nguyen, D.C.; Uyeki, T.M.; Jadhao, S.; Maines, T.; Shaw, M.; Matsuoka, Y.; Smith, C.; Rowe, T.; Lu, X.; Hall, H.; et al. Isolation and Characterization of Avian Influenza Viruses, Including Highly Pathogenic H5N1, from Poultry in Live Bird Markets in Hanoi, Vietnam, in 2001. J. Virol. 2005, 79, 4201–4212. [Google Scholar] [CrossRef] [PubMed]
- Klenk, H.D.; Rott, R.; Orlich, M.; Blödorn, J. Activation of influenza A viruses by trypsin treatment. Virology 1975, 68, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Meseko, C.A.; Oluwayelu, D.O. Avian influenza. In Transboundary Animal Diseases in Sahelian Africa and Connected Regions; Springer: Cham, Switzerland, 2019; ISBN 9783030253851. [Google Scholar]
- Stieneke-Grober, A.; Vey, M.; Angliker, H.; Shaw, E.; Thomas, G.; Roberts, C.; Klenk, H.D.; Garten, W. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 1992, 11, 2407–2414. [Google Scholar] [CrossRef]
- Swayne, D.E.; Boulianne, M.; Logue, C.M.; McDougald, L.R.; Nair, V.; Suarez, D.L.; De Wit, S.; Grimes, T.; Johnson, D.; Kromm, M.; et al. Diseases of Poultry, 14th ed.; John and Wiley and Sons: Hoboken, NJ, USA, 2019; ISBN 9781119371199. [Google Scholar]
- Post, J.; Burt, D.W.; Cornelissen, J.B.W.J.; Broks, V.; Van Zoelen, D.; Peeters, B.; Rebel, J.M.J. Systemic virus distribution and host responses in brain and intestine of chickens infected with low pathogenic or high pathogenic avian influenza virus. Virol. J. 2012, 9, 61. [Google Scholar] [CrossRef]
- Pantin-Jackwood, M.J.; Swayne, D.E. Pathogenesis and pathobiology of avian influenza virus infection in birds. OIE Rev. Sci. Tech. 2009, 28, 113–136. [Google Scholar] [CrossRef]
- Swayne, D.E. Understanding the complex pathobiology of high pathogenicity avian influenza viruses in birds. Avian Dis. 2007, 51, 242–249. [Google Scholar] [CrossRef]
- Kuiken, T.; van den Brand, J.; van Riel, D.; Pantin-Jackwood, M.; Swayne, D.E. Comparative pathology of select agent influenza a virus infections. Vet. Pathol. 2010, 47, 893–914. [Google Scholar] [CrossRef]
- Taubenberger, J.K.; Kash, J.C. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 2010, 7, 440–451. [Google Scholar] [CrossRef]
- Leigh Perkins, L.E.; Swayne, D.E. Pathogenicity of a Hong Kong-origin H5N1 highly pathogenic avian influenza virus for emus, geese, ducks, and pigeons. Avian Dis. 2002, 46, 53–63. [Google Scholar] [CrossRef]
- Kim, H.R.; Kwon, Y.K.; Jang, I.; Lee, Y.J.; Kang, H.M.; Lee, E.K.; Song, B.M.; Lee, H.S.; Joo, Y.S.; Lee, K.H.; et al. Pathologic changes in wild birds infected with highly pathogenic avian influenza A(H5N8) viruses, South Korea, 2014. Emerg. Infect. Dis. 2015, 21, 775–780. [Google Scholar] [CrossRef]
- Lee, D.H.; Kwon, J.H.; Noh, J.Y.; Park, J.K.; Yuk, S.S.; Erdene-Ochir, T.O.; Lee, J.B.; Park, S.Y.; Choi, I.S.; Lee, S.W.; et al. Pathogenicity of the Korean H5N8 highly pathogenic avian influenza virus in commercial domestic poultry species. Avian Pathol. 2016, 45, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, S.; Tosh, C.; Smith, D.K.; Peiris, J.S.M.; Murugkar, H.V.; Sridevi, R.; Kumar, M.; Katare, M.; Jain, R.; Syed, Z.; et al. Avian influenza (H5N1) virus of clade 2.3.2 in domestic poultry in India. PLoS ONE 2012, 7, e31844. [Google Scholar] [CrossRef]
- Pantin-Jackwood, M.J.; Costa-Hurtado, M.; Shepherd, E.; DeJesus, E.; Smith, D.; Spackman, E.; Kapczynski, D.R.; Suarez, D.L.; Stallknecht, D.E.; Swayne, D.E. Pathogenicity and Transmission of H5 and H7 Highly Pathogenic Avian Influenza Viruses in Mallards. J. Virol. 2016, 90, 9967–9982. [Google Scholar] [CrossRef] [PubMed]
- Fouchier, R.A.M.; Munster, V.J. Epidemiology of low pathogenic avian influenza viruses in wild birds. OIE Rev. Sci. Tech. 2009, 28, 49–58. [Google Scholar] [CrossRef]
- Pantin-Jackwood, M.J.; Swayne, D.E. Pathobiology of asian highly pathogenic avian influenza H5N1 virus infections in ducks. Avian Dis. 2007, 51, 250–259. [Google Scholar] [CrossRef]
- Gobbo, F.; Zanardello, C.; Bottinelli, M.; Budai, J.; Bruno, F.; De Nardi, R.; Patregnani, T.; Catania, S.; Terregino, C. Silent Infection of Highly Pathogenic Avian Influenza Virus (H5N1) Clade 2.3.4.4b in a Commercial Chicken Broiler Flock in Italy. Viruses 2022, 14, 1600. [Google Scholar] [CrossRef]
- Van Le, T.; Phan, L.T.; Ly, K.H.K.; Nguyen, L.T.; Nguyen, H.T.; Ho, N.T.T.; Trinh, T.X.; Tran Minh, N.N. Fatal avian influenza A(H5N1) infection in a 36-week pregnant woman survived by her newborn in Sóc Trăng Province, Vietnam, 2012. Influenza Other Respi. Viruses 2019, 13, 292–297. [Google Scholar] [CrossRef]
- Peiris, J.S.M.; Yu, W.C.; Leung, C.W.; Cheung, C.Y.; Ng, W.F.; Nicholls, J.M.; Ng, T.K.; Chan, K.H.; Lai, S.T.; Lim, W.L.; et al. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet 2004, 363, 617–619. [Google Scholar] [CrossRef] [PubMed]
- Ungchusak, K.; Auewarakul, P.; Dowell, S.F.; Kitphati, R.; Auwanit, W.; Puthavathana, P.; Uiprasertkul, M.; Boonnak, K.; Pittayawonganon, C.; Cox, N.J.; et al. Probable Person-to-Person Transmission of Avian Influenza A (H5N1). N. Engl. J. Med. 2005, 352, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Ku, A.S.W.; Chan, L.T.W. The first case of H5N1 avian influenza infection in a human with complications of adult respiratory distress syndrome and Reye’s syndrome. J. Paediatr. Child Health 1999, 35, 207–209. [Google Scholar] [CrossRef]
- To, K.F.; Chan, P.K.S.; Chan, K.F.; Lee, W.K.; Lam, W.Y.; Wong, K.F.; Tang, N.L.S.; Tsang, D.N.C.; Sung, R.Y.T.; Buckley, T.A.; et al. Pathology of fatal human infection associated with avian influenza A H5N1 virus. J. Med. Virol. 2001, 63, 242–246. [Google Scholar] [CrossRef] [PubMed]
- De Jong, M.D.; Simmons, C.P.; Thanh, T.T.; Hien, V.M.; Smith, G.J.D.; Chau, T.N.B.; Hoang, D.M.; Chau, N.V.V.; Khanh, T.H.; Dong, V.C.; et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 2006, 12, 1203–1207. [Google Scholar] [CrossRef]
- Zhou, J.; Law, H.K.W.; Cheung, C.Y.; Ng, I.H.Y.; Peiris, J.S.M.; Lau, Y.L. Differential expression of chemokines and their receptors in adult and neonatal macrophages infected with human or avian influenza viruses. J. Infect. Dis. 2006, 194, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Cheung, C.Y.; Poon, L.L.M.; Lau, A.S.; Luk, W.; Lau, Y.L.; Shortridge, K.F.; Gordon, S.; Guan, Y.; Peiris, J.S.M. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: A mechanism for the unusual severity of human disease? Lancet 2002, 360, 1831–1837. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.C.W.; Cheung, C.Y.; Chui, W.H.; Tsao, G.S.W.; Nicholls, J.M.; Chan, Y.O.; Chan, R.W.Y.; Long, H.T.; Poon, L.L.M.; Guan, Y.; et al. Proinflammatory cytokine responses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cells. Respir. Res. 2005, 6, 135. [Google Scholar] [CrossRef]
- Kash, J.C.; Tumpey, T.M.; Proll, S.C.; Carter, V.; Perwitasari, O.; Thomas, M.J.; Basler, C.F.; Palese, P.; Taubenberger, J.K.; García-Sastre, A.; et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 2006, 443, 578–581. [Google Scholar] [CrossRef]
- Zhou, J.; Law, H.K.W.; Cheung, C.Y.; Ng, I.H.Y.; Peiris, J.S.M.; Lau, Y.L. Functional tumor necrosis factor-related apoptosis-inducing ligand production by avian influenza virus-infected macrophages. J. Infect. Dis. 2006, 193, 945–953. [Google Scholar] [CrossRef] [PubMed]
- Mok, C.K.P.; Lee, D.C.W.; Cheung, C.Y.; Peiris, M.; Lau, A.S.Y. Differential onset of apoptosis in influenza A virus H5N1- and H1N1-infected human blood macrophages. J. Gen. Virol. 2007, 88, 1275–1280. [Google Scholar] [CrossRef]
- Uiprasertkul, M.; Kitphati, R.; Puthavathana, P.; Kriwong, R.; Kongchanagul, A.; Ungchusak, K.; Angkasekwinai, S.; Chokephaibulkit, K.; Srisook, K.; Vanprapar, N.; et al. Apoptosis and pathogenesis of avian influenza A (H5N1) virus in humans. Emerg. Infect. Dis. 2007, 13, 708–712. [Google Scholar] [CrossRef]
- Fornek, J.L.; Gillim-Ross, L.; Santos, C.; Carter, V.; Ward, J.M.; Cheng, L.I.; Proll, S.; Katze, M.G.; Subbarao, K. A Single-Amino-Acid Substitution in a Polymerase Protein of an H5N1 Influenza Virus Is Associated with Systemic Infection and Impaired T-Cell Activation in Mice. J. Virol. 2009, 83, 11102–11115. [Google Scholar] [CrossRef]
- Tumpey, T.M.; Lu, X.; Morken, T.; Zaki, S.R.; Katz, J.M. Depletion of Lymphocytes and Diminished Cytokine Production in Mice Infected with a Highly Virulent Influenza A (H5N1) Virus Isolated from Humans. J. Virol. 2000, 74, 6105–6116. [Google Scholar] [CrossRef]
- Boonnak, K.; Vogel, L.; Feldmann, F.; Feldmann, H.; Legge, K.L.; Subbarao, K. Lymphopenia Associated with Highly Virulent H5N1 Virus Infection Due to Plasmacytoid Dendritic Cell–Mediated Apoptosis of T Cells. J. Immunol. 2014, 192, 5906–5912. [Google Scholar] [CrossRef]
- Hsieh, S.-M.; Chang, S.-C. Cutting Edge: Insufficient Perforin Expression in CD8 + T Cells in Response to Hemagglutinin from Avian Influenza (H5N1) Virus. J. Immunol. 2006, 176, 4530–4533. [Google Scholar] [CrossRef]
- Guilliams, M.; Lambrecht, B.N.; Hammad, H. Division of labor between lung dendritic cells and macrophages in the defense against pulmonary infections. Mucosal Immunol. 2013, 6, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Gradil, G. Lethal Case of Viral-Bacterial Pneumonia with Relative Lymphopenia. Retrospective Evaluation. Inter Collegas 2019, 6, 106–111. [Google Scholar] [CrossRef]
- Pawestri, H.A.; Eggink, D.; Isfandari, S.; Thanh, T.T.; Rogier Van Doorn, H.; Setiawaty, V.; De Jong, M.D. Viral Factors Associated with the High Mortality Related to Human Infections with Clade 2.1 Influenza A/H5N1 Virus in Indonesia. Clin. Infect. Dis. 2020, 70, 1139–1146. [Google Scholar] [CrossRef] [PubMed]
- Perricone, C.; Bartoloni, E.; Bursi, R.; Cafaro, G.; Guidelli, G.M.; Shoenfeld, Y.; Gerli, R. COVID-19 as part of the hyperferritinemic syndromes: The role of iron depletion therapy. Immunol. Res. 2020, 68, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Chotpitayasunondh, T.; Ungchusak, K.; Hanshaoworakul, W.; Chunsuthiwat, S.; Sawanpanyalert, P.; Kijphati, R.; Lochindarat, S.; Srisan, P.; Suwan, P.; Osotthanakorn, Y.; et al. Human disease from influenza A (H5N1), Thailand, 2004. Emerg. Infect. Dis. 2005, 11, 201–209. [Google Scholar] [CrossRef]
- Chan, P.K.S. Outbreak of avian influenza A(H5N1) virus infection in Hong Kong in 1997. Clin. Infect. Dis. 2002, 34, S58–S64. [Google Scholar] [CrossRef]
- Tam, J.S. Influenza A (H5N1) in Hong Kong: An overview. Vaccine 2002, 20, S77–S81. [Google Scholar] [CrossRef]
- Yuen, K.Y.; Chan, P.K.S.; Peiris, M.; Tsang, D.N.C.; Que, T.L.; Shortridge, K.; Cheung, P.T.; To, W.K.; Ho, E.T.F.; Sung, R.; et al. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 1998, 351, 467–471. [Google Scholar] [CrossRef]
- Hien, T.T.; Liem, N.T.; Dung, N.T.; San, L.T.; Mai, P.P.; Chau, N.; van, V.; Suu, P.T.; Dong, V.C.; Mai, L.T.Q.; et al. Avian Influenza A (H5N1) in 10 Patients in Vietnam. N. Engl. J. Med. 2004, 350, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhong, H.; Song, T.; He, J.; Guo, L.; Tan, X.; Huang, G.; Kang, M. Epidemiological and clinical characteristics of humans with avian influenza A (H7N9) infection in Guangdong, China, 2013–2017. Int. J. Infect. Dis. 2017, 65, 148–155. [Google Scholar] [CrossRef]
- Guan, W.; Yang, Z.; Wu, N.C.; Lee, H.H.Y.; Li, Y.; Jiang, W.; Shen, L.; Wu, D.C.; Chen, R.; Zhong, N.; et al. Clinical correlations of transcriptional profile in patients infected with avian influenza H7N9 virus. J. Infect. Dis. 2018, 218, 1238–1248. [Google Scholar] [CrossRef]
- Philippon, D.A.M.; Wu, P.; Cowling, B.J.; Lau, E.H.Y. Avian influenza human infections at the human-animal interface. J. Infect. Dis. 2020, 222, 528–537. [Google Scholar] [CrossRef]
- Frasca, D.; DIaz, A.; Romero, M.; Garcia, D.; Blomberg, B.B. B Cell Immunosenescence. Annu. Rev. Cell Dev. Biol. 2020, 36, 551–574. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wong, G.; Yang, L.; Tan, S.; Li, J.; Bai, B.; Xu, Z.; Li, H.; Xu, W.; Zhao, X.; et al. Comparison between human infections caused by highly and low pathogenic H7N9 avian influenza viruses in Wave Five: Clinical and virological findings. J. Infect. 2019, 78, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.X.; Tang, S.J.; Yao, S.H.; Zhou, Y.Q.; Xiao, L.L.; Cheng, L.F.; Liu, F.M.; Wu, N.P.; Yao, H.P.; Li, L.J. The viral distribution and pathological characteristics of BALB/c mice infected with highly pathogenic Influenza H7N9 virus. Virol. J. 2021, 18, 1–11. [Google Scholar] [CrossRef]
- Pronier, C.; Gacouin, A.; Lagathu, G.; Le Tulzo, Y.; Tadié, J.M.; Thibault, V. Respiratory Influenza viral load as a marker of poor prognosis in patients with severe symptoms. J. Clin. Virol. 2021, 136, 104761. [Google Scholar] [CrossRef]
- Zheng, S.; Zou, Q.; Wang, X.; Bao, J.; Yu, F.; Guo, F.; Liu, P.; Shen, Y.; Wang, Y.; Yang, S.; et al. Factors associated with fatality due to avian influenza A(H7N9) infection in China. Clin. Infect. Dis. 2020, 71, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Morgan, D.J.; Casulli, J.; Chew, C.; Connolly, E.; Lui, S.; Brand, O.J.; Rahman, R.; Jagger, C.; Hussell, T. Innate Immune Cell Suppression and the Link With Secondary Lung Bacterial Pneumonia. Front. Immunol. 2018, 9, 2943. [Google Scholar] [CrossRef]
- Deng, L.S.; Yuan, J.; Ding, L.; Chen, Y.L.; Zhao, C.H.; Chen, G.Q.; Li, X.H.; Li, X.H.; Luo, W.T.; Lan, J.F.; et al. Comparison of patients hospitalized with COVID-19, H7N9 and H1N1. Infect. Dis. Poverty 2020, 9, 163. [Google Scholar] [CrossRef]
- Liu, R.; Zhao, B.; Li, Y.; Zhang, X.; Chen, S.; Chen, T. Clinical and epidemiological characteristics of a young child infected with avian influenza A (H9N2) virus in China. J. Int. Med. Res. 2018, 46, 3462–3467. [Google Scholar] [CrossRef]
- Pan, M.; Gao, R.; Lv, Q.; Huang, S.; Zhou, Z.; Yang, L.; Li, X.; Zhao, X.; Zou, X.; Tong, W.; et al. Human infection with a novel, highly pathogenic avian influenza A (H5N6) virus: Virological and clinical findings. J. Infect. 2016, 72, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, K.; Su, Q.; Chen, X.; Wang, X.; Li, Q.; Wang, W.; Mao, X.; Xu, J.; Zhou, X.; et al. Clinical features of the first critical case of acute encephalitis caused by the avian influenza A (H5N6) virus. Emerg. Microbes Infect. 2022, 11, 2437–2446. [Google Scholar] [CrossRef]
- Ostrowsky, B.; Huang, A.; Terry, W.; Anton, D.; Brunagel, B.; Traynor, L.; Abid, S.; Johnson, G.; Kacica, M.; Katz, J.; et al. Low pathogenic avian influenza A (H7N2) virus infection in immunocompromised adult, New York, USA, 2003. Emerg. Infect. Dis. 2012, 18, 1128–1131. [Google Scholar] [CrossRef]
- Koopmans, M.; Wilbrink, B.; Conyn, M.; Natrop, G.; Van Der Nat, H.; Vennema, H.; Meijer, A.; Van Steenbergen, J.; Fouchier, R.; Osterhaus, A.; et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet 2004, 363, 587–593. [Google Scholar] [CrossRef]
- Tweed, S.A.; Skowronski, D.M.; David, S.T.; Larder, A.; Petric, M.; Lees, W.; Li, Y.; Katz, J.; Krajden, M.; Tellier, R.; et al. Human illness from avian influenza H7N3, British Columbia. Emerg. Infect. Dis. 2004, 10, 2196–2199. [Google Scholar] [CrossRef]
- Prescott, K.; Kroona, S.; Quinlisk, P.; Gipple, D.; Garvey, A.; Desjardin, L.; Jirsa, S.; Benfer, J.; Gomez, T.; Finelli, L.; et al. Limited human-to-human transmission of Novel Influenza a (H3N2) virus—Iowa, November 2011. Morb. Mortal. Wkly. Rep. 2011, 60, 1615–1617. [Google Scholar]
- Charlton, B.; Crossley, B.; Hietala, S. Conventional and future diagnostics for avian influenza. Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Sakai-Tagawa, Y.; Ozawa, M.; Tamura, D.; Le, M.T.Q.; Nidom, C.A.; Sugaya, N.; Kawaoka, Y. Sensitivity of influenza rapid diagnostic tests to H5N1 and 2009 pandemic H1N1 viruses. J. Clin. Microbiol. 2010, 48, 2872–2877. [Google Scholar] [CrossRef] [PubMed]
- Fader, R.C. Comparison of the Binax NOW Flu A enzyme immunochromatographic assay and R-Mix shell vial culture for the 2003-2004 influenza season. J. Clin. Microbiol. 2005, 43, 6133–6135. [Google Scholar] [CrossRef] [PubMed]
- Hunter, A.O.I.E. Manual of Standards for Diagnostic Tests and Vaccines, List A and B Diseases of Mammals, Birds and Bees, 3rd ed.; Barbara Freischem: Paris, France, 1998; ISBN 9290443146. [Google Scholar]
- Saif, Y.M.; Fadly, A.M.; Glisson, J.R.; McDougald, L.R.; Nolan, L.K.; Swayne, D.E. (Eds.) Diseases of Poultry, 12th ed.; Iowa State University Press: Ames, IA, USA, 2008. [Google Scholar]
- Antarasena, C.; Sirimujalin, R.; Prommuang, P.; Promkuntod, N.; Prommuang, P.; Blacksell, S.D. The indirect immunofluorescence assay using cardiac tissue from chickens, quails and ducks for identification of influenza A virus during an outbreak of highly pathogenic avian influenza virus (H5N1): A rapid and simple screening tool for limited resource. Res. Vet. Sci. 2007, 83, 279–281. [Google Scholar] [CrossRef]
- Songserm, T.; Jam-On, R.; Sae-Heng, N.; Meemak, N.; Hulse-Post, D.J.; Sturm-Ramirez, K.M.; Webster, R.G. Domestic ducks and H5N1 influenza epidemic, Thailand. Emerg. Infect. Dis. 2006, 12, 575–581. [Google Scholar] [CrossRef]
- Hulse-Post, D.J.; Sturm-Ramirez, K.M.; Humberd, J.; Seiler, P.; Govorkova, E.A.; Krauss, S.; Scholtissek, C.; Puthavathana, P.; Buranathai, C.; Nguyen, T.D.; et al. Role of domestic ducks in the propagation and biological evolution of highly pathogenic H5N1 influenza viruses in Asia. Proc. Natl. Acad. Sci. USA 2005, 102, 10682–10687. [Google Scholar] [CrossRef]
- Brown, J.D.; Stallknecht, D.E.; Beck, J.R.; Suarez, D.L.; Swayne, D.E. Susceptibility of North American ducks and gulls to H5N1 highly pathogenic avian influenza viruses. Emerg. Infect. Dis. 2006, 12, 1663–1670. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, B.; Larsson, C.; Zweygberg, B.W. Simultaneous detection and typing of influenza viruses A and B by a nested reverse transcription-PCR: Comparison to virus isolation and antigen detection by immunofluorescence and optical immunoassay (FLU OIA). J. Clin. Microbiol. 2001, 39, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Stech, J.; Guan, Y.; Webster, R.G.; Perez, D.R. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 2001, 146, 2275–2289. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Donnelly, M.E.; Scholes, D.T.; St George, K.; Hatta, M.; Kawaoka, Y.; Wentworth, D.E. Single-Reaction Genomic Amplification Accelerates Sequencing and Vaccine Production for Classical and Swine Origin Human Influenza A Viruses. J. Virol. 2009, 83, 10309–10313. [Google Scholar] [CrossRef]
- Wright, K.E.; Wilson, G.A.R.; Novosad, D.; Dimock, C.; Tan, D.; Weber, J.M. Typing and subtyping of influenza viruses in clinical samples by PCR. J. Clin. Microbiol. 1995, 33, 1180–1184. [Google Scholar] [CrossRef]
- Spackman, E.; Senne, D.A.; Myers, T.J.; Bulaga, L.L.; Garber, L.P.; Perdue, M.L.; Lohman, K.; Daum, L.T.; Suarez, D.L. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 2002, 40, 3256–3260. [Google Scholar] [CrossRef]
- Lau, L.T.; Banks, J.; Aherne, R.; Brown, I.H.; Dillon, N.; Collins, R.A.; Chan, K.Y.; Fung, Y.W.W.; Xing, J.; Yu, A.C.H. Nucleic acid sequence-based amplification methods to detect avian influenza virus. Biochem. Biophys. Res. Commun. 2003, 313, 336–342. [Google Scholar] [CrossRef]
- Das, A.; Spackman, E.; Senne, D.; Pedersen, J.; Suarez, D.L. Development of an internal positive control for rapid diagnosis of avian influenza virus infections by real-time reverse transcription-PCR with lyophilized reagents. J. Clin. Microbiol. 2006, 44, 3065–3073. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Imai, M.; Ninomiya, A.; Minekawa, H.; Notomi, T.; Ishizaki, T.; Van Tu, P.; Tien, N.T.K.; Tashiro, M.; Odagiri, T. Rapid diagnosis of H5N1 avian influenza virus infection by newly developed influenza H5 hemagglutinin gene-specific loop-mediated isothermal amplification method. J. Virol. Methods 2007, 141, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Obenauar, J.C.; Denson, J.; Mehta, P.K.; Su, X.; Mukatira, S.; Finkelstein, D.B.; Xu, X.; Wang, J.; Ma, J.; Fan, Y.; et al. Large-scale sequence analysis of avian influenza isolates. Science 2006, 311, 1576–1580. [Google Scholar] [CrossRef] [PubMed]
- Pourmand, N.; Diamond, L.; Garten, R.; Erickson, J.P.; Kumm, J.; Donis, R.O.; Davis, R.W. Rapid and highly informative diagnostic assay for H5N1 influenza viruses. PLoS ONE 2006, 1, e95. [Google Scholar] [CrossRef] [PubMed]
- Pease, A.C.; Solas, D.; Sullivan, E.J.; Cronin, M.T.; Holmes, C.P.; Fodor, S.P.A. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc. Natl. Acad. Sci. USA 1994, 91, 5022–5026. [Google Scholar] [CrossRef] [PubMed]
- Schena, M.; Shalon, D.; Davis, R.W.; Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270, 467–470. [Google Scholar] [CrossRef]
- Sengupta, S.; Onodera, K.; Lai, A.; Melcher, U. Molecular detection and identification of influenza viruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 2003, 41, 4542–4550. [Google Scholar] [CrossRef]
- Dawson, E.D.; Moore, C.L.; Dankbar, D.M.; Mehlmann, M.; Townsend, M.B.; Smagala, J.A.; Smith, C.B.; Cox, N.J.; Kuchta, R.D.; Rowlen, K.L. Identification of A/H5N1 influenza viruses using a single gene diagnostic microarray. Anal. Chem. 2006, 79, 378–384. [Google Scholar] [CrossRef]
- Wang, Z.; Daum, L.T.; Vora, G.J.; Metzgar, D.; Walter, E.A.; Canas, L.C.; Malanoski, A.P.; Lin, B.; Stenger, D.A. Identifying influenza viruses with resequencing microarrays. Emerg. Infect. Dis. 2006, 12, 638–646. [Google Scholar] [CrossRef]
- Huang, Y.; Tang, H.; Duffy, S.; Hong, Y.; Norman, S.; Ghosh, M.; He, J.; Bose, M.; Henrickson, K.J.; Fan, J.; et al. Multiplex assay for simultaneously typing and subtyping influenza viruses by use of an electronic microarray. J. Clin. Microbiol. 2009, 47, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Ryabinin, V.A.; Kostina, E.V.; Maksakova, G.A.; Neverov, A.A.; Chumakov, K.M.; Sinyakov, A.N. Universal oligonucleotide microarray for sub-typing of Influenza A virus. PLoS ONE 2011, 6, e17529. [Google Scholar] [CrossRef] [PubMed]
- Abdelwhab, E.M.; Lüschow, D.; Harder, T.C.; Hafez, H.M. The use of FTA® filter papers for diagnosis of avian influenza virus. J. Virol. Methods 2011, 174, 120–122. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R.; Janeway, C. Innate immune recognition: Mechanisms and pathways. Immunol. Rev. 2000, 173, 89–97. [Google Scholar] [CrossRef]
- Medzhitov, R.; Janeway, C.A. An ancient system of host defense. Curr. Opin. Immunol. 1998, 10, 12–15. [Google Scholar] [CrossRef]
- Fearon, D.T.; Locksley, R.M. The instructive role of innate immunity in the acquired immune response. Science 1996, 272, 50–54. [Google Scholar] [CrossRef]
- Medzhitov, R.; Janeway, C.A. Innate immunity: Impact on the adaptive immune response. Curr. Opin. Immunol. 1997, 9, 4–9. [Google Scholar] [CrossRef]
- Matis, L. Specificity and selection of gamma-delta receptor-expressing T cells. Immunol. Res. 1991, 10, 5–14. [Google Scholar] [CrossRef]
- Vervelde, L.; Kapczynski, D.R. The innate and adaptive immune response to avian influenza virus. In Animal Influenza; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016. [Google Scholar]
- Wu, X.X.; Zhao, L.Z.; Tang, S.J.; Weng, T.H.; Wu, W.G.; Yao, S.H.; Wu, H.B.; Cheng, L.F.; Wang, J.; Hu, F.Y.; et al. Novel pathogenic characteristics of highly pathogenic avian influenza virus H7N9: Viraemia and extrapulmonary infection. Emerg. Microbes Infect. 2020, 9, 962–975. [Google Scholar] [CrossRef]
- Ichiyama, T.; Isumi, H.; Ozawa, H.; Matsubara, T.; Morishima, T.; Furukawa, S. Cerebrospinal fluid and serum levels of cytokines and soluble tumor necrosis factor receptor in influenza virus-associated encephalopathy. Scand. J. Infect. Dis. 2003, 35, 59–61. [Google Scholar] [CrossRef]
- Channappanavar, R.; Fehr, A.R.; Zheng, J.; Wohlford-Lenane, C.; Abrahante, J.E.; Mack, M.; Sompallae, R.; McCray, P.B.; Meyerholz, D.K.; Perlman, S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Investig. 2019, 129, 3625–3639. [Google Scholar] [CrossRef] [PubMed]
- Cilloniz, C.; Pantin-Jackwood, M.J.; Ni, C.; Goodman, A.G.; Peng, X.; Proll, S.C.; Carter, V.S.; Rosenzweig, E.R.; Szretter, K.J.; Katz, J.M.; et al. Lethal Dissemination of H5N1 Influenza Virus Is Associated with Dysregulation of Inflammation and Lipoxin Signaling in a Mouse Model of Infection. J. Virol. 2010, 84, 7613–7624. [Google Scholar] [CrossRef] [PubMed]
- Karki, R.; Sharma, B.R.; Tuladhar, S.; Williams, E.P.; Zalduondo, L.; Samir, P.; Zheng, M.; Sundaram, B.; Banoth, B.; Malireddi, R.K.S.; et al. Synergism of TNF-α and IFN-γ Triggers Inflammatory Cell Death, Tissue Damage, and Mortality in SARS-CoV-2 Infection and Cytokine Shock Syndromes. Cell 2021, 184, 149–168. [Google Scholar] [CrossRef] [PubMed]
- La Gruta, N.L.; Kedzierska, K.; Stambas, J.; Doherty, P.C. A question of self-preservation: Immunopathology in influenza virus infection. Immunol. Cell Biol. 2007, 85, 85–92. [Google Scholar] [CrossRef]
- Zhang, N.; Bao, Y.J.; Tong, A.H.Y.; Zuyderduyn, S.; Bader, G.D.; Malik Peiris, J.S.; Lok, S.; Lee, S.M.Y. Whole transcriptome analysis reveals differential gene expression profile reflecting macrophage polarization in response to influenza A H5N1 virus infection. BMC Med. Genomics 2018, 11, 1–14. [Google Scholar] [CrossRef]
- Geng, P.; Zhu, H.; Zhou, W.; Su, C.; Chen, M.; Huang, C.; Xia, C.; Huang, H.; Cao, Y.; Shi, X. Baicalin Inhibits Influenza A Virus Infection via Promotion of M1 Macrophage Polarization. Front. Pharmacol. 2020, 11, 01298. [Google Scholar] [CrossRef]
- Davis, M.J.; Tsang, T.M.; Qiu, Y.; Dayrit, J.K.; Freij, J.B.; Huffnagle, G.B.; Olszewski, M.A. Macrophage M1/M2 polarization dynamically adapts to changes in cytokine microenvironments in Cryptococcus neoformans infection. MBio 2013, 4, e00264-13. [Google Scholar] [CrossRef]
- Neerukonda, S.N.; Katneni, U. Avian pattern recognition receptor sensing and signaling. Vet. Sci. 2020, 7, 14. [Google Scholar] [CrossRef]
- Westenius, V.; Mäkelä, S.M.; Julkunen, I.; Österlund, P. Highly pathogenic H5N1 influenza A virus spreads efficiently in human primary monocyte-derived macrophages and dendritic cells. Front. Immunol. 2018, 9, 1664. [Google Scholar] [CrossRef]
- Liu, Q.; Yang, J.; Huang, X.; Liu, Y.; Han, K.; Zhao, D.; Zhang, L.; Li, Y. Transcriptomic profile of chicken bone marrow-derive dendritic cells in response to H9N2 avian influenza A virus. Vet. Immunol. Immunopathol. 2019, 220, 109992. [Google Scholar] [CrossRef]
- Mifsud, E.J.; Kuba, M.; Barr, I.G. Innate immune responses to influenza virus infections in the upper respiratory tract. Viruses 2021, 13, 2090. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019, 19, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Jiang, J.; Tien, P.; Liu, W.; Li, J. IFN-λ: A new spotlight in innate immunity against influenza virus infection. Protein Cell 2018, 9, 832–837. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Kappala, D. Avian infectious bronchitis virus. In Recent Advances in Animal Virology; Springer: Singapore, 2019; ISBN 9789811390739. [Google Scholar]
- Lamichhane, P.P.; Samarasinghe, A.E. The Role of Innate Leukocytes during Influenza Virus Infection. J. Immunol. Res. 2019, 2019, 8028725. [Google Scholar] [CrossRef]
- Marc, D. Influenza virus non-structural protein NS1: Interferon antagonism and beyond. J. Gen. Virol. 2014, 95, 2594–2611. [Google Scholar] [CrossRef]
- Zhao, Y.; Huang, F.; Zou, Z.; Bi, Y.; Yang, Y.; Zhang, C.; Liu, Q.; Shang, D.; Yan, Y.; Ju, X.; et al. Avian influenza viruses suppress innate immunity by inducing trans-transcriptional readthrough via SSU72. Cell. Mol. Immunol. 2022, 19, 702–714. [Google Scholar] [CrossRef] [PubMed]
- Berke, I.C.; Li, Y.; Modis, Y. Structural basis of innate immune recognition of viral RNA. Cell. Microbiol. 2013, 15, 386–394. [Google Scholar] [CrossRef]
- Hiscott, J.; Lin, R.; Nakhaei, P.; Paz, S. MasterCARD: A priceless link to innate immunity. Trends Mol. Med. 2006, 12, 53–56. [Google Scholar] [CrossRef]
- Ikushima, H.; Negishi, H.; Taniguchi, T. The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harb. Symp. Quant. Biol. 2013, 78, 105–116. [Google Scholar] [CrossRef]
- Hiscott, J.; Pitha, P.; Genin, P.; Nguyen, H.; Heylbroeck, C.; Mamane, Y.; Algarte, M.; Lin, R. Triggering the interferon response: The role of IRF-3 transcription factor. J. Interf. Cytokine Res. 1999, 19, 1–13. [Google Scholar] [CrossRef]
- Li, M.M.H.; MacDonald, M.R.; Rice, C.M. To translate, or not to translate: Viral and host mRNA regulation by interferon-stimulated genes. Trends Cell Biol. 2015, 25, 320–329. [Google Scholar] [CrossRef]
- Katze, M.G.; He, Y.; Gale, M. Viruses and interferon: A fight for supremacy. Nat. Rev. Immunol. 2002, 2, 675–687. [Google Scholar] [CrossRef]
- Thakar, J.; Hartmann, B.M.; Marjanovic, N.; Sealfon, S.C.; Kleinstein, S.H. Comparative analysis of anti-viral transcriptomics reveals novel effects of influenza immune antagonism. BMC Immunol. 2015, 16, 46. [Google Scholar] [CrossRef] [PubMed]
- Bean, W.J.; Simpson, R.W. Primary transcription of the influenza virus genome in permissive cells. Virology 1973, 56, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Gutman, N.R.; Manakhova, L.S. [Ability of influenza virus strains to induce interferon and their sensitivity to the action of exogenous interferon]. Vopr Virusol 1981, 2, 168–171. [Google Scholar]
- Portnoy, J.; Merigan, T.C. The effect of interferon and interferon inducers on avian influenza. J. Infect. Dis. 1971, 124, 545–552. [Google Scholar] [CrossRef]
- Sekellick, M.J.; Carra, S.A.; Bowman, A.; Hopkins, D.A.; Marcus, P.I. Transient resistance of influenza virus to interferon action attributed to random multiple packaging and activity of NS genes. J. Interf. Cytokine Res. 2000, 20, 963–970. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.-M.; Fornek, J.; Crochet, N.; Bajwa, G.; Perwitasari, O.; Martinez-Sobrido, L.; Akira, S.; Gill, M.A.; García-Sastre, A.; Katze, M.G.; et al. Distinct RIG-I and MDA5 Signaling by RNA Viruses in Innate Immunity. J. Virol. 2008, 82, 335–345. [Google Scholar] [CrossRef]
- Nakhaei, P.; Genin, P.; Civas, A.; Hiscott, J. RIG-I-like receptors: Sensing and responding to RNA virus infection. Semin. Immunol. 2009, 21, 215–222. [Google Scholar] [CrossRef]
- Jiang, H.; Yang, H.; Kapczynski, D.R. Chicken interferon alpha pretreatment reduces virus replication of pandemic H1N1 and H5N9 avian influenza viruses in lung cell cultures from different avian species. Virol. J. 2011, 8, 447. [Google Scholar] [CrossRef]
- Jia, D.; Rahbar, R.; Chan, R.W.Y.; Lee, S.M.Y.; Chan, M.C.W.; Wang, B.X.; Baker, D.P.; Sun, B.; Malik Peiris, J.S.; Nicholls, J.M.; et al. Influenza virus non-structural protein 1 (NS1) disrupts interferon signaling. PLoS ONE 2010, 5, e13927. [Google Scholar] [CrossRef]
- De Heer, H.J.; Hammad, H.; Kool, M.; Lambrecht, B.N. Dendritic cell subsets and immune regulation in the lung. Semin. Immunol. 2005, 17, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Reese, S.; Dalamani, G.; Kaspers, B. The avian lung-associated immune system: A review. Vet. Res. 2006, 37, 311–324. [Google Scholar] [CrossRef]
- Short, K.R.; Brooks, A.G.; Reading, P.C.; Londrigan, S.L. The fate of influenza A virus after infection of human macrophages and dendritic cells. J. Gen. Virol. 2012, 93, 2315–2325. [Google Scholar] [CrossRef]
- Beigel, J.H.; Farrar, J.; Han, A.M.; Hayden, F.G.; Hyer, R.; de Jong, M.D.; Lochindarat, S.; Nguyen, T.K.T.; Nguyen, T.H.; Tran, T.H.; et al. Avian Influenza A (H5N1) Infection in Humans. N. Engl. J. Med. 2005, 353, 1374–1385. [Google Scholar] [CrossRef] [PubMed]
- Denney, L.; Aitken, C.; Li, C.K.F.; Wilson-Davies, E.; Kok, W.L.; Clelland, C.; Rooney, K.; Young, D.; Dong, T.; McMichael, A.J.; et al. Reduction of natural killer but not effector CD8 t lymphoyctes in three consecutive cases of severe/lethal H1N1/09 influenza a virus infection. PLoS ONE 2010, 5, e10675. [Google Scholar] [CrossRef]
- Heltzer, M.L.; Coffin, S.E.; Maurer, K.; Bagashev, A.; Zhang, Z.; Orange, J.S.; Sullivan, K.E. Immune dysregulation in severe influenza. J. Leukoc. Biol. 2009, 85, 1036–1043. [Google Scholar] [CrossRef] [PubMed]
- Jansen, C.A.; De Geus, E.D.; Van Haarlem, D.A.; Van De Haar, P.M.; Löndt, B.Z.; Graham, S.P.; Göbel, T.W.; Van Eden, W.; Brookes, S.M.; Vervelde, L. Differential lung NK cell responses in avian influenza virus infected chickens correlate with pathogenicity. Sci. Rep. 2013, 3, srep02478. [Google Scholar] [CrossRef] [PubMed]
- Pritchett, T.J.; Paulson, J.C. Basis for the potent inhibition of influenza virus infection by equine and guinea pig alpha 2-macroglobulin. J. Biol. Chem. 1989, 264, 9850–9858. [Google Scholar] [CrossRef] [PubMed]
- Ryan-Poirier, K.A.; Kawaoka, Y. Alpha 2-macroglobulin is the major neutralizing inhibitor of influenza A virus in pig serum. Virology 1993, 193, 974–976. [Google Scholar] [CrossRef] [PubMed]
- Hillaire, M.L.B.; Haagsman, H.P.; Osterhaus, A.D.M.E.; Rimmelzwaan, G.F.; Van Eijk, M. Pulmonary surfactant protein D in first-line innate defence against influenza A virus infections. J. Innate Immun. 2013, 5, 197–208. [Google Scholar] [CrossRef]
- Ng, W.C.; Tate, M.D.; Brooks, A.G.; Reading, P.C. Soluble host defense lectins in innate immunity to influenza virus. J. Biomed. Biotechnol. 2012, 2012, 732191. [Google Scholar] [CrossRef] [PubMed]
- Ratcliffe, D.R.; Nolin, S.L.; Cramer, E.B. Neutrophil interaction with influenza-infected epithelial cells. Blood 1988, 72, 142–149. [Google Scholar] [CrossRef]
- van Strijp, J.A.; van Kessel, K.P.; Miltenburg, L.A.; Fluit, A.C.; Verhoef, J. Attachment of human polymorphonuclear leukocytes to herpes simplex virus-infected fibroblasts mediated by antibody-independent complement activation. J. Virol. 1988, 62, 847–850. [Google Scholar] [CrossRef]
- Tumpey, T.M.; García-Sastre, A.; Taubenberger, J.K.; Palese, P.; Swayne, D.E.; Pantin-Jackwood, M.J.; Schultz-Cherry, S.; Solórzano, A.; Van Rooijen, N.; Katz, J.M.; et al. Pathogenicity of Influenza Viruses with Genes from the 1918 Pandemic Virus: Functional Roles of Alveolar Macrophages and Neutrophils in Limiting Virus Replication and Mortality in Mice. J. Virol. 2005, 79, 14933–14944. [Google Scholar] [CrossRef]
- Dienz, O.; Rud, J.G.; Eaton, S.M.; Lanthier, P.A.; Burg, E.; Drew, A.; Bunn, J.; Suratt, B.T.; Haynes, L.; Rincon, M. Essential role of IL-6 in protection against H1N1 influenza virus by promoting neutrophil survival in the lung. Mucosal Immunol. 2012, 5, 258–266. [Google Scholar] [CrossRef]
- Lipatov, A.S.; Andreansky, S.; Webby, R.J.; Hulse, D.J.; Rehg, J.E.; Krauss, S.; Perez, D.R.; Doherty, P.C.; Webster, R.G.; Sangster, M.Y. Pathogenesis of Hong Kong H5N1 influenza virus NS gene reassortants in mice: The role of cytokines and B- and T-cell responses. J. Gen. Virol. 2005, 86, 1121–1130. [Google Scholar] [CrossRef]
- Cameron, C.M.; Cameron, M.J.; Bermejo-Martin, J.F.; Ran, L.; Xu, L.; Turner, P.V.; Ran, R.; Danesh, A.; Fang, Y.; Chan, P.-K.M.; et al. Gene Expression Analysis of Host Innate Immune Responses during Lethal H5N1 Infection in Ferrets. J. Virol. 2008, 82, 11308–11317. [Google Scholar] [CrossRef]
- Kobasa, D.; Jones, S.M.; Shinya, K.; Kash, J.C.; Copps, J.; Ebihara, H.; Hatta, Y.; Kim, J.H.; Halfmann, P.; Hatta, M.; et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 2007, 445, 319–323. [Google Scholar] [CrossRef]
- Zeng, H.; Belser, J.A.; Goldsmith, C.S.; Gustin, K.M.; Veguilla, V.; Katz, J.M.; Tumpey, T.M. A(H7N9) Virus Results in Early Induction of Proinflammatory Cytokine Responses in both Human Lung Epithelial and Endothelial Cells and Shows Increased Human Adaptation Compared with Avian H5N1 Virus. J. Virol. 2015, 89, 4655–4667. [Google Scholar] [CrossRef]
- Guo, J.; Huang, F.; Liu, J.; Chen, Y.; Wang, W.; Cao, B.; Zou, Z.; Liu, S.; Pan, J.; Bao, C.; et al. The serum profile of hypercytokinemia factors identified in H7N9-infected patients can predict fatal outcomes. Sci. Rep. 2015, 5, 10942. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, D.; Gao, R.; Zhao, B.; Song, J.; Qi, X.; Zhang, Y.; Shi, Y.; Yang, L.; Zhu, W.; et al. Biological features of novel avian influenza A (H7N9) virus. Nature 2013, 499, 500–503. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.Y.; Gardy, J.L.; Cheung, C.Y.; Cheung, T.K.W.; Hui, K.P.Y.; Ip, N.Y.; Guan, Y.; Hancock, R.E.W.; Malik Peiris, J.S. Systems-level comparison of host-responses elicited by avian H5N1 and seasonal H1N1 influenza viruses in primary human macrophages. PLoS ONE 2009, 4, e8072. [Google Scholar] [CrossRef]
- Peiris, J.S.M.; Cheung, C.Y.; Leung, C.Y.H.; Nicholls, J.M. Innate immune responses to influenza A H5N1: Friend or foe? Trends Immunol. 2009, 30, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Gingras, S.; Parganas, E.; De Pauw, A.; Ihle, J.N.; Murray, P.J. Re-examination of the role of suppressor of cytokine signaling 1 (SOCS1) in the regulation of Toll-like receptor signaling. J. Biol. Chem. 2004, 279, 54702–54707. [Google Scholar] [CrossRef] [PubMed]
- Mansell, A.; Smith, R.; Doyle, S.L.; Gray, P.; Fenner, J.E.; Crack, P.J.; Nicholson, S.E.; Hilton, D.J.; O’Neill, L.A.J.; Hertzog, P.J. Suppressor of cytokine signaling 1 negatively regulates Toll-like receptor signaling by mediating Mal degradation. Nat. Immunol. 2006, 7, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Pothlichet, J.; Chignard, M.; Si-Tahar, M. Cutting Edge: Innate Immune Response Triggered by Influenza A Virus Is Negatively Regulated by SOCS1 and SOCS3 through a RIG-I/IFNAR1-Dependent Pathway. J. Immunol. 2008, 180, 2034–2038. [Google Scholar] [CrossRef]
- Chen, S.; Cheng, A.; Wang, M. Innate sensing of viruses by pattern recognition receptors in birds. Vet. Res. 2013, 44, 82. [Google Scholar] [CrossRef]
- Takeda, K.; Akira, S. Toll-Like receptors. Curr. Protoc. Immunol. 2015, 109, 12–14. [Google Scholar] [CrossRef]
- Iwasaki, A.; Pillai, P.S. Innate immunity to influenza virus infection. Nat. Rev. Immunol. 2014, 14, 315–328. [Google Scholar] [CrossRef]
- Lee, S.M.Y.; Kok, K.H.; Jaume, M.; Cheung, T.K.W.; Yip, T.F.; Lai, J.C.C.; Guan, Y.; Webster, R.G.; Jin, D.Y.; Malik Peiris, J.S. Toll-like receptor 10 is involved in induction of innate immune responses to influenza virus infection. Proc. Natl. Acad. Sci. USA 2014, 111, 3793–3798. [Google Scholar] [CrossRef]
- Wei, L.; Jiao, P.; Yuan, R.; Song, Y.; Cui, P.; Guo, X.; Zheng, B.; Jia, W.; Qi, W.; Ren, T.; et al. Goose Toll-like receptor 7 (TLR7), myeloid differentiation factor 88 (MyD88) and antiviral molecules involved in anti-H5N1 highly pathogenic avian influenza virus response. Vet. Immunol. Immunopathol. 2013, 153, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Nang, N.; Lee, J.; Song, B.; Kang, Y.; Kim, H.; Seo, S. Induction of inflammatory cytokines and toll-like receptors in chickens infected with avian H9N2 influenza virus. Vet. Res. 2011, 42, 64. [Google Scholar] [CrossRef] [PubMed]
- Ranaware, P.B.; Mishra, A.; Vijayakumar, P.; Gandhale, P.N.; Kumar, H.; Kulkarni, D.D.; Raut, A.A. Genome wide host gene expression analysis in chicken lungs infected with avian influenza viruses. PLoS ONE 2016, 11, e0153671. [Google Scholar] [CrossRef] [PubMed]
- Barjesteh, N.; Behboudi, S.; Brisbin, J.T.; Villanueva, A.I.; Nagy, É.; Sharif, S. TLR ligands induce antiviral responses in chicken macrophages. PLoS ONE 2014, 9, e105713. [Google Scholar] [CrossRef] [PubMed]
- St Paul, M.; Brisbin, J.T.; Barjesteh, N.; Villaneueva, A.I.; Parvizi, P.; Read, L.R.; Nagy, É.; Sharif, S. Avian influenza virus vaccines containing toll-like receptors 2 and 5 ligand adjuvants promote protective immune responses in chickens. Viral Immunol. 2014, 27, 160–166. [Google Scholar] [CrossRef]
- St Paul, M.; Mallick, A.I.; Read, L.R.; Villanueva, A.I.; Parvizi, P.; Abdul-Careem, M.F.; Nagy, É.; Sharif, S. Prophylactic treatment with Toll-like receptor ligands enhances host immunity to avian influenza virus in chickens. Vaccine 2012, 30, 4524–4531. [Google Scholar] [CrossRef]
- Diebold, S.S.; Kaisho, T.; Hemmi, H.; Akira, S.; Reis e Sousa, C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004, 303, 1529–1531. [Google Scholar] [CrossRef]
- Lund, J.M.; Alexopoulou, L.; Sato, A.; Karow, M.; Adams, N.C.; Gale, N.W.; Iwasaki, A.; Flavell, R.A. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. USA 2004, 101, 5598–5603. [Google Scholar] [CrossRef]
- Wang, Z.; Wan, Y.; Qiu, C.; Quiñones-Parra, S.; Zhu, Z.; Loh, L.; Tian, D.; Ren, Y.; Hu, Y.; Zhang, X.; et al. Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8+ T cells. Nat. Commun. 2015, 6, 6833. [Google Scholar] [CrossRef]
- van de Sandt, C.E.; Kreijtz, J.H.C.M.; de Mutsert, G.; Geelhoed-Mieras, M.M.; Hillaire, M.L.B.; Vogelzang-van Trierum, S.E.; Osterhaus, A.D.M.E.; Fouchier, R.A.M.; Rimmelzwaan, G.F. Human Cytotoxic T Lymphocytes Directed to Seasonal Influenza A Viruses Cross-React with the Newly Emerging H7N9 Virus. J. Virol. 2014, 88, 1684–1693. [Google Scholar] [CrossRef]
- Henry Dunand, C.J.; Leon, P.E.; Huang, M.; Choi, A.; Chromikova, V.; Ho, I.Y.; Tan, G.S.; Cruz, J.; Hirsh, A.; Zheng, N.Y.; et al. Both Neutralizing and Non-Neutralizing Human H7N9 Influenza Vaccine-Induced Monoclonal Antibodies Confer Protection. Cell Host Microbe 2016, 19, 800–813. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Qin, K.; Wai, L.W.; Li, G.; Zhang, J.; Du, H.; Mun, H.N.; Shih, J.W.K.; Peiris, J.S.M.; Guan, Y.; et al. Broad cross-protection against H5N1 avian influenza virus infection by means of monoclonal antibodies that map to conserved viral epitopes. J. Infect. Dis. 2009, 199, 49–58. [Google Scholar] [CrossRef]
- Moriyama, M.; Ichinohe, T. High ambient temperature dampens adaptive immune responses to influenza A virus infection. Proc. Natl. Acad. Sci. USA 2019, 116, 3118–3125. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.C.Y.; Zhang, A.J.X.; Chu, H.; Li, C.; Zhu, H.; Mak, W.W.N.; Chen, Y.; Kok, K.H.; To, K.K.W.; Yuen, K.Y. H7N9 influenza A virus activation of necroptosis in human monocytes links innate and adaptive immune responses. Cell Death Dis. 2019, 10, 442. [Google Scholar] [CrossRef] [PubMed]
- Korenkov, D.; Isakova-Sivak, I.; Rudenko, L. Basics of CD8 T-cell immune responses after influenza infection and vaccination with inactivated or live attenuated influenza vaccine. Expert Rev. Vaccines 2018, 17, 977–987. [Google Scholar] [CrossRef]
- Yetter, R.A.; Barber, W.H.; Small, P.A. Heterotypic immunity to influenza in ferrets. Infect. Immun. 1980, 29, 650–653. [Google Scholar] [CrossRef]
- Van Reeth, K.; Braeckmans, D.; Cox, E.; Van Borm, S.; van den Berg, T.; Goddeeris, B.; De Vleeschauwer, A. Prior infection with an H1N1 swine influenza virus partially protects pigs against a low pathogenic H5N1 avian influenza virus. Vaccine 2009, 27, 6330–6339. [Google Scholar] [CrossRef]
- Lee, L.Y.H.; Ha, D.L.A.; Simmons, C.; De Jong, M.D.; Chau, N.V.V.; Schumacher, R.; Yan, C.P.; McMichael, A.J.; Farrar, J.J.; Smith, G.L.; et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Investig. 2008, 118, 3478–3490. [Google Scholar] [CrossRef]
- Scheible, K.; Zhang, G.; Baer, J.; Azadniv, M.; Lambert, K.; Pryhuber, G.; Treanor, J.J.; Topham, D.J. CD8+ T cell immunity to 2009 pandemic and seasonal H1N1 influenza viruses. Vaccine 2011, 29, 2159–2168. [Google Scholar] [CrossRef]
- Fraser, C.; Donnelly, C.A.; Cauchemez, S.; Hanage, W.P.; Van Kerkhove, M.D.; Hollingsworth, T.D.; Griffin, J.; Baggaley, R.F.; Jenkins, H.E.; Lyons, E.J.; et al. Pandemic potential of a strain of influenza A (H1N1): Early findings. Science 2009, 324, 1557–1561. [Google Scholar] [CrossRef] [PubMed]
- Smallman-Raynor, M.; Cliff, A.D. Avian influenza A (H5N1) age distribution in humans. Emerg. Infect. Dis. 2007, 13, 510–512. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhu, L.; Nguyen, T.H.O.; Wan, Y.; Sant, S.; Quiñones-Parra, S.M.; Crawford, J.C.; Eltahla, A.A.; Rizzetto, S.; Bull, R.A.; et al. Clonally diverse CD38+HLA-DR+CD8+ T cells persist during fatal H7N9 disease. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Dai, M.; Sun, H.; Zhao, L.; Wu, Q.; You, B.; Xu, F.; Liao, J.; Zhu, S.; Li, Z.; Yao, Y.; et al. Duck CD8(+) T Cell Response to H5N1 Highly Pathogenic Avian Influenza Virus Infection In Vivo and In Vitro. J. Immunol. 2022, 209, 979–990. [Google Scholar] [CrossRef]
- Hao, X.; Li, S.; Chen, L.; Dong, M.; Wang, J.; Hu, J.; Gu, M.; Wang, X.; Hu, S.; Peng, D.; et al. Establishing a multicolor flow cytometry to characterize cellular immune response in chickens following h7n9 avian influenza virus infection. Viruses 2020, 12, 1396. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, M.; Furlong, K.; Perez, J.T.; Manicassamy, S.; Manicassamy, B. Suppression of Cytotoxic T Cell Functions and Decreased Levels of Tissue-Resident Memory T Cells during H5N1 Infection. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Nüssing, S.; Sant, S.; Koutsakos, M.; Subbarao, K.; Nguyen, T.H.O.; Kedzierska, K. Innate and adaptive T cells in influenza disease. Front. Med. 2018, 12, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, T.M.; Li, C.K.F.; Chui, C.S.C.; Huang, A.K.Y.; Perkins, M.; Liebner, J.C.; Lambkin-Williams, R.; Gilbert, A.; Oxford, J.; Nicholas, B.; et al. Preexisting influenza-specific CD4 + T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 2012, 18, 274–280. [Google Scholar] [CrossRef]
- Loh, L.; Wang, Z.; Sant, S.; Koutsakos, M.; Jegaskanda, S.; Corbett, A.J.; Liu, L.; Fairlie, D.P.; Crowe, J.; Rossjohn, J.; et al. Human mucosal-associated invariant T cells contribute to antiviral influenza immunity via IL-18-dependent activation. Proc. Natl. Acad. Sci. USA 2016, 113, 10133–10138. [Google Scholar] [CrossRef] [PubMed]
- Clemens, E.B.; Van de Sandt, C.; Wong, S.S.; Wakim, L.M.; Valkenburg, S.A. Harnessing the power of T cells: The promising hope for a universal influenza vaccine. Vaccines 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Rimmelzwaan, G.F.; Katz, J.M. Immune responses to infection with H5N1 influenza virus. Virus Res. 2013, 178, 44–52. [Google Scholar] [CrossRef]
- Ye, J.; Shao, H.; Hickman, D.; Angel, M.; Xu, K.; Cai, Y.; Song, H.; Fouchier, R.A.M.; Qin, A.; Perez, D.R. Intranasal delivery of an IgA monoclonal antibody effective against sublethal H5N1 influenza virus infection in mice. Clin. Vaccine Immunol. 2010, 17, 1363–1370. [Google Scholar] [CrossRef] [PubMed]
- Katz, J.M.; Lim, W.; Bridges, C.B.; Rowe, T.; Hu-Primmer, J.; Lu, X.; Abernathy, R.A.; Clarke, M.; Conn, L.; Kwong, H.; et al. Antibody response in individuals infected with avian influenza A (H5N1) viruses and detection of anti-H5 antibody among household and social contacts. J. Infect. Dis. 1999, 180, 1763–1770. [Google Scholar] [CrossRef]
- Cheng, X.; Eisenbraun, M.; Xu, Q.; Zhou, H.; Kulkarni, D.; Subbarao, K.; Kemble, G.; Jin, H. H5N1 vaccine-specific B cell responses in ferrets primed with live attenuated seasonal influenza vaccines. PLoS ONE 2009, 4, e4436. [Google Scholar] [CrossRef]
- Sasaki, S.; Jaimes, M.C.; Holmes, T.H.; Dekker, C.L.; Mahmood, K.; Kemble, G.W.; Arvin, A.M.; Greenberg, H.B. Comparison of the Influenza Virus-Specific Effector and Memory B-Cell Responses to Immunization of Children and Adults with Live Attenuated or Inactivated Influenza Virus Vaccines. J. Virol. 2007, 81, 215–228. [Google Scholar] [CrossRef]
- Reemers, S.S.; van Leenen, D.; Groot Koerkamp, M.J.; van Haarlem, D.; van de Haar, P.; van Eden, W.; Vervelde, L. Early host responses to avian influenza A virus are prolonged and enhanced at transcriptional level depending on maturation of the immune system. Mol. Immunol. 2010, 47, 1675–1685. [Google Scholar] [CrossRef]
- Pantin-Jackwood, M.J.; Smith, D.M.; Wasilenko, J.L.; Cagle, C.; Shepherd, E.; Sarmento, L.; Kapczynski, D.R.; Afonso, C.L. Effect of age on the pathogenesis and innate immune responses in Pekin ducks infected with different H5N1 highly pathogenic avian influenza viruses. Virus Res. 2012, 167, 196–206. [Google Scholar] [CrossRef]
- Xing, Z.; Cardona, C.J.; Li, J.; Dao, N.; Tran, T.; Andrada, J. Modulation of the immune responses in chickens by low-pathogenicity avian influenza virus H9N2. J. Gen. Virol. 2008, 89, 1288–1299. [Google Scholar] [CrossRef]
- Jiang, H.; Yu, K.; Kapczynski, D.R. Transcription factor regulation and cytokine expression following in vitro infection of primary chicken cell culture with low pathogenic avian influenza virus. Virol. J. 2013, 10, 342. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Cardona, C.J.; Anunciacion, J.; Adams, S.; Dao, N. Roles of the ERK MAPK in the regulation of proinflammatory and apoptotic responses in chicken macrophages infected with H9N2 avian influenza virus. J. Gen. Virol. 2009, 91, 343–351. [Google Scholar] [CrossRef]
- Cui, Z.; Hu, J.; He, L.; Li, Q.; Gu, M.; Wang, X.; Hu, S.; Liu, H.; Liu, W.; Liu, X.; et al. Differential immune response of mallard duck peripheral blood mononuclear cells to two highly pathogenic avian influenza H5N1 viruses with distinct pathogenicity in mallard ducks. Arch. Virol. 2013, 159, 339–343. [Google Scholar] [CrossRef]
- Kumar, A.; Vijayakumar, P.; Gandhale, P.N.; Ranaware, P.B.; Kumar, H.; Kulkarni, D.D.; Raut, A.A.; Mishra, A. Genome-wide gene expression pattern underlying differential host response to high or low pathogenic H5N1 avian influenza virus in ducks. Acta Virol. 2017, 61, 66–76. [Google Scholar] [CrossRef]
- Barber, M.R.W.; Aldridge, J.R.; Webster, R.G.; Magor, K.E. Association of RIG-I with innate immunity of ducks to influenza. Proc. Natl. Acad. Sci. USA 2010, 107, 5913–5918. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liang, W.; Yang, S.; Wu, N.; Gao, H.; Sheng, J.; Yao, H.; Wo, J.; Fang, Q.; Cui, D.; et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: Clinical analysis and characterisation of viral genome. Lancet 2013, 381, 1916–1925. [Google Scholar] [CrossRef] [PubMed]
- Quiñones-Parra, S.; Grant, E.; Loh, L.; Nguyen, T.H.O.; Campbell, K.A.; Tong, S.Y.C.; Miller, A.; Doherty, P.C.; Vijaykrishna, D.; Rossjohn, J.; et al. Preexisting CD8+ T-cell immunity to the H7N9 influenza a virus varies across ethnicities. Proc. Natl. Acad. Sci. USA 2014, 111, 1049–1054. [Google Scholar] [CrossRef]
- Kerstetter, L.J.; Buckley, S.; Bliss, C.M.; Coughlan, L. Adenoviral Vectors as Vaccines for Emerging Avian Influenza Viruses. Front. Immunol. 2021, 11, 607333. [Google Scholar] [CrossRef]
- Kreijtz, J.H.C.M.; Bodewes, R.; van den Brand, J.M.A.; de Mutsert, G.; Baas, C.; van Amerongen, G.; Fouchier, R.A.M.; Osterhaus, A.D.M.E.; Rimmelzwaan, G.F. Infection of mice with a human influenza A/H3N2 virus induces protective immunity against lethal infection with influenza A/H5N1 virus. Vaccine 2009, 27, 4983–4989. [Google Scholar] [CrossRef]
- Gillim-Ross, L.; Subbarao, K. Emerging respiratory viruses: Challenges and vaccine strategies. Clin. Microbiol. Rev. 2006, 19, 614–636. [Google Scholar] [CrossRef]
- De Jong, M.D.; Hien, T.T. Avian influenza A (H5N1). J. Clin. Virol. 2006, 35, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Couch, R.B.; Kasel, J.A. Immunity to influenza in man. Annu. Rev. Microbiol. 1983, 37, 529–549. [Google Scholar] [CrossRef] [PubMed]
- Potter, C.W.; Oxford, J.S. Determinants of immunity to influenza infection in man. Br. Med. Bull. 1979, 35, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Gerhard, W. The role of the antibody response in influenza virus infection. Curr. Top. Microbiol. Immunol. 2001, 260, 171–190. [Google Scholar] [CrossRef]
- Kilbourne, E.D.; Laver, W.G.; Schulman, J.L.; Webster, R.G. Antiviral Activity of Antiserum Specific for an Influenza Virus Neuraminidase. J. Virol. 1968, 2, 281–288. [Google Scholar] [CrossRef]
- Murphy, B.R.; Kasel, J.A.; Chanock, R.M. Association of Serum Anti-Neuraminidase Antibody with Resistance to Influenza in Man. N. Engl. J. Med. 1972, 286, 1329–1332. [Google Scholar] [CrossRef]
- Altenburg, A.F.; Rimmelzwaan, G.F.; de Vries, R.D. Virus-specific T cells as correlate of (cross-)protective immunity against influenza. Vaccine 2015, 33, 500–506. [Google Scholar] [CrossRef]
- Schmidt, A.; Lapuente, D. T cell immunity against influenza: The long way from animal models towards a real-life universal flu vaccine. Viruses 2021, 13, 199. [Google Scholar] [CrossRef]
- Zhao, M.; Liu, K.; Luo, J.; Tan, S.; Quan, C.; Zhang, S.; Chai, Y.; Qi, J.; Li, Y.; Bi, Y.; et al. Heterosubtypic protections against human-infecting avian influenza viruses correlate to biased cross-T-cell responses. MBio 2018, 9, e01408-18. [Google Scholar] [CrossRef]
- Wang, Y.; Deng, L.; Kang, S.M.; Wang, B.Z. Universal influenza vaccines: From viruses to nanoparticles. Expert Rev. Vaccines 2018, 17, 967–976. [Google Scholar] [CrossRef]
- Ren, Z.; Ji, X.; Meng, L.; Wei, Y.; Wang, T.; Feng, N.; Zheng, X.; Wang, H.; Li, N.; Gao, X.; et al. H5N1 influenza virus-like particle vaccine protects mice from heterologous virus challenge better than whole inactivated virus. Virus Res. 2015, 200, 9–18. [Google Scholar] [CrossRef]
- Subbarao, K.; Joseph, T. Scientific barriers to developing vaccines against avian influenza viruses. Nat. Rev. Immunol. 2007, 7, 267–278. [Google Scholar] [CrossRef]
- Furuya, Y. Return of inactivated whole-virus vaccine for superior efficacy. Immunol. Cell Biol. 2011, 90, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Nelson, S.A.; Sant, A.J. Imprinting and editing of the human CD4 T cell response to influenza virus. Front. Immunol. 2019, 10, 932. [Google Scholar] [CrossRef] [PubMed]
- Treanor, J.J.; Campbell, J.D.; Zangwill, K.M.; Rowe, T.; Wolff, M. Safety and Immunogenicity of an Inactivated Subvirion Influenza A (H5N1) Vaccine. N. Engl. J. Med. 2006, 354, 1343–1351. [Google Scholar] [CrossRef]
- Bresson, J.L.; Perronne, C.; Launay, O.; Gerdil, C.; Saville, M.; Wood, J.; Höschler, K.; Zambon, M.C. Safety and immunogenicity of an inactivated split-virion influenza A/Vietnam/1194/2004 (H5N1) vaccine: Phase I randomised trial. Lancet 2006, 367, 1657–1664. [Google Scholar] [CrossRef]
- Stephenson, I.; Nicholson, K.G.; Colegate, A.; Podda, A.; Wood, J.; Ypma, E.; Zambon, M. Boosting immunity to influenza H5N1 with MF59-adjuvanted H5N3 A/Duck/Singapore/97 vaccine in a primed human population. Vaccine 2002, 21, 1687–1693. [Google Scholar] [CrossRef]
- Baz, M.; Luke, C.J.; Cheng, X.; Jin, H.; Subbarao, K. H5N1 vaccines in humans. Virus Res. 2013, 178, 78–98. [Google Scholar] [CrossRef] [PubMed]
- Gurwith, M.; Lock, M.; Taylor, E.M.; Ishioka, G.; Alexander, J.; Mayall, T.; Ervin, J.E.; Greenberg, R.N.; Strout, C.; Treanor, J.J.; et al. Safety and immunogenicity of an oral, replicating adenovirus serotype 4 vector vaccine for H5N1 influenza: A Randomised, double-blind, placebo-controlled, phase 1 study. Lancet Infect. Dis. 2013, 13, 238–250. [Google Scholar] [CrossRef]
- Ledgerwood, J.E.; Wei, C.J.; Hu, Z.; Gordon, I.J.; Enama, M.E.; Hendel, C.S.; McTamney, P.M.; Pearce, M.B.; Yassine, H.M.; Boyington, J.C.; et al. DNA priming and influenza vaccine immunogenicity: Two phase 1 open label randomised clinical trials. Lancet Infect. Dis. 2011, 11, 916–924. [Google Scholar] [CrossRef]
- Belshe, R.B.; Gruber, W.C.; Mendelman, P.M.; Mehta, H.B.; Mahmood, K.; Reisinger, K.; Treanor, J.; Zangwill, K.; Hayden, F.G.; Bernstein, D.I.; et al. Correlates of immune protection induced by live, attenuated, cold- adapted, trivalent, intranasal influenza virus vaccine. J. Infect. Dis. 2000, 181, 1133–1137. [Google Scholar] [CrossRef] [PubMed]
- Forrest, B.D.; Pride, M.W.; Dunning, A.J.; Capeding, M.R.Z.; Chotpitayasunondh, T.; Tam, J.S.; Rappaport, R.; Eldridge, J.H.; Gruber, W.C. Correlation of cellular immune responses with protection against culture-confirmed influenza virus in young children. Clin. Vaccine Immunol. 2008, 15, 1042–1053. [Google Scholar] [CrossRef] [PubMed]
- Mohn, K.G.I.; Zhou, F.; Brokstad, K.A.; Sridhar, S.; Cox, R.J. Boosting of Cross-Reactive and Protection-Associated T Cells in Children after Live Attenuated Influenza Vaccination. J. Infect. Dis. 2017, 215, 1527–1535. [Google Scholar] [CrossRef]
- Pitisuttithum, P.; Boonnak, K.; Chamnanchanunt, S.; Puthavathana, P.; Luvira, V.; Lerdsamran, H.; Kaewkungwal, J.; Lawpoolsri, S.; Thanachartwet, V.; Silachamroon, U.; et al. Safety and immunogenicity of a live attenuated influenza H5 candidate vaccine strain A/17/turkey/Turkey/05/133 H5N2 and its priming effects for potential pre-pandemic use: A randomised, double-blind, placebo-controlled trial. Lancet Infect. Dis. 2017, 17, 833–842. [Google Scholar] [CrossRef] [PubMed]
- Baz, M.; Boonnak, K.; Paskel, M.; Santos, C.; Powell, T.; Townsend, A.; Subbarao, K. Nonreplicating influenza a virus vaccines confer broad protection against lethal challenge. MBio 2015, 6, e01487-15. [Google Scholar] [CrossRef]
- Wang, B.X.; Fish, E.N. Global virus outbreaks: Interferons as 1st responders. Semin. Immunol. 2019, 43, 101300. [Google Scholar] [CrossRef]
- Coelingh, K.L.; Luke, C.J.; Jin, H.; Talaat, K.R. Development of live attenuated influenza vaccines against pandemic influenza strains. Expert Rev. Vaccines 2014, 13, 855–871. [Google Scholar] [CrossRef]
- Rudenko, L.; Isakova-Sivak, I. Pandemic preparedness with live attenuated influenza vaccines based on A/Leningrad/134/17/57 master donor virus. Expert Rev. Vaccines 2015, 14, 395–412. [Google Scholar] [CrossRef]
- Donelan, N.R.; Basler, C.F.; García-Sastre, A. A Recombinant Influenza A Virus Expressing anRNA-Binding-Defective NS1 Protein Induces High Levels of BetaInterferon and Is Attenuated inMice. J. Virol. 2003, 77, 13257–13266. [Google Scholar] [CrossRef]
- Wang, X.; Li, M.; Zheng, H.; Muster, T.; Palese, P.; Beg, A.A.; García-Sastre, A. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J. Virol. 2000, 74, 11566–11573. [Google Scholar] [CrossRef]
- Peng, Y.C.; Wang, B.; Talaat, K.; Karron, R.; Powell, T.J.; Zeng, H.; Dong, D.; Luke, C.J.; McMichael, A.; Subbarao, K.; et al. Boosted influenza-specific T cell responses after H5N1 pandemic live attenuated influenza virus vaccination. Front. Immunol. 2015, 6, 287. [Google Scholar] [CrossRef]
- Sayedahmed, E.E.; Elkashif, A.; Alhashimi, M.; Sambhara, S.; Mittal, S.K. Adenoviral vector-based vaccine platforms for developing the next generation of influenza vaccines. Vaccines 2020, 8, 574. [Google Scholar] [CrossRef]
- De Vries, R.D.; Altenburg, A.F.; Nieuwkoop, N.J.; De Bruin, E.; Van Trierum, S.E.; Pronk, M.R.; Lamers, M.M.; Richard, M.; Nieuwenhuijse, D.F.; Koopmans, M.P.G.; et al. Induction of cross-clade antibody and T-Cell responses by a modified vaccinia virus Ankara-based influenza A(H5N1) vaccine in a randomized phase 1/2a Clinical Trial. J. Infect. Dis. 2018, 218, 614–623. [Google Scholar] [CrossRef] [PubMed]
- Pittman, P.R.; Hahn, M.; Lee, H.S.; Koca, C.; Samy, N.; Schmidt, D.; Hornung, J.; Weidenthaler, H.; Heery, C.R.; Meyer, T.P.H.; et al. Phase 3 Efficacy Trial of Modified Vaccinia Ankara as a Vaccine against Smallpox. N. Engl. J. Med. 2019, 381, 1897–1908. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, S.; Lambe, T. Clinical advances in viral-vectored influenza vaccines. Vaccines 2018, 6, 29. [Google Scholar] [CrossRef]
- de Vries, R.D.; Rimmelzwaan, G.F. Viral vector-based influenza vaccines. Hum. Vaccines Immunother. 2016, 12, 2881–2901. [Google Scholar] [CrossRef] [PubMed]
- Erbelding, E.J.; Post, D.J.; Stemmy, E.J.; Roberts, P.C.; Augustine, A.D.; Ferguson, S.; Paules, C.I.; Graham, B.S.; Fauci, A.S. A universal influenza vaccine: The strategic plan for the national institute of allergy and infectious diseases. J. Infect. Dis. 2018, 218, 347–354. [Google Scholar] [CrossRef]
- Paules, C.I.; Sullivan, S.G.; Subbarao, K.; Fauci, A.S. Chasing Seasonal Influenza—The Need for a Universal Influenza Vaccine. N. Engl. J. Med. 2018, 378, 7–9. [Google Scholar] [CrossRef]
- Ichihashi, T.; Yoshida, R.; Sugimoto, C.; Takada, A.; Kajino, K. Cross-protective peptide vaccine against influenza A viruses developed in HLA-A*2402 human immunity model. PLoS ONE 2011, 6, e24626. [Google Scholar] [CrossRef]
- Yan, J.; Villarreal, D.O.; Racine, T.; Chu, J.S.; Walters, J.N.; Morrow, M.P.; Khan, A.S.; Sardesai, N.Y.; Kim, J.J.; Kobinger, G.P.; et al. Protective immunity to H7N9 influenza viruses elicited by synthetic DNA vaccine. Vaccine 2014, 32, 2833–2842. [Google Scholar] [CrossRef]
- Xu, Z.; Chokkalingam, N.; Tello-Ruiz, E.; Walker, S.; Kulp, D.W.; Weiner, D.B. Incorporation of a Novel CD4+ Helper Epitope Identified from Aquifex aeolicus Enhances Humoral Responses Induced by DNA and Protein Vaccinations. iScience 2020, 23, 101399. [Google Scholar] [CrossRef] [PubMed]
- Freyn, A.W.; Ramos da Silva, J.; Rosado, V.C.; Bliss, C.M.; Pine, M.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; de Souza Ferreira, L.C.; Weissman, D.; et al. A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol. Ther. 2020, 28, 1569–1584. [Google Scholar] [CrossRef] [PubMed]
- Yassine, H.M.; Boyington, J.C.; McTamney, P.M.; Wei, C.J.; Kanekiyo, M.; Kong, W.P.; Gallagher, J.R.; Wang, L.; Zhang, Y.; Joyce, M.G.; et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015, 21, 1065–1070. [Google Scholar] [CrossRef]
- Xu, Z.; Wise, M.C.; Chokkalingam, N.; Walker, S.; Tello-Ruiz, E.; Elliott, S.T.C.; Perales-Puchalt, A.; Xiao, P.; Zhu, X.; Pumroy, R.A.; et al. In Vivo Assembly of Nanoparticles Achieved through Synergy of Structure-Based Protein Engineering and Synthetic DNA Generates Enhanced Adaptive Immunity. Adv. Sci. 2020, 7, 1902802. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.M.; Kim, M.C.; Compans, R.W. Virus-like particles as universal influenza vaccines. Expert Rev. Vaccines 2012, 11, 995–1007. [Google Scholar] [CrossRef] [PubMed]
- Coughlan, L. Factors Which Contribute to the Immunogenicity of Non-replicating Adenoviral Vectored Vaccines. Front. Immunol. 2020, 11, 909. [Google Scholar] [CrossRef] [PubMed]
- Giles, B.M.; Crevar, C.J.; Carter, D.M.; Bissel, S.J.; Schultz-Cherry, S.; Wiley, C.A.; Ross, T.M. A computationally optimized hemagglutinin virus-like particle vaccine elicits broadly reactive antibodies that protect nonhuman primates from H5N1 infection. J. Infect. Dis. 2012, 205, 1562–1570. [Google Scholar] [CrossRef] [PubMed]
- Theiler, J.; Korber, B. Graph-based optimization of epitope coverage for vaccine antigen design. Stat. Med. 2018, 37, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Padilla-Quirarte, H.O.; Lopez-Guerrero, D.V.; Gutierrez-Xicotencatl, L.; Esquivel-Guadarrama, F. Protective antibodies against influenza proteins. Front. Immunol. 2019, 10, 1677. [Google Scholar] [CrossRef] [PubMed]
- Krause, J.C.; Tsibane, T.; Tumpey, T.M.; Huffman, C.J.; Basler, C.F.; Crowe, J.E. A Broadly Neutralizing Human Monoclonal Antibody That Recognizes a Conserved, Novel Epitope on the Globular Head of the Influenza H1N1 Virus Hemagglutinin. J. Virol. 2011, 85, 10905–10908. [Google Scholar] [CrossRef]
- Zost, S.J.; Lee, J.; Gumina, M.E.; Parkhouse, K.; Henry, C.; Wu, N.C.; Lee, C.C.D.; Wilson, I.A.; Wilson, P.C.; Bloom, J.D.; et al. Identification of Antibodies Targeting the H3N2 Hemagglutinin Receptor Binding Site following Vaccination of Humans. Cell Rep. 2019, 29, 4460–4470. [Google Scholar] [CrossRef]
- Yoshida, R.; Igarashi, M.; Ozaki, H.; Kishida, N.; Tomabechi, D.; Kida, H.; Ito, K.; Takada, A. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. 2009, 5, e1000350. [Google Scholar] [CrossRef]
- Bajic, G.; Maron, M.J.; Adachi, Y.; Onodera, T.; McCarthy, K.R.; McGee, C.E.; Sempowski, G.D.; Takahashi, Y.; Kelsoe, G.; Kuraoka, M.; et al. Influenza Antigen Engineering Focuses Immune Responses to a Subdominant but Broadly Protective Viral Epitope. Cell Host Microbe 2019, 25, 827–835. [Google Scholar] [CrossRef]
- Ekiert, D.C.; Kashyap, A.K.; Steel, J.; Rubrum, A.; Bhabha, G.; Khayat, R.; Lee, J.H.; Dillon, M.A.; O’Neil, R.E.; Faynboym, A.M.; et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 2012, 489, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, E.; Wang, W.; McAuliffe, J.M.; Palmer-Hill, F.J.; Kallewaard, N.L.; Chen, Z.; Suzich, J.A.; Blair, W.S.; Jin, H.; Zhu, Q. A Broadly Neutralizing Human Monoclonal Antibody Directed against a Novel Conserved Epitope on the Influenza Virus H3 Hemagglutinin Globular Head. J. Virol. 2014, 88, 6743–6750. [Google Scholar] [CrossRef] [PubMed]
- Raymond, D.D.; Bajic, G.; Ferdman, J.; Suphaphiphat, P.; Settembre, E.C.; Moody, M.A.; Schmidt, A.G.; Harrison, S.C. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc. Natl. Acad. Sci. USA 2018, 115, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Gostic, K.M.; Ambrose, M.; Worobey, M.; Lloyd-Smith, J.O. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 2016, 354, 722–726. [Google Scholar] [CrossRef]
- Gouma, S.; Kim, K.; Weirick, M.E.; Gumina, M.E.; Branche, A.; Topham, D.J.; Martin, E.T.; Monto, A.S.; Cobey, S.; Hensley, S.E. Middle-aged individuals may be in a perpetual state of H3N2 influenza virus susceptibility. Nat. Commun. 2020, 11, 1–8. [Google Scholar] [CrossRef]
- Ng, S.; Nachbagauer, R.; Balmaseda, A.; Stadlbauer, D.; Ojeda, S.; Patel, M.; Rajabhathor, A.; Lopez, R.; Guglia, A.F.; Sanchez, N.; et al. Novel correlates of protection against pandemic H1N1 influenza A virus infection. Nat. Med. 2019, 25, 962–967. [Google Scholar] [CrossRef]
- Meade, P.; Kuan, G.; Strohmeier, S.; Maier, H.E.; Amanat, F.; Balmaseda, A.; Ito, K.; Kirkpatrick, E.; Javier, A.; Gresh, L.; et al. Influenza virus infection induces a narrow antibody response in children but a broad recall response in adults. MBio 2020, 11. [Google Scholar] [CrossRef]
- Nolan, T.M.; Richmond, P.C.; Skeljo, M.V.; Pearce, G.; Hartel, G.; Formica, N.T.; Höschler, K.; Bennet, J.; Ryan, D.; Papanaoum, K.; et al. Phase I and II randomised trials of the safety and immunogenicity of a prototype adjuvanted inactivated split-virus influenza A (H5N1) vaccine in healthy adults. Vaccine 2008, 26, 4160–4167. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, H.J.; Müller, M.; Oh, H.M.L.; Tambyah, P.A.; Joukhadar, C.; Montomoli, E.; Fisher, D.; Berezuk, G.; Fritsch, S.; Löw-Baselli, A.; et al. A Clinical Trial of a Whole-Virus H5N1 Vaccine Derived from Cell Culture. N. Engl. J. Med. 2008, 358, 2573–2584. [Google Scholar] [CrossRef]
- Beran, J.; Abdel-Messih, I.A.; Raupachova, J.; Hobzova, L.; Fragapane, E. A phase III, randomized, open-label study to assess the tolerability and immunogenicity of an H5N1 influenza vaccine administered to healthy adults with a 1-, 2-, 3-, or 6-week interval between first and second doses. Clin. Ther. 2010, 32, 2186–2197. [Google Scholar] [CrossRef]
- Couch, R.B.; Patel, S.M.; Wade-Bowers, C.L.; Niño, D. A Randomized Clinical Trial of an Inactivated Avian Influenza A (H7N7) Vaccine. PLoS ONE 2012, 7, e49704. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, K.; Huang, J.; Zhou, T.; Sheng, Z.; Kang, B.H.; Ishida, E.; Griesman, T.; Stuccio, S.; Bolkhovitinov, L.; Wohlbold, T.J.; et al. Prolonged evolution of the memory B cell response induced by a replicating adenovirus-influenza H5 vaccine. Sci. Immunol. 2019, 4, eaau2710. [Google Scholar] [CrossRef]
- Duong, T.N.; Thiem, V.D.; Anh, D.D.; Cuong, N.P.; Thang, T.C.; Huong, V.M.; Chien, V.C.; Phuong, N.T.L.; Montomoli, E.; Holt, R.; et al. A Phase 2/3 double blinded, randomized, placebo-controlled study in healthy adult participants in Vietnam to examine the safety and immunogenicity of an inactivated whole virion, alum adjuvanted, A(H5N1) influenza vaccine (IVACFLU-A/H5N1). Vaccine 2019, 38, 1541–1550. [Google Scholar] [CrossRef] [PubMed]
- Trombetta, C.M.; Marchi, S.; Manini, I.; Lazzeri, G.; Montomoli, E. Challenges in the development of egg-independent vaccines for influenza. Expert Rev. Vaccines 2019, 18, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Berlanda Scorza, F.; Tsvetnitsky, V.; Donnelly, J.J. Universal influenza vaccines: Shifting to better vaccines. Vaccine 2016, 34, 2926–2933. [Google Scholar] [CrossRef]
- ZENG, X.; HE, X.; MENG, F.; MA, Q.; WANG, Y.; BAO, H.; LIU, Y.; DENG, G.; SHI, J.; LI, Y.; et al. Protective efficacy of an H5/H7 trivalent inactivated vaccine (H5-Re13, H5-Re14, and H7-Re4 strains) in chickens, ducks, and geese against newly detected H5N1, H5N6, H5N8, and H7N9 viruses. J. Integr. Agric. 2022, 21, 2086–2094. [Google Scholar] [CrossRef]
- Europa.eu. Available online: https://european-union.europa.eu/select-language?destination=/node/1 (accessed on 2 December 2022).
- Banzhoff, A.; Pellegrini, M.; Del Giudice, G.; Fragapane, E.; Groth, N.; Podda, A. MF59® -adjuvanted vaccines for seasonal and pandemic influenza prophylaxis. Influenza Other Respi. Viruses 2008, 2, 243–249. [Google Scholar] [CrossRef]
- Yang, W.H.; Dionne, M.; Kyle, M.; Aggarwal, N.; Li, P.; Madariaga, M.; Godeaux, O.; Vaughn, D.W. Long-term immunogenicity of an AS03-adjuvanted influenza A(H1N1)pdm09 vaccine in young and elderly adults: An observer-blind, randomized trial. Vaccine 2013, 31, 4389–4397. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, L.; Kiseleva, I.; Krutikova, E.; Stepanova, E.; Isakova-Sivak, I.; Donina, S.; Rekstin, A.; Pisareva, M.; Bazhenova, E.; Kotomina, T.; et al. Two live attenuated vaccines against recent low–and highly pathogenic H7N9 influenza viruses are safe and immunogenic in ferrets. Vaccines 2018, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Astill, J.; Alkie, T.; Yitbarek, A.; Taha-Abdelaziz, K.; Bavananthasivam, J.; Nagy, É.; Petrik, J.J.; Sharif, S. Examination of the effects of virus inactivation methods on the induction of antibody- and cell-mediated immune responses against whole inactivated H9N2 avian influenza virus vaccines in chickens. Vaccine 2018, 36, 3908–3916. [Google Scholar] [CrossRef]
- Ferreira, H.L.; Pirlot, J.F.; Reynard, F.; Van Den Berg, T.; Bublot, M.; Lambrecht, B. Immune responses and protection against H5N1 highly pathogenic avian influenza virus induced by the newcastle disease virus H5 vaccine in ducks. Avian Dis. 2012, 56, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Sanofi Pasteur. Influenza Virus Vaccine, {H5N1}. FDA. Available online: https://www.fda.gov/vaccines-blood-biologics/vaccines/influenza-virus-vaccine-h5n1-national-stockpile (accessed on 2 December 2022).
- EMA Pandemic influenza vaccine {H5N1} {AstraZeneca} (previously Pandemic influenza vaccine {H5N1} Medimmune). Eur. Med. Agency 2018.
- Babu, T.M.; Levine, M.; Fitzgerald, T.; Luke, C.; Sangster, M.Y.; Jin, H.; Topham, D.; Katz, J.; Treanor, J.; Subbarao, K. Live attenuated H7N7 influenza vaccine primes for a vigorous antibody response to inactivated H7N7 influenza vaccine. Vaccine 2014, 32, 6798–6804. [Google Scholar] [CrossRef]
- Lee, D.H.; Park, J.K.; Lee, Y.N.; Song, J.M.; Kang, S.M.; Lee, J.B.; Park, S.Y.; Choi, I.S.; Song, C.S. H9N2 avian influenza virus-like particle vaccine provides protective immunity and a strategy for the differentiation of infected from vaccinated animals. Vaccine 2011, 29, 4003–4007. [Google Scholar] [CrossRef]

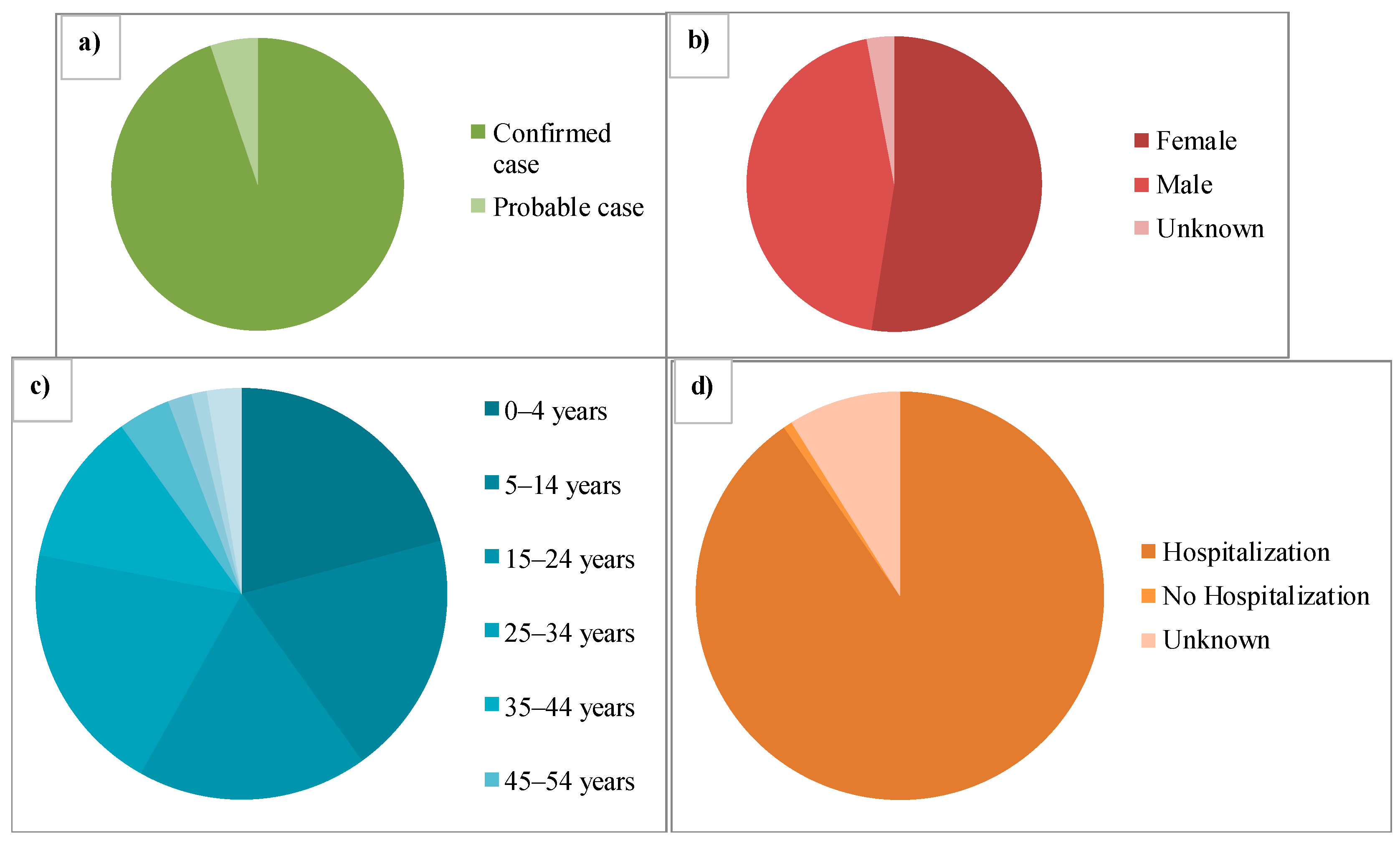
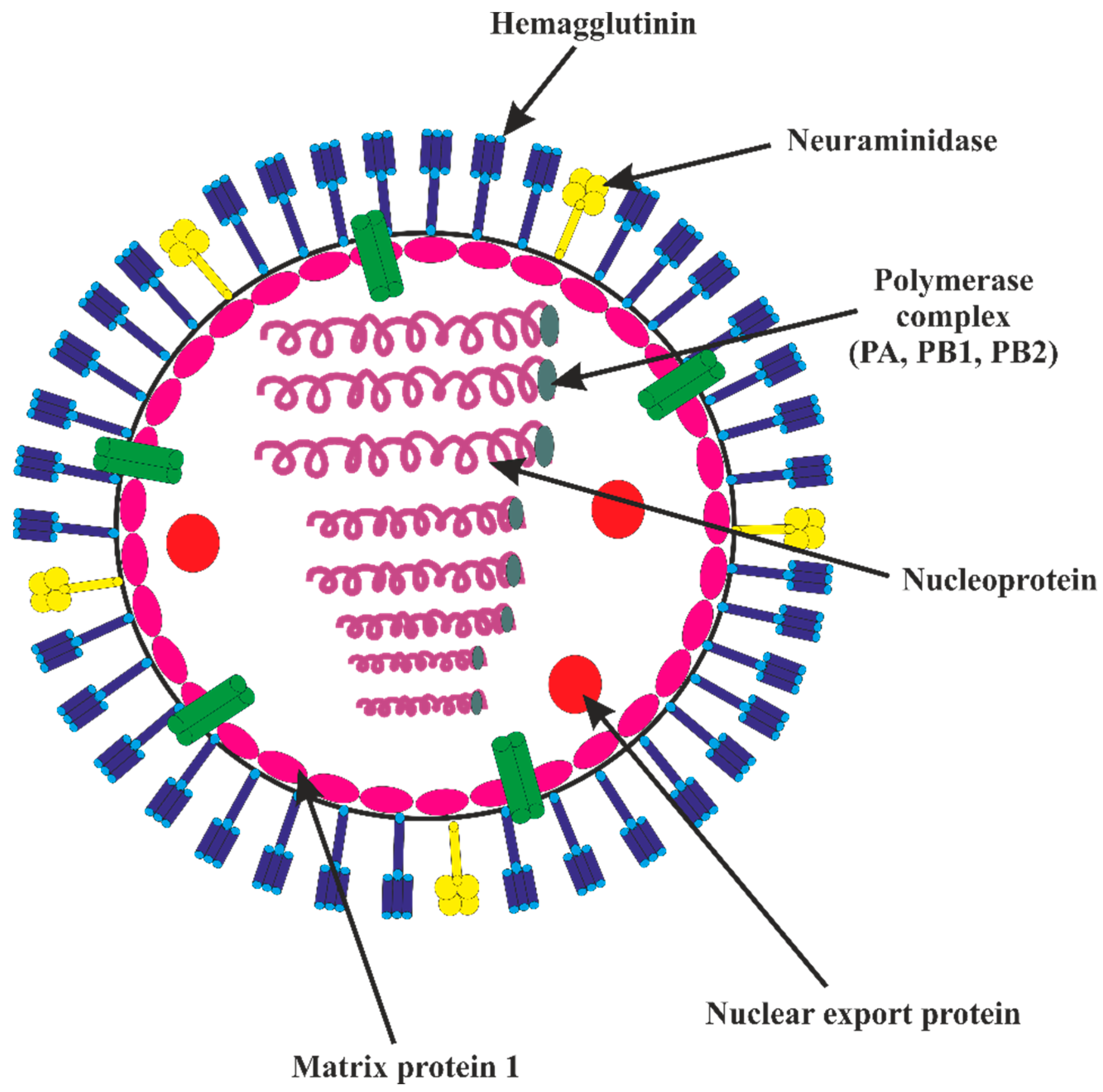
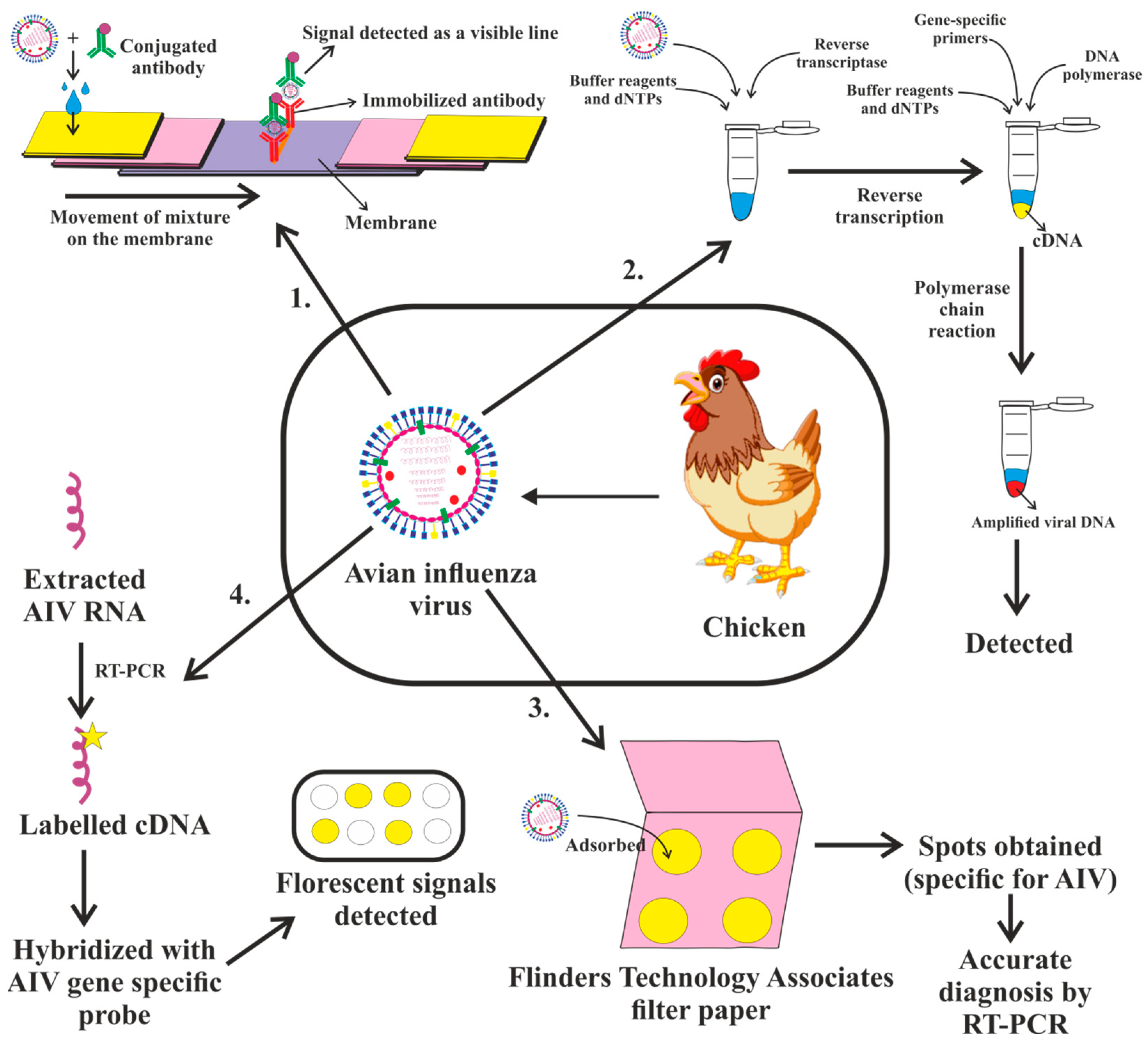
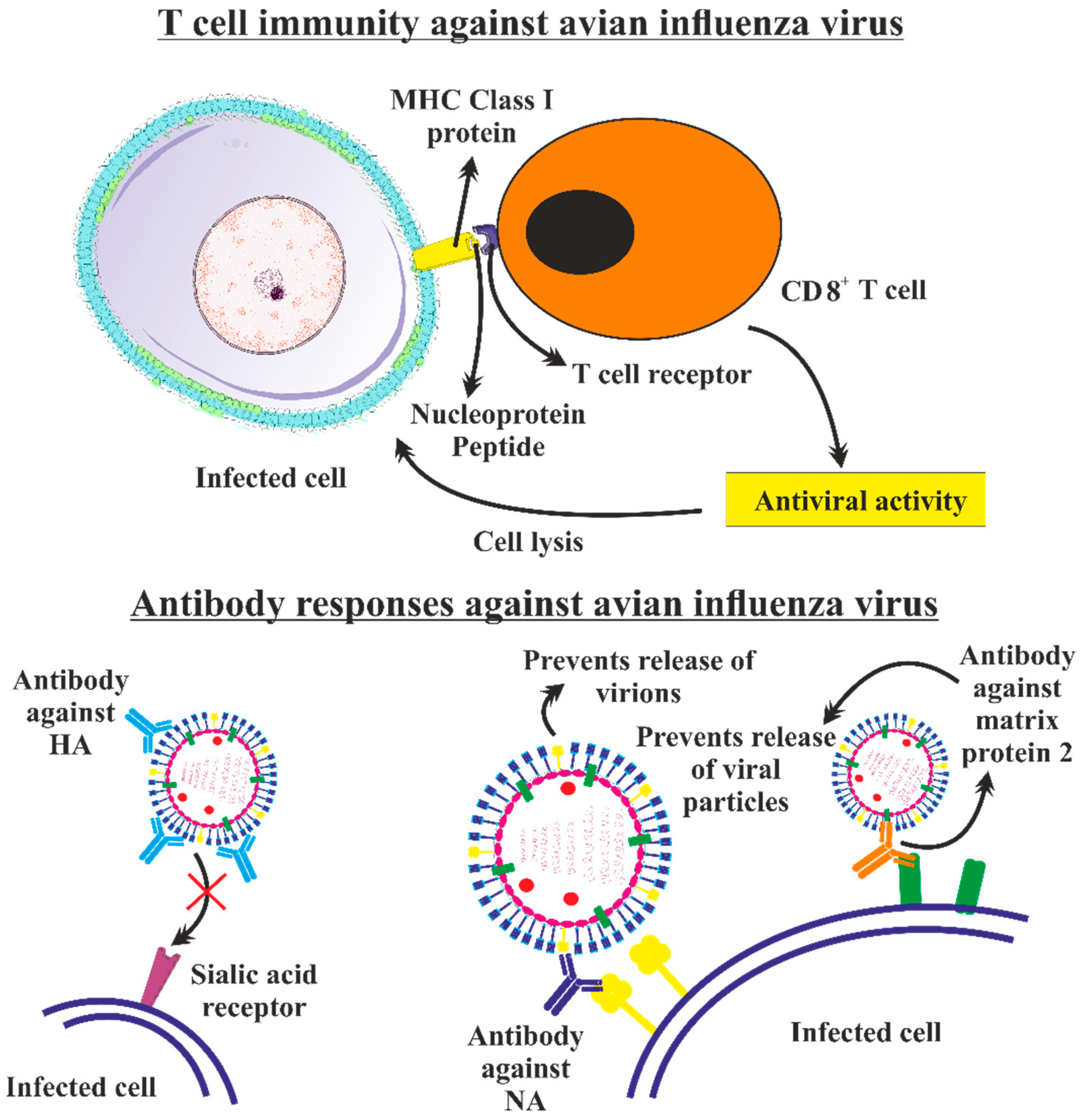
| S. No. | Name of Vaccine | Type of Vaccine | Immune Response | Virus Subtype | Reference |
|---|---|---|---|---|---|
| 1 | H5-Re13, H5-Re-14, H7-Re-4 | Inactivated virus vaccine | Antibody-mediated response | H5N1, H5N6, H5N8, H7N9 | [348] |
| 2 | H5 candidate vaccine strain A/17/turkey/Turkey/05/133 H5N2 | Live-attenuated influenza virus | Antibody-mediated response | H5N2 | [303] |
| 3 | Nobilis Influenza H5N2 vaccine | Inactivated virus vaccine | Antibody-mediated response | H5N2 | [349] |
| 4 | MF59-adjuvanted seasonal influenza vaccine (Fluad®) Novartis Vaccines and Diagnostics Inc., MA, USA | Trivalent inactivated vaccine | Antibody, Cell-mediated response | H5N1 | [350] |
| 5 | AS03-adjuvanted prepandemic H5N1 influenza vaccine | Inactivated virus vaccine | Antibody and cell- mediated response | H5N1 | [351] |
| 6 | H7N9 LAIV | Live-attenuated influenza virus | Antibody-mediated response | N7N9 | [352] |
| 7 | Beta-propriolactone whole-inactivated virus | Inactivated virus vaccine | Antibody and cell-mediated responses | H9N2 | [353] |
| 8 | H5N1 pandemic live-attenuated influenza virus vaccination | Live-attenuated influenza virus | T-cell-mediated response | H5N1 | [310] |
| 9 | Newcastle Disease Virus H5 vaccine | Vector-based vaccine | Antibody, mucosal and cell-mediated response | H5N1 | [354] |
| 10 | H5N1 influenza virus vaccine (Manufactured by: Sanofi Pasteur, Inc.) | Inactivated monovalent influenza virus vaccine | Antibody-mediated response | H5N1 | [355] |
| 11 | Pandemic influenza vaccine H5N1 Astrazeneca | Live-attenuated influenza virus | Antibody-mediated response | H5N1 | [356] |
| 12 | H7 pandemic live-attenuated influenza vaccines (pLAIV) | Live-attenuated influenza virus | Antibody-mediated response | H7N7 | [357] |
| 13 | H9N2 avian influenza virus-like particle vaccine | Virus-like particle vaccine | Antibody and cell-mediated responses | H9N2 | [358] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dey, P.; Ahuja, A.; Panwar, J.; Choudhary, P.; Rani, S.; Kaur, M.; Sharma, A.; Kaur, J.; Yadav, A.K.; Sood, V.; et al. Immune Control of Avian Influenza Virus Infection and Its Vaccine Development. Vaccines 2023, 11, 593. https://doi.org/10.3390/vaccines11030593
Dey P, Ahuja A, Panwar J, Choudhary P, Rani S, Kaur M, Sharma A, Kaur J, Yadav AK, Sood V, et al. Immune Control of Avian Influenza Virus Infection and Its Vaccine Development. Vaccines. 2023; 11(3):593. https://doi.org/10.3390/vaccines11030593
Chicago/Turabian StyleDey, Piyush, Akanksha Ahuja, Jaishal Panwar, Poonam Choudhary, Shital Rani, Mandeep Kaur, Akanksha Sharma, Jatinder Kaur, Ashok Kumar Yadav, Vikas Sood, and et al. 2023. "Immune Control of Avian Influenza Virus Infection and Its Vaccine Development" Vaccines 11, no. 3: 593. https://doi.org/10.3390/vaccines11030593
APA StyleDey, P., Ahuja, A., Panwar, J., Choudhary, P., Rani, S., Kaur, M., Sharma, A., Kaur, J., Yadav, A. K., Sood, V., Suresh Babu, A. R., Bhadada, S. K., Singh, G., & Barnwal, R. P. (2023). Immune Control of Avian Influenza Virus Infection and Its Vaccine Development. Vaccines, 11(3), 593. https://doi.org/10.3390/vaccines11030593






