Establish a Pregnant Sow–Neonate Model to Assess Maternal Immunity of a Candidate Influenza Vaccine
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Pig Experiment
2.3. Quantitative RT-PCR
2.4. Hemagglutination Inhibition (HAI)
2.5. Cellular Cytokine Gene Detection
2.6. Flow Cytometry Analysis
2.7. Statistical Analysis
3. Results
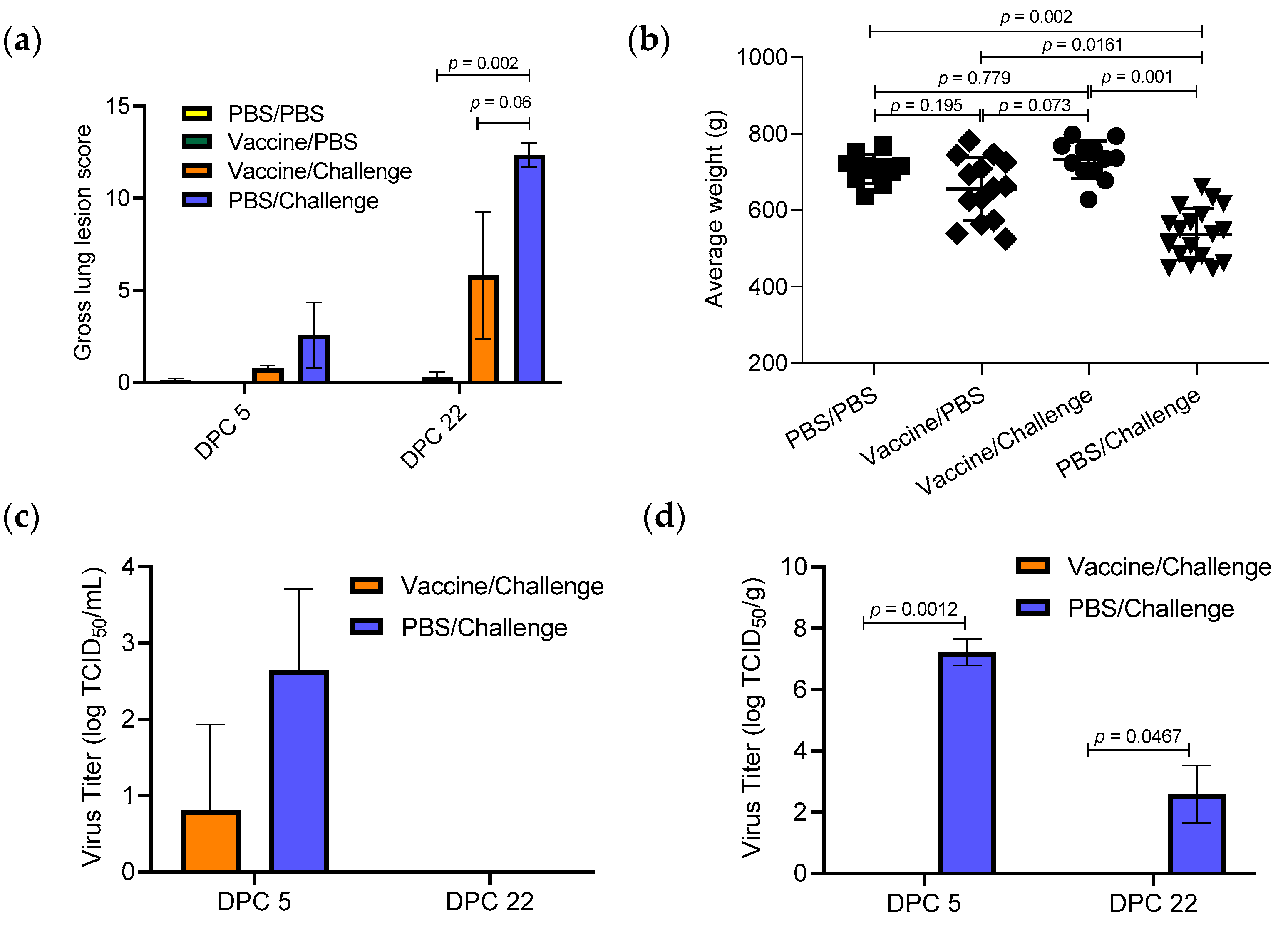
3.1. Effect of TX98-129 Vaccine on Protecting Pregnant Sows against Influenza Virus Infection
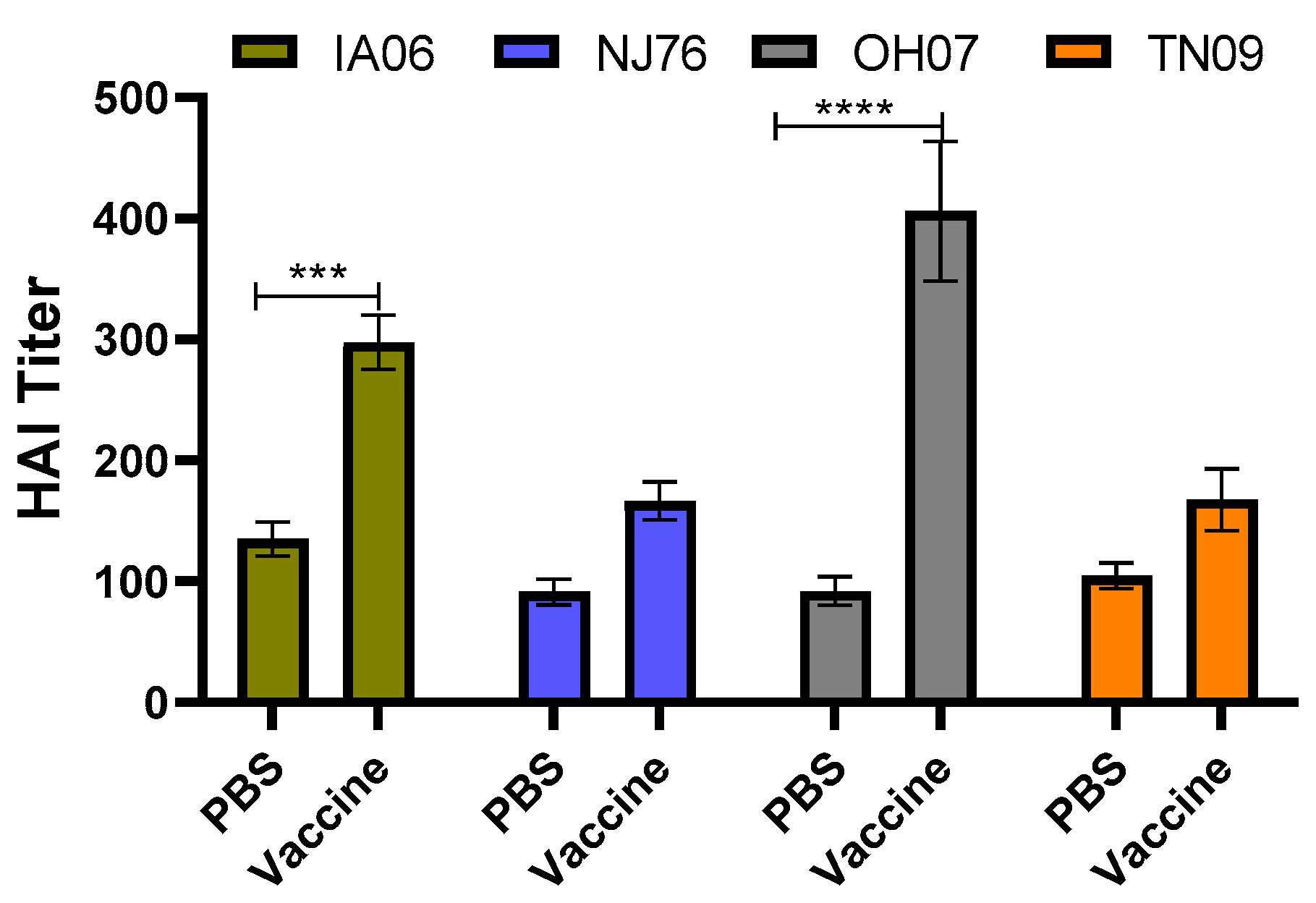
3.2. Protective Immunity Induced by TX98-129 Vaccine in Pregnant Sows
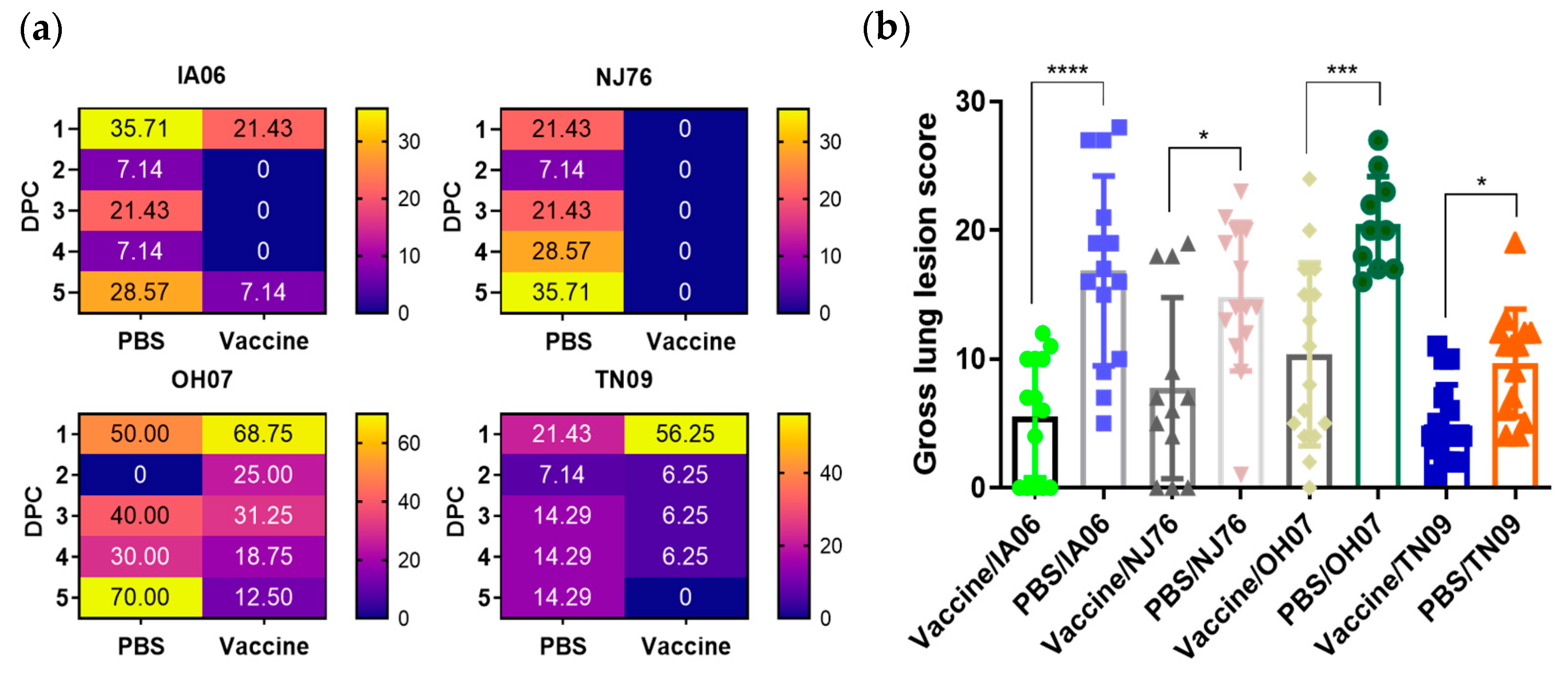
3.3. Effect of Passive Maternal Immunity on Protecting Neonatal Piglets against Influenza Virus Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019, 19, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Mor, G.; Cardenas, I. The immune system in pregnancy: A unique complexity. Am. J. Reprod. Immunol 2010, 63, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Moniz, M.H.; Beigi, R.H. Maternal immunization. Clinical experiences, challenges, and opportunities in vaccine acceptance. Hum. Vaccines Immunother. 2014, 10, 2562–2570. [Google Scholar] [CrossRef] [PubMed]
- van Riel, D.; Mittrücker, H.-W.; Engels, G.; Klingel, K.; Markert, U.R.; Gabriel, G. Influenza pathogenicity during pregnancy in women and animal models. Semin. Immunopathol. 2016, 38, 719–726. [Google Scholar] [CrossRef]
- Rasmussen, S.A.; Jamieson, D.J. 2009 H1N1 Influenza and Pregnancy—5 Years Later. N. Engl. J. Med. 2014, 371, 1373–1375. [Google Scholar] [CrossRef]
- Zaman, K.; Roy, E.; Arifeen, S.E.; Rahman, M.; Raqib, R.; Wilson, E.; Omer, S.B.; Shahid, N.S.; Breiman, R.F.; Steinhoff, M.C. Effectiveness of Maternal Influenza Immunization in Mothers and Infants. N. Engl. J. Med. 2008, 359, 1555–1564. [Google Scholar] [CrossRef]
- Tamma, P.D.; Ault, K.A.; del Rio, C.; Steinhoff, M.C.; Halsey, N.A.; Omer, S.B. Safety of influenza vaccination during pregnancy. Am. J. Obstet. Gynecol. 2009, 201, 547–552. [Google Scholar] [CrossRef]
- Corbeau, M.; Mulliez, A.; Chenaf, C.; Eschalier, B.; Lesens, O.; Vorilhon, P. Trends of influenza vaccination coverage in pregnant women: A ten-year analysis from a French healthcare database. Sci. Rep. 2022, 12, 7153. [Google Scholar] [CrossRef]
- Rasmussen, S.A.; Jamieson, D.J.; Bresee, J.S. Pandemic influenza and pregnant women. Emerg. Infect. Dis. 2008, 14, 95–100. [Google Scholar] [CrossRef]
- Meurens, F.; Summerfield, A.; Nauwynck, H.; Saif, L.; Gerdts, V. The pig: A model for human infectious diseases. Trends Microbiol. 2012, 20, 50–57. [Google Scholar] [CrossRef]
- McCormick, K.; Jiang, Z.; Zhu, L.; Lawson, S.R.; Langenhorst, R.; Ransburgh, R.; Brunick, C.; Tracy, M.C.; Hurtig, H.R.; Mabee, L.M.; et al. Construction and Immunogenicity Evaluation of Recombinant Influenza A Viruses Containing Chimeric Hemagglutinin Genes Derived from Genetically Divergent Influenza A H1N1 Subtype Viruses. PLoS ONE 2015, 10, e0127649. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Neumann, G.; Kawaoka, Y.; Hobom, G.; Webster Robert, G. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 2000, 97, 6108–6113. [Google Scholar] [CrossRef] [PubMed]
- Chaussee, M.S.; Sandbulte, H.R.; Schuneman, M.J.; DePaula, F.P.; Addengast, L.A.; Schlenker, E.H.; Huber, V.C. Inactivated and live, attenuated influenza vaccines protect mice against influenza:Streptococcus pyogenes super-infections. Vaccine 2011, 29, 3773–3781. [Google Scholar] [CrossRef] [PubMed]
- Huber, V.C.; McCullers, J.A. Live Attenuated Influenza Vaccine Is Safe and Immunogenic in Immunocompromised Ferrets. J. Infect. Dis. 2006, 193, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Halbur, P.G.; Paul, P.S.; Frey, M.L.; Landgraf, J.; Eernisse, K.; Meng, X.J.; Lum, M.A.; Andrews, J.J.; Rathje, J.A. Comparison of the Pathogenicity of Two US Porcine Reproductive and Respiratory Syndrome Virus Isolates with that of the Lelystad Virus. Vet. Pathol. 1995, 32, 648–660. [Google Scholar] [CrossRef]
- Zhang, J.; Harmon, K.M. RNA Extraction from Swine Samples and Detection of Influenza A Virus in Swine by Real-Time RT-PCR. In Animal Influenza Virus; Spackman, E., Ed.; Springer New York: New York, NY, USA, 2014; pp. 277–293. [Google Scholar]
- Cwach, K.T.; Sandbulte, H.R.; Klonoski, J.M.; Huber, V.C. Contribution of murine innate serum inhibitors toward interference within influenza virus immune assays. Influenza Other Respir Viruses 2012, 6, 127–135. [Google Scholar] [CrossRef]
- Li, Y.; Shyu, D.-L.; Shang, P.; Bai, J.; Ouyang, K.; Dhakal, S.; Hiremath, J.; Binjawadagi, B.; Renukaradhya, G.J.; Fang, Y. Mutations in a Highly Conserved Motif of nsp1β Protein Attenuate the Innate Immune Suppression Function of Porcine Reproductive and Respiratory Syndrome Virus. J. Virol. 2016, 90, 3584–3599. [Google Scholar] [CrossRef]
- Erbelding, E.J.; Post, D.J.; Stemmy, E.J.; Roberts, P.C.; Augustine, A.D.; Ferguson, S.; Paules, C.I.; Graham, B.S.; Fauci, A.S. A Universal Influenza Vaccine: The Strategic Plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 2018, 218, 347–354. [Google Scholar] [CrossRef]
- Yap, K.L.; Ada, G.L.; McKenzie, I.F.C. Transfer of specific cytotoxic T lymphocytes protects mice inoculated with influenza virus. Nature 1978, 273, 238–239. [Google Scholar] [CrossRef]
- DiPiazza, A.; Richards, K.; Batarse, F.; Lockard, L.; Zeng, H.; García-Sastre, A.; Albrecht Randy, A.; Sant Andrea, J.; Schultz-Cherry, S. Flow Cytometric and Cytokine ELISpot Approaches To Characterize the Cell-Mediated Immune Response in Ferrets following Influenza Virus Infection. J. Virol. 2016, 90, 7991–8004. [Google Scholar] [CrossRef]
- Vanderven, H.A.; Ana-Sosa-Batiz, F.; Jegaskanda, S.; Rockman, S.; Laurie, K.; Barr, I.; Chen, W.; Wines, B.; Hogarth, P.M.; Lambe, T. What lies beneath: Antibody dependent natural killer cell activation by antibodies to internal influenza virus proteins. EBioMedicine 2016, 8, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Jegaskanda, S.; Co, M.D.T.; Cruz, J.; Subbarao, K.; Ennis, F.A.; Terajima, M. Induction of H7N9-Cross-Reactive Antibody-Dependent Cellular Cytotoxicity Antibodies by Human Seasonal Influenza A Viruses that are Directed Toward the Nucleoprotein. J. Infect. Dis. 2017, 215, 818–823. [Google Scholar] [CrossRef] [PubMed]
- Hartshorn, K.L. Innate Immunity and Influenza A Virus Pathogenesis: Lessons for COVID-19. Front. Cell. Infect. Microbiol. 2020, 10, 563850. [Google Scholar] [CrossRef] [PubMed]
- García-Sastre, A. Mechanisms of inhibition of the host interferon α/β-mediated antiviral responses by viruses. Microbes Infect. 2002, 4, 647–655. [Google Scholar] [CrossRef] [PubMed]
- García-Sastre, A.; Durbin Russell, K.; Zheng, H.; Palese, P.; Gertner, R.; Levy David, E.; Durbin Joan, E. The Role of Interferon in Influenza Virus Tissue Tropism. J. Virol. 1998, 72, 8550–8558. [Google Scholar] [CrossRef]
- Koerner, I.; Kochs, G.; Kalinke, U.; Weiss, S.; Staeheli, P. Protective Role of Beta Interferon in Host Defense against Influenza A Virus. J. Virol. 2007, 81, 2025–2030. [Google Scholar] [CrossRef]
- Iwasaki, A.; Pillai, P.S. Innate immunity to influenza virus infection. Nat. Rev. Immunol. 2014, 14, 315–328. [Google Scholar] [CrossRef]
- Forbes, R.L.; Wark, P.A.B.; Murphy, V.E.; Gibson, P.G. Pregnant Women Have Attenuated Innate Interferon Responses to 2009 Pandemic Influenza A Virus Subtype H1N1. J. Infect. Dis. 2012, 206, 646–653. [Google Scholar] [CrossRef]
- Tavares, L.P.; Teixeira, M.M.; Garcia, C.C. The inflammatory response triggered by Influenza virus: A two edged sword. Inflamm. Res. 2017, 66, 283–302. [Google Scholar] [CrossRef]
- Gu, Y.; Zuo, X.; Zhang, S.; Ouyang, Z.; Jiang, S.; Wang, F.; Wang, G. The Mechanism behind Influenza Virus Cytokine Storm. Viruses 2021, 13, 1362. [Google Scholar] [CrossRef]
- Loo, Y.-M.; Gale, M. Fatal immunity and the 1918 virus. Nature 2007, 445, 267–268. [Google Scholar] [CrossRef] [PubMed]
- Kobasa, D.; Jones, S.M.; Shinya, K.; Kash, J.C.; Copps, J.; Ebihara, H.; Hatta, Y.; Hyun Kim, J.; Halfmann, P.; Hatta, M.; et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 2007, 445, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Sperling, R.S.; Engel, S.M.; Wallenstein, S.; Kraus, T.A.; Garrido, J.; Singh, T.; Kellerman, L.; Moran, T.M. Immunogenicity of trivalent inactivated influenza vaccination received during pregnancy or postpartum. Obs. Gynecol. 2012, 119, 631–639. [Google Scholar] [CrossRef]
- Healy, C.M.; Rench, M.A.; Swaim, L.S.; Smith, E.O.B.; Sangi-Haghpeykar, H.; Mathis, M.H.; Martin, M.D.; Baker, C.J. Association Between Third-Trimester Tdap Immunization and Neonatal Pertussis Antibody Concentration. JAMA 2018, 320, 1464–1470. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Hisano, M.; Isojima, S.; Irie, S.; Arata, N.; Watanabe, N.; Kubo, T.; Kato, T.; Murashima, A. Relationship of Th1/Th2 cell balance with the immune response to influenza vaccine during pregnancy. J. Med. Virol. 2009, 81, 1923–1928. [Google Scholar] [CrossRef] [PubMed]
- Gozde Kanmaz, H.; Erdeve, O.; Suna Oğz, S.; Uras, N.; Çelen, Ş.; Korukluoglu, G.; Zergeroglu, S.; Kara, A.; Dilmen, U. Placental Transmission of Novel Pandemic Influenza a Virus. Fetal Pediatr. Pathol. 2011, 30, 280–285. [Google Scholar] [CrossRef]
- Liong, S.; Oseghale, O.; To Eunice, E.; Brassington, K.; Erlich Jonathan, R.; Luong, R.; Liong, F.; Brooks, R.; Martin, C.; O’Toole, S.; et al. Influenza A virus causes maternal and fetal pathology via innate and adaptive vascular inflammation in mice. Proc. Natl. Acad. Sci. USA 2020, 117, 24964–24973. [Google Scholar] [CrossRef]
- Littauer, E.Q.; Esser, E.S.; Antao, O.Q.; Vassilieva, E.V.; Compans, R.W.; Skountzou, I. H1N1 influenza virus infection results in adverse pregnancy outcomes by disrupting tissue-specific hormonal regulation. PLoS Pathog. 2017, 13, e1006757. [Google Scholar] [CrossRef]
- Lieberman, R.W.; Bagdasarian, N.; Thomas, D.; Van De Ven, C. Seasonal influenza A (H1N1) infection in early pregnancy and second trimester fetal demise. Emerg. Infect. Dis. 2011, 17, 107–109. [Google Scholar] [CrossRef]
- Beato, M.S.; Capua, I. Transboundary spread of highly pathogenic avian influenza through poultry commodities and wild birds: A review. Rev. Sci. Tech. Off. Int. Epizoot. 2011, 30, 51–61. [Google Scholar] [CrossRef]



| Group | Vaccine (60, 75 DG) | Challenge Virus (90 DG) | Fetus Collection (DG) |
|---|---|---|---|
| A (n = 4) | PBS | PBS | 95 (n = 2) 112 (n = 2) |
| B (n = 4) | TX98-129 | PBS | 95 (n = 2) 112 (n = 2) |
| C (n = 4) | TX98-129 | IL13 | 95 (n = 2) 112 (n = 2) |
| D (n = 4) | PBS | IL13 | 95 (n = 2) 112 (n = 2) |
| Group | Vaccine in Sows (70, 90 DG) | Number of Piglets after Farrowing (115 or 116 DG) | Challenge Virus in Neonatal Piglets (6 DPF) |
|---|---|---|---|
| 1 | PBS | 14 | TX98-IA06 |
| 2 | PBS | 14 | TX98-NJ76 |
| 3 | PBS | 10 | TX98-OH07 |
| 4 | PBS | 14 | TX98-TN09 |
| 5 | TX98-129 | 14 | TX98-IA06 |
| 6 | TX98-129 | 12 | TX98-NJ76 |
| 7 | TX98-129 | 16 | TX98-OH07 |
| 8 | TX98-129 | 16 | TX98-TN09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, F.; Schieber, T.; Stein, T.L.; Sestak, R.M.; Olson, C.J.; Chen, C.; Huber, V.C.; Lechtenberg, K.; McGill, J.; Fang, Y. Establish a Pregnant Sow–Neonate Model to Assess Maternal Immunity of a Candidate Influenza Vaccine. Vaccines 2023, 11, 646. https://doi.org/10.3390/vaccines11030646
Yuan F, Schieber T, Stein TL, Sestak RM, Olson CJ, Chen C, Huber VC, Lechtenberg K, McGill J, Fang Y. Establish a Pregnant Sow–Neonate Model to Assess Maternal Immunity of a Candidate Influenza Vaccine. Vaccines. 2023; 11(3):646. https://doi.org/10.3390/vaccines11030646
Chicago/Turabian StyleYuan, Fangfeng, Teresa Schieber, Tara L. Stein, Rachel M. Sestak, Callie J. Olson, Chi Chen, Victor C. Huber, Kelly Lechtenberg, Jodi McGill, and Ying Fang. 2023. "Establish a Pregnant Sow–Neonate Model to Assess Maternal Immunity of a Candidate Influenza Vaccine" Vaccines 11, no. 3: 646. https://doi.org/10.3390/vaccines11030646
APA StyleYuan, F., Schieber, T., Stein, T. L., Sestak, R. M., Olson, C. J., Chen, C., Huber, V. C., Lechtenberg, K., McGill, J., & Fang, Y. (2023). Establish a Pregnant Sow–Neonate Model to Assess Maternal Immunity of a Candidate Influenza Vaccine. Vaccines, 11(3), 646. https://doi.org/10.3390/vaccines11030646





