An Overview of Indian Biomedical Research on the Chikungunya Virus with Particular Reference to Its Vaccine, an Unmet Medical Need
Abstract
:1. Introduction
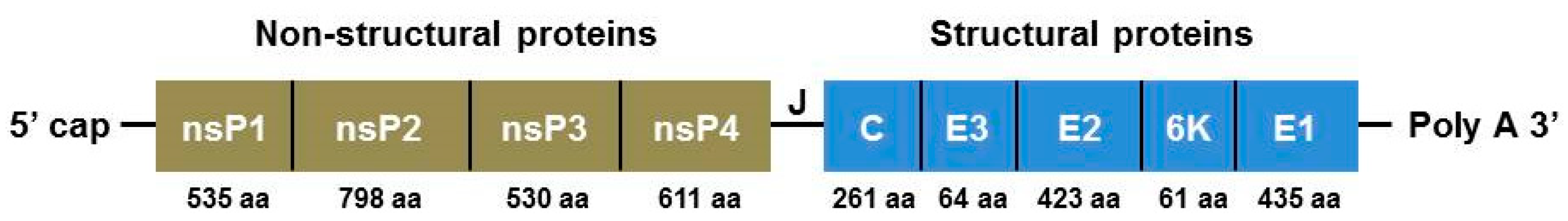
2. Organization of the CHIKV Genome
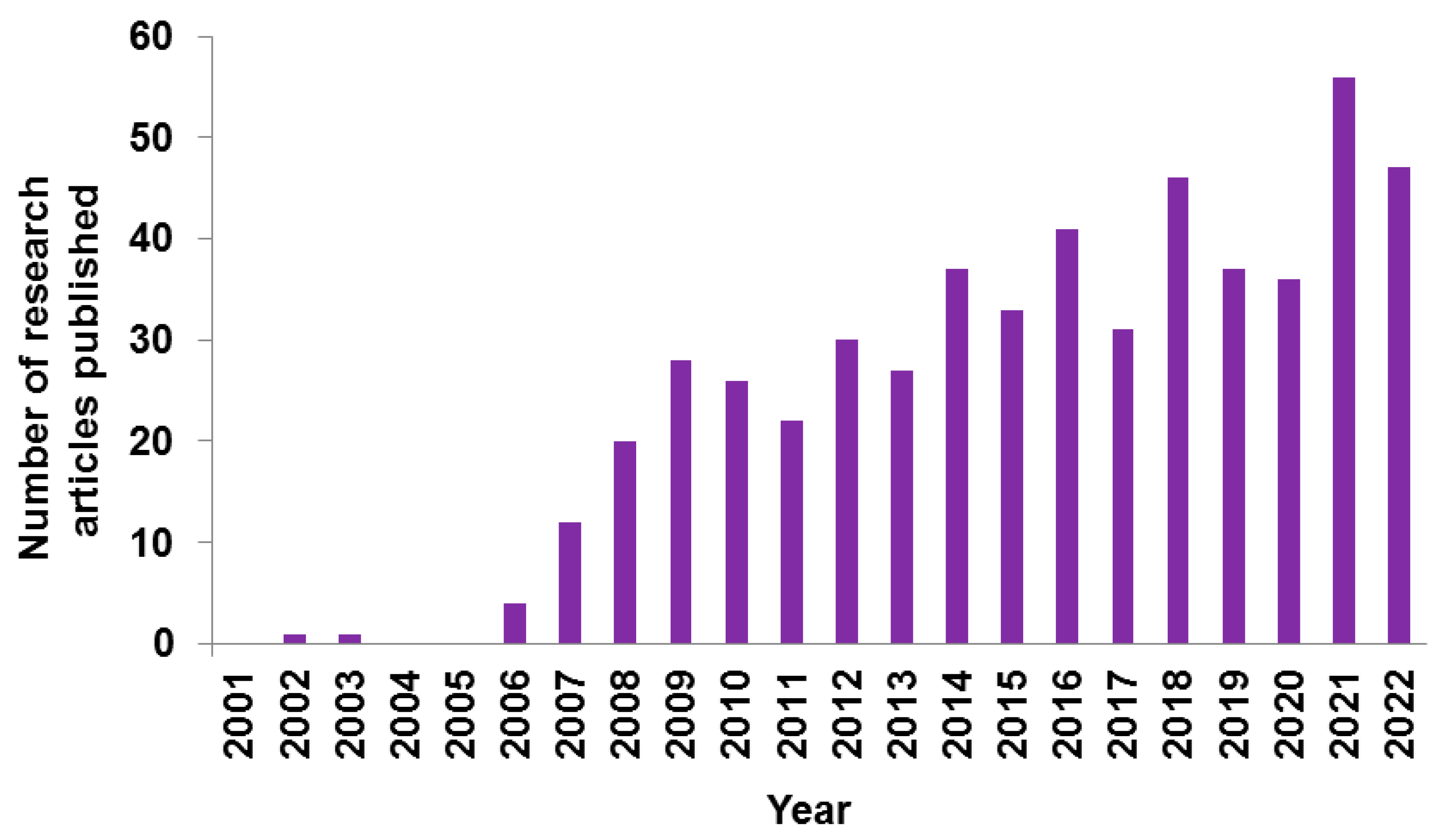
3. CHIKV Research: Indian Scenario
3.1. Epidemiological Profile of CHIKV in India
3.2. Genomic and Proteomic Aspects of CHIKV
3.3. Diagnostics, Treatment, and Vaccines for CHIKV
3.3.1. Diagnostics
3.3.2. Treatment
3.3.3. Vaccine
3.4. Biotechnological Aspects of CHIKV
4. Research Priorities and Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schmidt, C.; Schnierle, B.S. Chikungunya Vaccine Candidates: Current Landscape and Future Prospects. Drug. Des. Dev. Ther. 2022, 16, 3663–3673. [Google Scholar] [CrossRef]
- Montalvo Zurbia-Flores, G.; Reyes-Sandoval, A.; Kim, Y.C. Chikungunya Virus: Priority Pathogen or Passing Trend? Vaccines 2023, 11, 568. [Google Scholar] [CrossRef]
- Sergon, K.; Njuguna, C.; Kalani, R.; Ofula, V.; Onyango, C.; Konongoi, L.S.; Bedno, S.; Burke, H.; Dumilla, A.M.; Konde, J. Seroprevalence of chikungunya virus (CHIKV) infection on Lamu Island, Kenya, October 2004. Am. J. Trop. Med. Hyg. 2008, 78, 333–337. [Google Scholar] [CrossRef]
- Amdekar, S.; Parashar, D.; Alagarasu, K. Chikungunya virus-induced arthritis: Role of host and viral factors in the pathogenesis. Viral Immunol. 2017, 30, 691–702. [Google Scholar] [CrossRef]
- Saxena, S.K.; Swamy, M.A.; Verma, S.K.; Saxena, R. The intolerable burden of Chikungunya: What’s new, what’s needed? Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2012, 82, 153–165. [Google Scholar] [CrossRef]
- Saxena, S.K. Re-emergence of the knotty chikungunya virus: Facts, fear or fiction. Future Virol. 2007, 2, 121. [Google Scholar] [CrossRef]
- Teixeira, M.G.; Andrade, A.M.; Maria da Conceição, N.C.; Castro, J.S.; Oliveira, F.L.; Goes, C.S.; Maia, M.; Santana, E.B.; Nunes, B.T.; Vasconcelos, P.F. East/Central/South african genotype chikungunya virus, Brazil, 2014. Emerg. Infect. Dis. 2015, 21, 906. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.C. An epidemic of virus disease in Southern Province, Tanganyika territory, in 1952–1953. Trans. R. Soc. Trop. Med. Hyg. 1955, 49, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Burt, F.J.; Chen, W.; Miner, J.J.; Lenschow, D.J.; Merits, A.; Schnettler, E.; Kohl, A.; Rudd, P.A.; Taylor, A.; Herrero, L.J. Chikungunya virus: An update on the biology and pathogenesis of this emerging pathogen. Lancet Infect. Dis. 2017, 17, e107–e117. [Google Scholar] [CrossRef] [PubMed]
- Eisenhut, M.; Schwarz, T.; Hegenscheid, B. Seroprevalence of dengue, chikungunya and Sindbis virus infections in German aid workers. Infection 1999, 27, 82. [Google Scholar] [CrossRef]
- Kuniholm, M.H.; Wolfe, N.D.; Huang, C.Y.-h.; Mpoudi-Ngole, E.; Tamoufe, U.; Burke, D.S.; Gubler, D.J. Seroprevalence and distribution of Flaviviridae, Togaviridae, and Bunyaviridae arboviral infections in rural Cameroonian adults. Am. J. Trop. Med. Hyg. 2006, 74, 1078–1083. [Google Scholar] [CrossRef]
- Peyrefitte, C.N.; Rousset, D.; Pastorino, B.A.; Pouillot, R.; Bessaud, M.; Tock, F.; Mansaray, H.; Merle, O.L.; Pascual, A.M.; Paupy, C. Chikungunya virus, Cameroon, 2006. Emerg. Infect. Dis. 2007, 13, 768. [Google Scholar] [CrossRef] [PubMed]
- Peyrefitte, C.N.; Bessaud, M.; Pastorino, B.A.; Gravier, P.; Plumet, S.; Merle, O.L.; Moltini, I.; Coppin, E.; Tock, F.; Daries, W. Circulation of Chikungunya virus in Gabon, 2006–2007. J. Med. Virol. 2008, 80, 430–433. [Google Scholar] [CrossRef] [PubMed]
- Jentes, E.S.; Robinson, J.; Johnson, B.W.; Conde, I.; Sakouvougui, Y.; Iverson, J.; Beecher, S.; Diakite, F.; Coulibaly, M.; Bausch, D.G. Acute arboviral infections in Guinea, west Africa, 2006. Am. J. Trop. Med. Hyg. 2010, 83, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Pistone, T.; Ezzedine, K.; Schuffenecker, I.; Receveur, M.-C.; Malvy, D. An imported case of Chikungunya fever from Madagascar: Use of the sentinel traveller for detecting emerging arboviral infections in tropical and European countries. Travel. Med. Infect. Dis. 2009, 7, 52–54. [Google Scholar] [CrossRef]
- Moore, D.á.; Causey, O.; Carey, D.; Reddy, S.; Cooke, A.; Akinkugbe, F.; David-West, T.; Kemp, G. Arthropod-borne viral infections of man in Nigeria, 1964–1970. Ann. Trop. Med. Parasitol. 1975, 69, 49–64. [Google Scholar] [CrossRef]
- Pistone, T.; Ezzedine, K.; Boisvert, M.; Receveur, M.C.; Schuffenecker, I.; Zeller, H.; Lafon, M.E.; Fleury, H.; Malvy, D. Cluster of chikungunya virus infection in travelers returning from Senegal, 2006. J. Travel Med. 2009, 16, 286–288. [Google Scholar] [CrossRef]
- Lahariya, C.; Pradhan, S. Emergence of chikungunya virus in Indian subcontinent after 32 years: A review. J. Vector Borne Dis. 2006, 43, 151. [Google Scholar]
- Mackenzie, J.; Chua, K.; Daniels, P.; Eaton, B.; Field, H.; Hall, R.; Halpin, K.; Johansen, C.; Kirkland, P.; Lam, S. Emerging viral diseases of Southeast Asia and the Western Pacific. Emerg. Infect. Dis. 2001, 7, 497. [Google Scholar] [CrossRef]
- Pavri, K.; Banerjee, G.; Anderson, C.; Aikat, B. Virological and serological studies of cases of haemorrhagic fever in Calcutta: Material collected by the Institute of Post-graduate Medical Education and Research (IPGME), Calcutta. Indian J. Med. Res. 1964, 52, 692–697. [Google Scholar]
- Wangchuk, S.; Chinnawirotpisan, P.; Dorji, T.; Tobgay, T.; Dorji, T.; Yoon, I.-K.; Fernandez, S. Chikungunya fever outbreak, Bhutan, 2012. Emerg. Infect. Dis. 2013, 19, 1681. [Google Scholar] [CrossRef] [PubMed]
- Rezza, G.; El-Sawaf, G.; Faggioni, G.; Vescio, F.; Al Ameri, R.; De Santis, R.; Helaly, G.; Pomponi, A.; Metwally, D.; Fantini, M. Co-circulation of dengue and chikungunya viruses, Al Hudaydah, Yemen, 2012. Emerg. Infect. Dis. 2014, 20, 1351. [Google Scholar] [CrossRef]
- Hussain, R.; Alomar, I.; Memish, Z.A. Chikungunya virus: Emergence of an arthritic arbovirus in Jeddah, Saudi Arabia. East. Mediterr. Health J. 2013, 19, 506–508. [Google Scholar] [CrossRef]
- Tsetsarkin, K.A.; Vanlandingham, D.L.; McGee, C.E.; Higgs, S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007, 3, e201. [Google Scholar] [CrossRef] [PubMed]
- Townson, H.; Nathan, M.B. Resurgence of chikungunya. Trans. R. Soc. Trop. Med. Hyg. 2008, 102, 308–309. [Google Scholar] [CrossRef]
- Parola, P.; De Lamballerie, X.; Jourdan, J.; Rovery, C.; Vaillant, V.; Minodier, P.; Brouqui, P.; Flahault, A.; Raoult, D.; Charrel, R.N. Novel chikungunya virus variant in travelers returning from Indian Ocean islands. Emerg. Infect. Dis. 2006, 12, 1493. [Google Scholar] [CrossRef]
- Weaver, S.C.; Forrester, N.L. Chikungunya: Evolutionary history and recent epidemic spread. Antivir. Res. 2015, 120, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Sudeep, A.; Parashar, D. Chikungunya: An overview. J. Biosci. 2008, 33, 443–449. [Google Scholar] [CrossRef]
- Mavalankar, D.; Shastri, P.; Raman, P. Chikungunya epidemic in India: A major public-health disaster. Lancet Infect. Dis. 2007, 7, 306–307. [Google Scholar] [CrossRef]
- Nunes, M.R.T.; Faria, N.R.; de Vasconcelos, J.M.; Golding, N.; Kraemer, M.U.; de Oliveira, L.F.; da Silva Azevedo, R.d.S.; da Silva, D.E.A.; da Silva, E.V.P.; da Silva, S.P. Emergence and potential for spread of Chikungunya virus in Brazil. BMC Med. 2015, 13, 102. [Google Scholar] [CrossRef]
- Van Genderen, F.T.; Krishnadath, I.; Sno, R.; Grunberg, M.G.; Zijlmans, W.; Adhin, M.R. First chikungunya outbreak in Suriname; clinical and epidemiological features. PLoS Negl. Trop. Dis. 2016, 10, e0004625. [Google Scholar] [CrossRef]
- Madariaga, M.; Ticona, E.; Resurrecion, C. Chikungunya: Bending over the Americas and the rest of the world. Braz. J. Infect. Dis. 2016, 20, 91–98. [Google Scholar] [CrossRef]
- Carbajo, A.E.; Vezzani, D. Waiting for chikungunya fever in Argentina: Spatio-temporal risk maps. Memórias Do Inst. Oswaldo Cruz 2015, 110, 259–262. [Google Scholar] [CrossRef]
- Mattar, S.; Miranda, J.; Pinzon, H.; Tique, V.; Bolaños, A.; Aponte, J.; Arrieta, G.; Gonzalez, M.; Barrios, K.; Contreras, H. Outbreak of Chikungunya virus in the north Caribbean area of Colombia: Clinical presentation and phylogenetic analysis. J. Infect. Dev. Ctries. 2015, 9, 1126–1132. [Google Scholar] [CrossRef] [PubMed]
- Díaz, Y.; Carrera, J.-P.; Cerezo, L.; Arauz, D.; Guerra, I.; Cisneros, J.; Armién, B.; Botello, A.M.; Araúz, A.B.; Gonzalez, V. Chikungunya virus infection: First detection of imported and autochthonous cases in Panama. Am. J. Trop. Med. Hyg. 2015, 92, 482–485. [Google Scholar] [CrossRef]
- Nava-Frías, M.; Searcy-Pavía, R.E.; Juárez-Contreras, C.A.; Valencia-Bautista, A. Chikungunya fever: Current status in Mexico. Boletín Médico Hosp. Infant. México 2016, 73, 67–74. [Google Scholar]
- Wahid, B.; Ali, A.; Rafique, S.; Idrees, M. Global expansion of chikungunya virus: Mapping the 64-year history. Int. J. Infect. Dis. 2017, 58, 69–76. [Google Scholar] [CrossRef] [PubMed]
- National Institute of Allergy and Infectious Diseases. NIAID Emerging Infectious Diseases/Pathogens. Available online: https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens (accessed on 17 April 2023).
- World Health Organization. Annual Review of Diseases Prioritized under the Research and Development Blueprint. Available online: https://www.who.int/docs/default-source/blue-print/first-annual-review-of-diseases-prioritized-under-r-and-d-blueprint.pdf?sfvrsn=1f6b5da0_4 (accessed on 27 April 2023).
- Silva, L.A.; Dermody, T.S. Chikungunya virus: Epidemiology, replication, disease mechanisms, and prospective intervention strategies. J. Clin. Investig. 2017, 127, 737–749. [Google Scholar] [CrossRef]
- Powers, A.M. Chikungunya. Clin. Lab. Med. 2010, 30, 209–219. [Google Scholar] [CrossRef]
- Konishi, E.; Hotta, S. Studies on Structural Proteins of Chikungunya Virus: I. Separation of Three Species of Proteins and Their Preliminary Characterization. Microbiol. Immunol. 1980, 24, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Simizu, B.; Yamamoto, K.; Hashimoto, K.; Ogata, T. Structural proteins of Chikungunya virus. J. Virol. 1984, 51, 254–258. [Google Scholar] [CrossRef]
- Fros, J.J.; Liu, W.J.; Prow, N.A.; Geertsema, C.; Ligtenberg, M.; Vanlandingham, D.L.; Schnettler, E.; Vlak, J.M.; Suhrbier, A.; Khromykh, A.A. Chikungunya virus nonstructural protein 2 inhibits type I/II interferon-stimulated JAK-STAT signaling. J. Virol. 2010, 84, 10877–10887. [Google Scholar] [CrossRef]
- Strauss, J.H.; Strauss, E.G. The alphaviruses: Gene expression, replication, and evolution. Microbiol. Mol. Biol. Rev. 1994, 58, 491–562. [Google Scholar] [CrossRef] [PubMed]
- Metz, S.W.; Gardner, J.; Geertsema, C.; Le, T.T.; Goh, L.; Vlak, J.M.; Suhrbier, A.; Pijlman, G.P. Effective chikungunya virus-like particle vaccine produced in insect cells. PLoS Negl. Trop. Dis. 2013, 7, e2124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kielian, M.; Rey, F.A. Virus membrane-fusion proteins: More than one way to make a hairpin. Nat. Rev. Microbiol. 2006, 4, 67. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, R. Togaviridae: The viruses and their replication. Fields Virol. 2007, 1, 1001–1022. [Google Scholar]
- Metz, S.W.; Geertsema, C.; Martina, B.E.; Andrade, P.; Heldens, J.G.; van Oers, M.M.; Goldbach, R.W.; Vlak, J.M.; Pijlman, G.P. Functional processing and secretion of Chikungunya virus E1 and E2 glycoproteins in insect cells. Virol. J. 2011, 8, 353. [Google Scholar] [CrossRef]
- Jose, J.; Snyder, J.E.; Kuhn, R.J. A structural and functional perspective of alphavirus replication and assembly. Future Microbiol. 2009, 4, 837–856. [Google Scholar] [CrossRef]
- Yao, J.S.; Strauss, E.G.; Strauss, J.H. Interactions between PE2, E1, and 6K required for assembly of alphaviruses studied with chimeric viruses. J. Virol. 1996, 70, 7910–7920. [Google Scholar] [CrossRef]
- Lusa, S.; Garoff, H.; Liueström, P. Fate of the 6K membrane protein of Semliki Forest virus during virus assembly. Virology 1991, 185, 843–846. [Google Scholar] [CrossRef]
- Hahon, N.; Zimmerman, W.D. Chikungunya virus infection of cell monolayers by cell-to-cell and extracellular transmission. Appl. Environ. Microbiol. 1970, 19, 389–391. [Google Scholar] [CrossRef] [PubMed]
- Myers, R.; Carey, D.; Reuben, R.; Jesudass, E.; De Ranitz, C.; Jadhav, M. The 1964 epidemic of dengue-like fever in South India: Isolation of chikungunya virus from human sera and from mosquitoes. Indian J. Med. Res. 1965, 53. [Google Scholar]
- Pavri, K. Disappearance of chikungunya virus from India and South East Asia. Trans. R. Soc. Trop. Med. Hyg. 1986, 80, 491. [Google Scholar] [CrossRef] [PubMed]
- Yergolkar, P.N.; Tandale, B.V.; Arankalle, V.A.; Sathe, P.S.; AB, S.; Gandhe, S.S.; Gokhle, M.D.; Jacob, G.P.; Hundekar, S.L.; Mishra, A.C. Chikungunya outbreaks caused by African genotype, India. Emerg. Infect. Dis. 2006, 12, 1580. [Google Scholar] [CrossRef]
- Padbidri, V.; Gnaneswar, T. Epidemiological investigations of chikungunya epidemic at Barsi, Maharashtra state, India. J. Hyg. Epidemiol. Microbiol. Immunol. 1979, 23, 445–451. [Google Scholar]
- Schuffenecker, I.; Iteman, I.; Michault, A.; Murri, S.; Frangeul, L.; Vaney, M.-C.; Lavenir, R.; Pardigon, N.; Reynes, J.-M.; Pettinelli, F. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006, 3, e263. [Google Scholar] [CrossRef]
- Mourya, D.; Yadav, P. Vector biology of dengue & chikungunya viruses. Indian J. Med. Res. 2006, 124, 475. [Google Scholar]
- Naresh Kumar, C.; Sai Gopal, D. Reemergence of chikungunya virus in Indian subcontinent. Indian. J. Virol. 2010, 21, 8–17. [Google Scholar] [CrossRef]
- Kalantri, S.; Joshi, R.; Riley, L.W. Chikungunya epidemic: An Indian perspective. Natl. Med. J. India 2006, 19, 315. [Google Scholar]
- Translational Research Consortia (TRC) for Chikungunya Virus in India. Current Status of Chikungunya in India. Front. Microbiol. 2021, 12, 695173. [Google Scholar] [CrossRef]
- Muniaraj, M. Fading chikungunya fever from India: Beginning of the end of another episode? Indian J. Med. Res. 2014, 139, 468. [Google Scholar]
- Jain, J.; Kushwah, R.B.S.; Singh, S.S.; Sharma, A.; Adak, T.; Singh, O.P.; Bhatnagar, R.K.; Subbarao, S.K.; Sunil, S. Evidence for natural vertical transmission of chikungunya viruses in field populations of Aedes aegypti in Delhi and Haryana states in India—A preliminary report. Acta Trop. 2016, 162, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Dhiman, R.C. Climate change and human health: Indian context. J. Vector Borne Dis. 2012, 49, 55. [Google Scholar]
- Kakarla, S.G.; Mopuri, R.; Mutheneni, S.R.; Bhimala, K.R.; Kumaraswamy, S.; Kadiri, M.R.; Gouda, K.C.; Upadhyayula, S.M. Temperature dependent transmission potential model for chikungunya in India. Sci. Total Environ. 2019, 647, 66–74. [Google Scholar] [CrossRef]
- Department of Health and Family Welfare. Annual Report of Department of Health and Family Welfare 2017–2018, Chapter 5. Available online: https://main.mohfw.gov.in/sites/default/files/05Chapter.pdf (accessed on 20 May 2023).
- Ministry of Health and Family Welfare; Government of India. Integrated Disease Surveillance Programme (IDSP). Available online: https://idsp.mohfw.gov.in/ (accessed on 10 December 2010).
- National Center for Vector Borne Diseases Control. Chikungunya Cases in the Country since 2017. Available online: https://ncvbdc.mohfw.gov.in/index4.php?lang=1&level=0&linkid=486&lid=3765 (accessed on 1 June 2023).
- Kumar, M.S.; Kamaraj, P.; Khan, S.A.; Allam, R.R.; Barde, P.V.; Dwibedi, B.; Kanungo, S.; Mohan, U.; Mohanty, S.S.; Roy, S.; et al. Seroprevalence of chikungunya virus infection in India, 2017: A cross-sectional population-based serosurvey. Lancet Microbe 2021, 2, e41–e47. [Google Scholar] [CrossRef]
- Sreekumar, E.; Issac, A.; Nair, S.; Hariharan, R.; Janki, M.; Arathy, D.; Regu, R.; Mathew, T.; Anoop, M.; Niyas, K. Genetic characterization of 2006–2008 isolates of Chikungunya virus from Kerala, South India, by whole genome sequence analysis. Virus Genes 2010, 40, 14–27. [Google Scholar] [CrossRef]
- Arankalle, V.A.; Shrivastava, S.; Cherian, S.; Gunjikar, R.S.; Walimbe, A.M.; Jadhav, S.M.; Sudeep, A.; Mishra, A.C. Genetic divergence of Chikungunya viruses in India (1963–2006) with special reference to the 2005–2006 explosive epidemic. J. Gen. Virol. 2007, 88, 1967–1976. [Google Scholar] [CrossRef]
- Cherian, S.S.; Walimbe, A.M.; Jadhav, S.M.; Gandhe, S.S.; Hundekar, S.L.; Mishra, A.C.; Arankalle, V.A. Evolutionary rates and timescale comparison of Chikungunya viruses inferred from the whole genome/E1 gene with special reference to the 2005–07 outbreak in the Indian subcontinent. Infect. Genet. Evol. 2009, 9, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Mittal, V.; Rizvi, M.A.; Bhattacharya, D.; Chhabra, M.; Rawat, D.S.; Ichhpujani, R.L.; Chauhan, L.S.; Rai, A. Northward movement of East Central South African genotype of Chikungunya virus causing an epidemic between 2006–2010 in India. J. Infect. Dev. Ctries. 2012, 6, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Sharma, P.; Kumar, S.; Chhabra, M.; Rizvi, M.A.; Mittal, V.; Bhattacharya, D.; Venkatesh, S.; Rai, A. Continued persistence of ECSA genotype with replacement of K211E in E1 gene of Chikungunya virus in Delhi from 2010 to 2014. Asian Pac. J. Trop. Dis. 2016, 6, 564–566. [Google Scholar] [CrossRef]
- Kumar, N.P.; Mitha, M.M.; Krishnamoorthy, N.; Kamaraj, T.; Joseph, R.; Jambulingam, P. Genotyping of virus involved in the 2006 Chikungunya outbreak in South India (Kerala and Puducherry). Curr. Sci. 2007, 1412–1416. [Google Scholar]
- Kumar, N.P.; Joseph, R.; Kamaraj, T.; Jambulingam, P. A226V mutation in virus during the 2007 chikungunya outbreak in Kerala, India. J. Gen. Virol. 2008, 89, 1945–1948. [Google Scholar] [CrossRef] [PubMed]
- Raghavendhar, B.S.; Ray, P.; Ratagiri, V.H.; Sharma, B.; Kabra, S.K.; Lodha, R. Evaluation of chikungunya virus infection in children from India during 2009–2010: A cross sectional observational study. J. Med. Virol. 2016, 88, 923–930. [Google Scholar] [CrossRef]
- Kumar, N.P.; Sabesan, S.; Krishnamoorthy, K.; Jambulingam, P. Detection of Chikungunya virus in wild populations of Aedes albopictus in Kerala State, India. Vector-Borne Zoonotic Dis. 2012, 12, 907–911. [Google Scholar] [CrossRef]
- Santhosh, S.; Dash, P.; Parida, M.; Khan, M.; Tiwari, M.; Rao, P.L. Comparative full genome analysis revealed E1: A226V shift in 2007 Indian Chikungunya virus isolates. Virus Res. 2008, 135, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Sharma, A.K.; Sukumaran, D.; Parida, M.; Dash, P.K. Two novel epistatic mutations (E1: K211E and E2: V264A) in structural proteins of Chikungunya virus enhance fitness in Aedes aegypti. Virology 2016, 497, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Dash, P.K.; Singh, A.K.; Sharma, S.; Gopalan, N.; Rao, P.V.L.; Parida, M.M.; Reiter, P. Evidence of experimental vertical transmission of emerging novel ECSA genotype of chikungunya virus in Aedes aegypti. PLoS Negl. Trop. Dis. 2014, 8, e2990. [Google Scholar] [CrossRef]
- Shrinet, J.; Jain, S.; Sharma, A.; Singh, S.S.; Mathur, K.; Rana, V.; Bhatnagar, R.K.; Gupta, B.; Gaind, R.; Deb, M. Genetic characterization of Chikungunya virus from New Delhi reveal emergence of a new molecular signature in Indian isolates. Virol. J. 2012, 9, 100. [Google Scholar] [CrossRef]
- Abraham, R.; Manakkadan, A.; Mudaliar, P.; Joseph, I.; Sivakumar, K.C.; Nair, R.R.; Sreekumar, E. Correlation of phylogenetic clade diversification and in vitro infectivity differences among Cosmopolitan genotype strains of Chikungunya virus. Infect. Genet. Evol. 2016, 37, 174–184. [Google Scholar] [CrossRef]
- Harsha, P.K.; Reddy, V.; Rao, D.; Pattabiraman, C.; Mani, R.S. Continual circulation of ECSA genotype and identification of a novel mutation I317V in the E1 gene of Chikungunya viral strains in southern India during 2015–2016. J. Med. Virol. 2020, 92, 1007–1012. [Google Scholar] [CrossRef]
- Muruganandam, N.; Chaaithanya, I.; Senthil, G.; Shriram, A.; Bhattacharya, D.; JeevaBharathi, G.; Sudeep, A.; PradeepKumar, N.; Vijayachari, P. Isolation and molecular characterization of Chikungunya virus from the Andaman and Nicobar archipelago, India: Evidence of an East, Central, and South African genotype. Can. J. Microbiol. 2011, 57, 1073–1077. [Google Scholar] [CrossRef] [PubMed]
- Paramasivan, R.; Samuel, P.P.; Thenmozhi, V.; Rajendran, R.; Leo, S.V.J.; Dhananjeyan, K.J.; Krishnamoorthi, R.; Arunachalam, N.; Tyagi, B.K. Chikungunya virus isolated in Lakshadweep islands in the Indian Ocean: Evidence of the Central/East African genotype. Jpn. J. Infect. Dis. 2009, 62, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.K.; Pal, T.; Saha, B.; Mandal, S.; Tripathi, A. Copy number variation of chikungunya ECSA virus with disease symptoms among Indian patients. J. Med. Virol. 2014, 86, 1386–1392. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.K.; Bhattacharya, T.; Tripathi, A. Chikungunya virus: Genomic microevolution in Eastern India and its in-silico epitope prediction. 3 Biotech. 2018, 8, 318. [Google Scholar] [CrossRef]
- Dutta, S.K.; Tripathi, A. Association of toll-like receptor polymorphisms with susceptibility to chikungunya virus infection. Virology 2017, 511, 207–213. [Google Scholar] [CrossRef]
- Chaaithanya, I.K.; Muruganandam, N.; Anwesh, M.; Rajesh, R.; Ghosal, S.R.; Kartick, C.; Prasad, K.N.; Muthumani, K.; Vijayachari, P. HLA class II allele polymorphism in an outbreak of chikungunya fever in M Iddle A Ndaman, I Ndia. Immunology 2013, 140, 202–210. [Google Scholar] [CrossRef]
- Chaaithanya, I.K.; Muruganandam, N.; Surya, P.; Anwesh, M.; Alagarasu, K.; Vijayachari, P. Association of Oligoadenylate Synthetase Gene Cluster and DC-SIGN (CD209) gene polymorphisms with clinical symptoms in Chikungunya virus infection. DNA Cell. Biol. 2016, 35, 44–50. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, A.; Mamidi, P.; Tiwari, A.; Kumar, S.; Mayavannan, A.; Mudulli, S.; Singh, A.K.; Subudhi, B.B.; Chattopadhyay, S. Chikungunya virus nsP1 interacts directly with nsP2 and modulates its ATPase activity. Sci. Rep. 2018, 8, 1045. [Google Scholar] [CrossRef]
- Niyas, K.P.; Abraham, R.; Unnikrishnan, R.N.; Mathew, T.; Nair, S.; Manakkadan, A.; Issac, A.; Sreekumar, E. Molecular characterization of Chikungunya virus isolates from clinical samples and adult Aedes albopictus mosquitoes emerged from larvae from Kerala, South India. Virol. J. 2010, 7, 189. [Google Scholar] [CrossRef]
- Jain, J.; Kaur, N.; Haller, S.L.; Kumar, A.; Rossi, S.L.; Narayanan, V.; Kumar, D.; Gaind, R.; Weaver, S.C.; Auguste, A.J. Chikungunya outbreaks in India: A prospective study comparing neutralization and sequelae during two outbreaks in 2010 and 2016. Am. J. Trop. Med. Hyg. 2020, 102, 857. [Google Scholar] [CrossRef]
- Chaaithanya, I.K.; Muruganandam, N.; Sundaram, S.G.; Kawalekar, O.; Sugunan, A.P.; Manimunda, S.P.; Ghosal, S.R.; Muthumani, K.; Vijayachari, P. Role of proinflammatory cytokines and chemokines in chronic arthropathy in CHIKV infection. Viral Immunol. 2011, 24, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Fulsundar, S.R.; Roy, S.; Manimunda, S.P.; Singh, S.S.; Sugunan, A.; Vijayachari, P. Investigations on possible role of MIF gene polymorphism in progression of chikungunya infection into cases of acute flaccid paralysis and chronic arthropathy. J. Genet. 2009, 88, 123–125. [Google Scholar] [CrossRef]
- Agarwal, A.; Joshi, G.; Nagar, D.P.; Sharma, A.K.; Sukumaran, D.; Pant, S.C.; Parida, M.M.; Dash, P.K. Mosquito saliva induced cutaneous events augment Chikungunya virus replication and disease progression. Infect. Genet. Evol. 2016, 40, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.R.; Abraham, R.; Sundaram, S.; Sreekumar, E. Interferon regulated gene (IRG) expression-signature in a mouse model of chikungunya virus neurovirulence. J. Neurovirol. 2017, 23, 886–902. [Google Scholar] [CrossRef]
- Abraham, R.; Mudaliar, P.; Padmanabhan, A.; Sreekumar, E. Induction of cytopathogenicity in human glioblastoma cells by chikungunya virus. PLoS ONE 2013, 8, e75854. [Google Scholar] [CrossRef] [PubMed]
- Mathur, K.; Anand, A.; Dubey, S.K.; Sanan-Mishra, N.; Bhatnagar, R.K.; Sunil, S. Analysis of chikungunya virus proteins reveals that non-structural proteins nsP2 and nsP3 exhibit RNA interference (RNAi) suppressor activity. Sci. Rep. 2016, 6, 38065. [Google Scholar] [CrossRef]
- Kumar, R.; Srivastava, P.; Mathur, K.; Shrinet, J.; Dubey, S.K.; Chinnappan, M.; Kaur, I.; Nayak, D.; Chattopadhyay, S.; Bhatnagar, R.K.; et al. Chikungunya virus non-structural protein nsP3 interacts with Aedes aegypti DEAD-box helicase RM62F. Virusdisease 2021, 32, 657–665. [Google Scholar] [CrossRef]
- Dubey, S.K.; Shrinet, J.; Jain, J.; Ali, S.; Sunil, S. Aedes aegypti microRNA miR-2b regulates ubiquitin-related modifier to control chikungunya virus replication. Sci. Rep. 2017, 7, 17666. [Google Scholar] [CrossRef]
- Dubey, S.K.; Mehta, D.; Chaudhary, S.; Hasan, A.; Sunil, S. An E3 Ubiquitin Ligase Scaffolding Protein Is Proviral during Chikungunya Virus Infection in Aedes aegypti. Microbiol. Spectr. 2022, 10, e0059522. [Google Scholar] [CrossRef]
- Shrinet, J.; Srivastava, P.; Sunil, S. Transcriptome analysis of Aedes aegypti in response to mono-infections and co-infections of dengue virus-2 and chikungunya virus. Biochem. Biophys. Res. Commun. 2017, 492, 617–623. [Google Scholar] [CrossRef]
- Shrinet, J.; Bhavesh, N.S.; Sunil, S. Understanding oxidative stress in Aedes during chikungunya and dengue virus infections using integromics analysis. Viruses 2018, 10, 314. [Google Scholar] [CrossRef]
- Jain, J.; Mathur, K.; Shrinet, J.; Bhatnagar, R.K.; Sunil, S. Analysis of coevolution in nonstructural proteins of chikungunya virus. Virol. J. 2016, 13, 86. [Google Scholar] [CrossRef]
- Parashar, D.; Paingankar, M.S.; More, A.; Patil, P.; Amdekar, S. Altered microRNA expression signature in Chikungunya-infected mammalian fibroblast cells. Virus Genes 2018, 54, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Patil, J.; More, A.; Patil, P.; Jadhav, S.; Newase, P.; Agarwal, M.; Amdekar, S.; Raut, C.; Parashar, D.; Cherian, S.S. Genetic characterization of chikungunya viruses isolated during the 2015-2017 outbreaks in different states of India, based on their E1 and E2 genes. Arch. Virol. 2018, 163, 3135–3140. [Google Scholar] [CrossRef]
- Soni, A.; Pandey, K.M.; Ray, P.; Jayaram, B. Genomes to hits in Silico-a country path today, a highway tomorrow: A case study of Chikungunya. Curr. Pharm. Des. 2013, 19, 4687–4700. [Google Scholar] [CrossRef]
- Soni, A.; Dhingra, P.; Mishra, A.; Singh, T.; Mukherjee, G.; Jayaram, B. 107 Genomes to hit molecules In Silico: A country path today, a highway tomorrow: A case study of Chikungunya. J. Biomol. Struct. Dyn. 2013, 31, 67–68. [Google Scholar] [CrossRef]
- Jain, N.; Bhat, R.; Jayaram, B. Development of Novel Antivirals for Chikungunya Virus. Stud. Indian. Place Names 2020, 40, 1196–1198. [Google Scholar]
- McPherson, R.L.; Abraham, R.; Sreekumar, E.; Ong, S.-E.; Cheng, S.-J.; Baxter, V.K.; Kistemaker, H.A.; Filippov, D.V.; Griffin, D.E.; Leung, A.K. ADP-ribosylhydrolase activity of Chikungunya virus macrodomain is critical for virus replication and virulence. Proc. Natl. Acad. Sci. USA 2017, 114, 1666–1671. [Google Scholar] [CrossRef]
- Abraham, R.; Mudaliar, P.; Jaleel, A.; Srikanth, J.; Sreekumar, E. High throughput proteomic analysis and a comparative review identify the nuclear chaperone, Nucleophosmin among the common set of proteins modulated in Chikungunya virus infection. J. Proteom. 2015, 120, 126–141. [Google Scholar] [CrossRef] [PubMed]
- Abraham, R.; Singh, S.; Nair, S.R.; Hulyalkar, N.V.; Surendran, A.; Jaleel, A.; Sreekumar, E. Nucleophosmin (NPM1)/B23 in the proteome of human astrocytic cells restricts chikungunya virus replication. J. Proteome Res. 2017, 16, 4144–4155. [Google Scholar] [CrossRef]
- Dhanwani, R.; Khan, M.; Alam, S.I.; Rao, P.V.L.; Parida, M. Differential proteome analysis of Chikungunya virus-infected new-born mice tissues reveal implication of stress, inflammatory and apoptotic pathways in disease pathogenesis. Proteomics 2011, 11, 1936–1951. [Google Scholar] [CrossRef] [PubMed]
- Dhanwani, R.; Khan, M.; Lomash, V.; Rao, P.V.L.; Ly, H.; Parida, M. Characterization of chikungunya virus induced host response in a mouse model of viral myositis. PLoS ONE 2014, 9, e92813. [Google Scholar] [CrossRef] [PubMed]
- Puttamallesh, V.N.; Sreenivasamurthy, S.K.; Singh, P.K.; Harsha, H.; Ganjiwale, A.; Broor, S.; Pandey, A.; Narayana, J.; Prasad, T.K. Proteomic profiling of serum samples from chikungunya-infected patients provides insights into host response. Clin. Proteom. 2013, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Kalawat, U.; Sharma, K.K.; Reddy, S.G. Prevalence of dengue and chickungunya fever and their co-infection. Indian. J. Pathol. Microbiol. 2011, 54, 844. [Google Scholar]
- Verma, A.; Mudhigeti, N.; Kommireddy, S.; Anagoni, S.; Sharma, K.; Nallapireddy, U.; Kalawat, U. Seroprevalence of Anti-Chikungunya IgG Antibodies among Rheumatoid Arthritis (RA) Patients. J. Clin. Diagn. Res. 2019, 13, 6–9. [Google Scholar] [CrossRef]
- Manimunda, S.P.; Vijayachari, P.; Uppoor, R.; Sugunan, A.P.; Singh, S.S.; Rai, S.K.; Sudeep, A.B.; Muruganandam, N.; Chaitanya, I.K.; Guruprasad, D.R. Clinical progression of chikungunya fever during acute and chronic arthritic stages and the changes in joint morphology as revealed by imaging. Trans. R. Soc. Trop. Med. Hyg. 2010, 104, 392–399. [Google Scholar] [CrossRef]
- Singh, S.; Manimunda, S.; Sugunan, A.; Vijayachari, P. Four cases of acute flaccid paralysis associated with chikungunya virus infection. Epidemiol. Infect. 2008, 136, 1277–1280. [Google Scholar] [CrossRef]
- Chaaithanya, I.K.; Muruganandam, N.; Raghuraj, U.; Sugunan, A.P.; Rajesh, R.; Anwesh, M.; Rai, S.K.; Vijayachari, P. Chronic inflammatory arthritis with persisting bony erosions in patients following chikungunya infection. Indian J. Med. Res. 2014, 140, 142. [Google Scholar]
- Lalitha, P.; Rathinam, S.; Banushree, K.; Maheshkumar, S.; Vijayakumar, R.; Sathe, P. Ocular involvement associated with an epidemic outbreak of chikungunya virus infection. Am. J. Ophthalmol. 2007, 144, 552–556. [Google Scholar] [CrossRef]
- Mahendradas, P.; Ranganna, S.K.; Shetty, R.; Balu, R.; Narayana, K.M.; Babu, R.B.; Shetty, B.K. Ocular manifestations associated with chikungunya. Ophthalmology 2008, 115, 287–291. [Google Scholar] [CrossRef]
- Mahendradas, P. Chikungunya and the Eye. In The Uveitis Atlas; Springer: New Delhi, India, 2020; pp. 363–367. [Google Scholar]
- Mahendradas, P. Chikungunya. In Emerging Infectious Uveitis; Springer: Berlin/Heidelberg, Germany, 2017; pp. 119–124. [Google Scholar]
- Khairallah, M.; Mahendradas, P.; Curi, A.; Khochtali, S.; Cunningham, E.T., Jr. Emerging viral infections causing anterior uveitis. Ocul. Immunol. Inflamm. 2019, 27, 219–228. [Google Scholar] [CrossRef]
- Mahendradas, P.; Avadhani, K.; Shetty, R. Chikungunya and the eye: A review. J. Ophthalmic Inflamm. Infect. 2013, 3, 35. [Google Scholar] [CrossRef] [PubMed]
- Mahendradas, P.; Shetty, R.; Malathi, J.; Madhavan, H. Chikungunya virus iridocyclitis in Fuchs’ heterochromic iridocyclitis. Indian. J. Ophthalmol. 2010, 58, 545. [Google Scholar] [CrossRef] [PubMed]
- Mittal, A.; Mittal, S.; Bharati, M.J.; Ramakrishnan, R.; Saravanan, S.; Sathe, P.S. Optic neuritis associated with chikungunya virus infection in South India. Arch. Ophthalmol. 2007, 125, 1381–1386. [Google Scholar] [CrossRef] [PubMed]
- Mittal, A.; Mittal, S.; Bharathi, J.M.; Ramakrishnan, R.; Sathe, P.S. Uveitis during outbreak of Chikungunya fever. Ophthalmology 2007, 114, 1798. [Google Scholar] [CrossRef] [PubMed]
- Rose, N.; Anoop, T.; John, A.P.; Jabbar, P.; George, K. Acute optic neuritis following infection with chikungunya virus in southern rural India. Int. J. Infect. Dis. 2011, 15, e147–e150. [Google Scholar] [CrossRef]
- Riyaz, N.; Riyaz, A.; Latheef, E.A.; Anitha, P.; Aravindan, K.; Nair, A.S.; Shameera, P. Cutaneous manifestations of chikungunya during a recent epidemic in Calicut, north Kerala, south India. Indian. J. Dermatol. Venereol. Leprol. 2010, 76, 671. [Google Scholar] [CrossRef]
- Pakran, J.; George, M.; Riyaz, N.; Arakkal, R.; George, S.; Rajan, U.; Khader, A.; Thomas, S.; Abdurahman, R.; Sasidharanpillai, S. Purpuric macules with vesiculobullous lesions: A novel manifestation of Chikungunya. Int. J. Dermatol. 2011, 50, 61–69. [Google Scholar] [CrossRef]
- Parish, L.C.; Virendra, N.; Sehgal, M.D.; Najeeba Riyaz, M.D. Commentary Chikungunya. SKINmed 2015, 13, 423–426. [Google Scholar]
- Bandyopadhyay, D.; Ghosh, S.K. Mucocutaneous manifestations of Chikungunya fever. Indian J. Dermatol. 2010, 55, 64. [Google Scholar] [CrossRef]
- Bandyopadhyay, D. Dengue and chikungunya fever: Resurgent viral infections with prominent mucocutaneous features. Indian J. Dermatol. 2010, 55, 53. [Google Scholar] [CrossRef]
- Bandyopadhyay, D.; Ghosh, S.K. Mucocutaneous features of Chikungunya fever: A study from an outbreak in West Bengal, India. Int. J. Dermatol. 2008, 47, 1148–1152. [Google Scholar] [CrossRef]
- Nayak, K.; Jain, V.; Kaur, M.; Khan, N.; Gottimukkala, K.; Aggarwal, C.; Sagar, R.; Gupta, S.; Rai, R.C.; Dixit, K.; et al. Antibody response patterns in chikungunya febrile phase predict protection versus progression to chronic arthritis. JCI Insight 2020, 5, e130509. [Google Scholar] [CrossRef]
- Suryawanshi, S.; Dube, A.; Khadse, R.; Jalgaonkar, S.; Sathe, P.; Zawar, S.; Holay, M. Clinical profile of chikungunya fever in patients in a tertiary care centre in Maharashtra, India. Indian J. Med. Res. 2009, 129, 438. [Google Scholar]
- Sebastian, M.R.; Lodha, R.; Kabra, S. Chikungunya infection in children. Indian J. Pediatr. 2009, 76, 185. [Google Scholar] [CrossRef]
- Dhochak, N.; Kabra, S.K.; Lodha, R. Dengue and Chikungunya infections in children. Indian J. Pediatr. 2019, 86, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.K.; Kabra, S.K.; Lodha, R.; Ratageri, V.H.; Ray, P. Virus load and clinical features during the acute phase of Chikungunya infection in children. PLoS ONE 2019, 14, e0211036. [Google Scholar]
- Ray, P.; Ratagiri, V.H.; Kabra, S.K.; Lodha, R.; Sharma, S.; Sharma, B.; Kalaivani, M.; Wig, N. Chikungunya infection in India: Results of a prospective hospital based multi-centric study. PLoS ONE 2012, 7, e30025. [Google Scholar] [CrossRef]
- Patil, H.P.; Rane, P.S.; Gosavi, M.; Mishra, A.C.; Arankalle, V.A. Standardization of ELISA for anti-chikungunya-IgG antibodies and age-stratified prevalence of anti-chikungunya-IgG antibodies in Pune, India. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2020, 39, 1925–1932. [Google Scholar] [CrossRef] [PubMed]
- Gosavi, M.; Patil, H.P. Evaluation of monophosphoryl lipid A as an adjuvanted for inactivated chikungunya virus. Vaccine 2022, 40, 5060–5068. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.; Mani, R.S.; Desai, A.; Ravi, V. Correlation of plasma viral loads and presence of Chikungunya IgM antibodies with cytokine/chemokine levels during acute Chikungunya virus infection. J. Med. Virol. 2014, 86, 1393–1401. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.; Ravi, V.; Desai, A.; Parida, M.; Powers, A.M.; Johnson, B.W. Utility of IgM ELISA, TaqMan real-time PCR, reverse transcription PCR, and RT-LAMP assay for the diagnosis of Chikungunya fever. J. Med. Virol. 2012, 84, 1771–1778. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, V.; Neeraja, M.; Subbalaxmi, M.; Parida, M.; Dash, P.; Santhosh, S.; Rao, P. Clinical features and molecular diagnosis of Chikungunya fever from South India. Clin. Infect. Dis. 2008, 46, 1436–1442. [Google Scholar] [CrossRef] [PubMed]
- Cecilia, D.; Kakade, M.; Alagarasu, K.; Patil, J.; Salunke, A.; Parashar, D.; Shah, P. Development of a multiplex real-time RT-PCR assay for simultaneous detection of dengue and chikungunya viruses. Arch. Virol. 2015, 160, 323–327. [Google Scholar] [CrossRef]
- Parashar, D.; Paingankar, M.S.; Sudeep, A.; More, A.; Shinde, S.B.; Arankalle, V.A. Assessment of qPCR, nested RT-PCR and ELISA techniques in diagnosis of Chikungunya. Curr. Sci. 2014, 107, 2011–2013. [Google Scholar]
- Choudhary, S.; Neetu, N.; Singh, V.A.; Kumar, P.; Chaudhary, M.; Tomar, S. Chikungunya virus titration, detection and diagnosis using N-Acetylglucosamine (GlcNAc) specific lectin based virus capture assay. Virus Res. 2021, 302, 198493. [Google Scholar] [CrossRef]
- Singh, V.A.; Kumar, C.S.; Khare, B.; Kuhn, R.J.; Banerjee, M.; Tomar, S. Surface decorated reporter-tagged chikungunya virus-like particles for clinical diagnostics and identification of virus entry inhibitors. Virology 2023, 578, 92–102. [Google Scholar] [CrossRef]
- Jain, J.; Okabayashi, T.; Kaur, N.; Nakayama, E.; Shioda, T.; Gaind, R.; Kurosu, T.; Sunil, S. Evaluation of an immunochromatography rapid diagnosis kit for detection of chikungunya virus antigen in India, a dengue-endemic country. Virol. J. 2018, 15, 84. [Google Scholar] [CrossRef]
- Jain, J.; Nayak, K.; Tanwar, N.; Gaind, R.; Gupta, B.; Shastri, J.; Bhatnagar, R.K.; Kaja, M.K.; Chandele, A.; Sunil, S. Clinical, serological, and virological analysis of 572 chikungunya patients from 2010 to 2013 in India. Clin. Infect. Dis. 2017, 65, 133–140. [Google Scholar] [CrossRef]
- Londhey, V.; Agrawal, S.; Vaidya, N.; Kini, S.; Shastri, J.; Sunil, S. Dengue and chikungunya virus co-infections: The inside story. J. Assoc. Physicians India 2016, 64, 36–40. [Google Scholar]
- Verma, A.K.; Chandele, A.; Kaja, M.-K.; Arulandu, A.; Ray, P. Cloning, expression and purification of Chikungunya virus E2 recombinant protein in E. coli. BMC Infect. Dis. 2014, 14, P65. [Google Scholar] [CrossRef]
- Verma, A.; Chandele, A.; Nayak, K.; Kaja, M.K.; Arulandu, A.; Lodha, R.; Ray, P. High yield expression and purification of Chikungunya virus E2 recombinant protein and its evaluation for serodiagnosis. J. Virol. Methods 2016, 235, 73–79. [Google Scholar] [CrossRef]
- Raghavenhdar, S.; Kabra, S.; Ray, P. Evaluation of chikungunya virus infection and screening of antibodies. Int. J. Infect. Dis. 2016, 45, 240. [Google Scholar] [CrossRef]
- Verma, A.; Nayak, K.; Chandele, A.; Singla, M.; Ratageri, V.H.; Lodha, R.; Kabra, S.K.; Murali-Krishna, K.; Ray, P. Chikungunya-specific IgG and neutralizing antibody responses in natural infection of Chikungunya virus in children from India. Arch. Virol. 2021, 166, 1913–1920. [Google Scholar] [CrossRef]
- Saxena, T.; Tandon, B.; Sharma, S.; Chameettachal, S.; Ray, P.; Ray, A.R.; Kulshreshtha, R. Combined miRNA and mRNA signature identifies key molecular players and pathways involved in chikungunya virus infection in human cells. PLoS ONE 2013, 8, e79886. [Google Scholar] [CrossRef]
- Shukla, J.; Khan, M.; Tiwari, M.; Sannarangaiah, S.; Sharma, S.; Rao, P.V.L.; Parida, M. Development and evaluation of antigen capture ELISA for early clinical diagnosis of chikungunya. Diagn. Microbiol. Infect. Dis. 2009, 65, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Parida, M.; Santhosh, S.; Dash, P.; Lakshmana Rao, P. Rapid and real-time assays for detection and quantification of chikungunya virus. Future Med. 2008, 3. [Google Scholar] [CrossRef]
- Sharma, S.; Dash, P.K.; Santhosh, S.; Shukla, J.; Parida, M.; Rao, P.L. Development of a Quantitative Competitive Reverse Transcription Polymerase Chain Reaction (QC-RT–PCR) for Detection and Quantitation of Chikungunya Virus. Mol. Biotechnol. 2010, 45, 49–55. [Google Scholar] [CrossRef]
- Khan, M.; Dhanwani, R.; Kumar, J.S.; Rao, P.L.; Parida, M. Comparative evaluation of the diagnostic potential of recombinant envelope proteins and native cell culture purified viral antigens of Chikungunya virus. J. Med. Virol. 2014, 86, 1169–1175. [Google Scholar] [CrossRef]
- Kumar, J.S.; Khan, M.; Gupta, G.; Bhoopati, M.; Lakshmana Rao, P.; Parida, M. Production, characterization, and application of monoclonal antibodies specific to recombinant (E2) structural protein in antigen-capture ELISA for clinical diagnosis of Chikungunya virus. Viral Immunol. 2012, 25, 153–160. [Google Scholar] [CrossRef]
- Priya, R.; Khan, M.; Rao, M.K.; Parida, M. Cloning, expression and evaluation of diagnostic potential of recombinant capsid protein based IgM ELISA for chikungunya virus. J. Virol. Methods 2014, 203, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Parida, M.; Santhosh, S.; Dash, P.; Tripathi, N.; Lakshmi, V.; Mamidi, N.; Shrivastva, A.; Gupta, N.; Saxena, P.; Babu, J.P. Rapid and real-time detection of Chikungunya virus by reverse transcription loop-mediated isothermal amplification assay. J. Clin. Microbiol. 2007, 45, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Santhosh, S.; Parida, M.; Dash, P.; Pateriya, A.; Pattnaik, B.; Pradhan, H.; Tripathi, N.; Ambuj, S.; Gupta, N.; Saxena, P. Development and evaluation of SYBR Green I-based one-step real-time RT-PCR assay for detection and quantification of Chikungunya virus. J. Clin. Virol. 2007, 39, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Acharya, B.N.; Priya, R.; Tripathi, N.K.; Shrivastava, A.; Rao, M.K.; Kesari, P.; Narwal, M.; Tomar, S.; Bhagyawant, S.S. Development of nsP2 protease based cell free high throughput screening assay for evaluation of inhibitors against emerging Chikungunya virus. Sci. Rep. 2018, 8, 10831. [Google Scholar] [CrossRef] [PubMed]
- Wellems, T.E.; Plowe, C.V. Chloroquine-resistant malaria. J. Infect. Dis. 2001, 184, 770–776. [Google Scholar] [CrossRef]
- Khan, M.; Santhosh, S.; Tiwari, M.; Lakshmana Rao, P.; Parida, M. Assessment of in vitro prophylactic and therapeutic efficacy of chloroquine against Chikungunya virus in vero cells. J. Med. Virol. 2010, 82, 817–824. [Google Scholar] [CrossRef]
- Saha, A.; Bhagyawant, S.S.; Parida, M.; Dash, P.K. Vector-delivered artificial miRNA effectively inhibited replication of Chikungunya virus. Antivir. Res. 2016, 134, 42–49. [Google Scholar] [CrossRef]
- Khan, M.; Dhanwani, R.; Patro, I.; Rao, P.; Parida, M. Cellular IMPDH enzyme activity is a potential target for the inhibition of Chikungunya virus replication and virus induced apoptosis in cultured mammalian cells. Antivir. Res. 2011, 89, 1–8. [Google Scholar] [CrossRef]
- Dash, R.N.; Prabhudutta, M.; De, S.; Swain, R.P.; Moharana, A.K.; Subudhi, B.B.; Chattopadhyay, S. Conjugates of ibuprofen inhibit CHIKV infection and inflammation. Mol. Divers. 2023. [Google Scholar] [CrossRef]
- Parashar, D.; Paingankar, M.S.; Kumar, S.; Gokhale, M.D.; Sudeep, A.; Shinde, S.B.; Arankalle, V. Administration of E2 and NS1 siRNAs inhibit chikungunya virus replication in vitro and protects mice infected with the virus. PLoS Negl. Trop. Dis. 2013, 7, e2405. [Google Scholar] [CrossRef]
- Paingankar, M.S.; Arankalle, V.A. Identification of chikungunya virus interacting proteins in mammalian cells. J. Biosci. 2014, 39, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mamidi, P.; Das, I.; Nayak, T.K.; Kumar, S.; Chhatai, J.; Chattopadhyay, S.; Suryawanshi, A.R.; Chattopadhyay, S. A novel 2006 Indian outbreak strain of Chikungunya virus exhibits different pattern of infection as compared to prototype strain. PLoS ONE 2014, 9, e85714. [Google Scholar] [CrossRef] [PubMed]
- Das, I.; Basantray, I.; Mamidi, P.; Nayak, T.K.; Pratheek, B.; Chattopadhyay, S.; Chattopadhyay, S. Heat shock protein 90 positively regulates Chikungunya virus replication by stabilizing viral non-structural protein nsP2 during infection. PLoS ONE 2014, 9, e100531. [Google Scholar] [CrossRef] [PubMed]
- Nayak, T.K.; Mamidi, P.; Kumar, A.; Singh, L.P.K.; Sahoo, S.S.; Chattopadhyay, S.; Chattopadhyay, S. Regulation of viral replication, apoptosis and pro-inflammatory responses by 17-AAG during Chikungunya virus infection in macrophages. Viruses 2017, 9, 3. [Google Scholar] [CrossRef]
- Dhindwal, S.; Kesari, P.; Singh, H.; Kumar, P.; Tomar, S. Conformer and pharmacophore based identification of peptidomimetic inhibitors of chikungunya virus nsP2 protease. J. Biomol. Struct. Dyn. 2017, 35, 3522–3539. [Google Scholar] [CrossRef]
- Bhatia, P.; Singh, V.A.; Rani, R.; Nath, M.; Tomar, S. Cellular uptake of metal oxide-based nanocomposites and targeting of chikungunya virus replication protein nsP3. J. Trace Elem. Med. Biol. 2023, 78, 127176. [Google Scholar] [CrossRef]
- Mudgal, R.; Mahajan, S.; Tomar, S. Inhibition of Chikungunya virus by an adenosine analog targeting the SAM-dependent nsP1 methyltransferase. FEBS Lett. 2020, 594, 678–694. [Google Scholar] [CrossRef]
- Singh, H.; Mudgal, R.; Narwal, M.; Kaur, R.; Singh, V.A.; Malik, A.; Chaudhary, M.; Tomar, S. Chikungunya virus inhibition by peptidomimetic inhibitors targeting virus-specific cysteine protease. Biochimie 2018, 149, 51–61. [Google Scholar] [CrossRef]
- Narwal, M.; Singh, H.; Pratap, S.; Malik, A.; Kuhn, R.J.; Kumar, P.; Tomar, S. Crystal structure of chikungunya virus nsP2 cysteine protease reveals a putative flexible loop blocking its active site. Int. J. Biol. Macromol. 2018, 116, 451–462. [Google Scholar] [CrossRef]
- Kumar, R.; Nehul, S.; Singh, A.; Tomar, S. Identification and evaluation of antiviral potential of thymoquinone, a natural compound targeting Chikungunya virus capsid protein. Virology 2021, 561, 36–46. [Google Scholar] [CrossRef]
- Aggarwal, M.; Sharma, R.; Kumar, P.; Parida, M.; Tomar, S. Kinetic characterization of trans-proteolytic activity of Chikungunya virus capsid protease and development of a FRET-based HTS assay. Sci. Rep. 2015, 5, 14753. [Google Scholar] [CrossRef] [PubMed]
- Fatma, B.; Kumar, R.; Singh, V.A.; Nehul, S.; Sharma, R.; Kesari, P.; Kuhn, R.J.; Tomar, S. Alphavirus capsid protease inhibitors as potential antiviral agents for Chikungunya infection. Antivir. Res. 2020, 179, 104808. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Fatma, B.; Saha, A.; Bajpai, S.; Sistla, S.; Dash, P.K.; Parida, M.; Kumar, P.; Tomar, S. Inhibition of chikungunya virus by picolinate that targets viral capsid protein. Virology 2016, 498, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, M.; Kaur, R.; Saha, A.; Mudgal, R.; Yadav, R.; Dash, P.K.; Parida, M.; Kumar, P.; Tomar, S. Evaluation of antiviral activity of piperazine against Chikungunya virus targeting hydrophobic pocket of alphavirus capsid protein. Antivir. Res. 2017, 146, 102–111. [Google Scholar] [CrossRef]
- Kaur, R.; Mudgal, R.; Jose, J.; Kumar, P.; Tomar, S. Glycan-dependent chikungunya viral infection divulged by antiviral activity of NAG specific chi-like lectin. Virology 2019, 526, 91–98. [Google Scholar] [CrossRef]
- Islamuddin, M.; Afzal, O.; Khan, W.H.; Hisamuddin, M.; Altamimi, A.S.A.; Husain, I.; Kato, K.; Alamri, M.A.; Parveen, S. Inhibition of Chikungunya Virus Infection by 4-Hydroxy-1-Methyl-3-(3-morpholinopropanoyl)quinoline-2(1H)-one (QVIR) Targeting nsP2 and E2 Proteins. ACS Omega 2021, 6, 9791–9803. [Google Scholar] [CrossRef]
- Deeba, F.; Malik, M.Z.; Naqvi, I.H.; Haider, M.S.H.; Shafat, Z.; Sinha, P.; Ishrat, R.; Ahmed, A.; Parveen, S. Potential entry inhibitors of the envelope protein (E2) of Chikungunya virus: In silico structural modeling, docking and molecular dynamic studies. VirusDisease 2017, 28, 39–49. [Google Scholar] [CrossRef]
- Khan, N.; Bhat, R.; Patel, A.K.; Ray, P. Discovery of small molecule inhibitors of chikungunya virus proteins (nsP2 and E1) using in silico approaches. J. Biomol. Struct. Dyn. 2020, 39, 2607–2616. [Google Scholar] [CrossRef]
- Dilip, C.; Saraswathi, R.; Krishnan, P.; Azeem, A. Comparitive evaluation of different systems of medicines and the present scenario of chikungunya in Kerala. Asian Pac. J. Trop. Med. 2010, 3, 443–447. [Google Scholar] [CrossRef]
- Kothandan, S.; Swaminathan, R. Evaluation of in vitro antiviral activity of Vitex Negundo, L., Hyptis suaveolens (L.) poit., Decalepis hamiltonii Wight & Arn., to Chikungunya virus. Asian Pac. J. Trop. Dis. 2014, 4, S111–S115. [Google Scholar]
- Jain, J.; Pai, S.; Sunil, S. Standardization of in vitro assays to evaluate the activity of polyherbal siddha formulations against Chikungunya virus infection. Virusdisease 2018, 29, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Jain, J.; Kumar, A.; Narayanan, V.; Ramaswamy, R.; Sathiyarajeswaran, P.; Devi, M.S.; Kannan, M.; Sunil, S. Antiviral activity of ethanolic extract of Nilavembu Kudineer against dengue and chikungunya virus through in vitro evaluation. J. Ayurveda Integr. Med. 2019, 11, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Jain, J.; Narayanan, V.; Chaturvedi, S.; Pai, S.; Sunil, S. In Vivo Evaluation of Withania somnifera–Based Indian Traditional Formulation (Amukkara Choornam), Against Chikungunya Virus–Induced Morbidity and Arthralgia. J. Evid.-Based Integr. Med. 2018, 23, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hasan, A.; Devi Ms, S.; Sharma, G.; Narayanan, V.; Sathiyarajeswaran, P.; Vinayak, S.; Sunil, S. Vathasura Kudineer, an Andrographis based polyherbal formulation exhibits immunomodulation and inhibits chikungunya virus (CHIKV) under invitro conditions. J. Ethnopharmacol. 2023, 302, 115762. [Google Scholar] [CrossRef] [PubMed]
- Raghavendhar, S.; Tripati, P.K.; Ray, P.; Patel, A.K. Evaluation of medicinal herbs for Anti-CHIKV activity. Virology 2019, 533, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.K.; Soni, A.; Yadav, S.P.S.; Kumar, A.; Gaurav, N.; Siva, R.; Sharma, P.; Sunil, S.; Jayaram, B.; Patel, A.K. Evaluation of novobiocin and telmisartan for anti-CHIKV activity. Virology 2020, 548, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.K.; Singh, J.; Gaurav, N.; Garg, D.K.; Patel, A.K. In-silico and biophysical investigation of biomolecular interaction between naringin and nsP2 of the chikungunya virus. Int. J. Biol. Macromol. 2020, 160, 1061–1066. [Google Scholar] [CrossRef]
- Pratheek, B.; Suryawanshi, A.R.; Chattopadhyay, S.; Chattopadhyay, S. In silico analysis of MHC-I restricted epitopes of Chikungunya virus proteins: Implication in understanding anti-CHIKV CD8+ T cell response and advancement of epitope based immunotherapy for CHIKV infection. Infect. Genet. Evol. 2015, 31, 118–126. [Google Scholar] [CrossRef]
- Kumar, S.; Mamidi, P.; Kumar, A.; Basantray, I.; Bramha, U.; Dixit, A.; Maiti, P.K.; Singh, S.; Suryawanshi, A.R.; Chattopadhyay, S. Development of novel antibodies against non-structural proteins nsP1, nsP3 and nsP4 of chikungunya virus: Potential use in basic research. Arch. Virol. 2015, 160, 2749–2761. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Kumar, A.; Mamidi, P.; Nayak, T.K.; Das, I.; Chhatai, J.; Basantray, I.; Bramha, U.; Maiti, P.K.; Singh, S. Development and characterization of monoclonal antibody against non-structural protein-2 of Chikungunya virus and its application. J. Virol. Methods 2014, 199, 86–94. [Google Scholar] [CrossRef]
- Tripathy, A.S.; Tandale, B.V.; Balaji, S.S.; Hundekar, S.L.; Ramdasi, A.Y.; Arankalle, V.A. Envelope specific T cell responses & cytokine profiles in chikungunya patients hospitalized with different clinical presentations. Indian J. Med. Res. 2015, 141, 205. [Google Scholar] [PubMed]
- Patil, H.P.; Gosavi, M.; Mishra, A.C.; Arankalle, V.A. Age-Dependent Evaluation of Immunoglobulin G Response after Chikungunya Virus Infection. Am. J. Trop. Med. Hyg. 2021, 104, 1438–1443. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Sudeep, A.; Arankalle, V.A. Evaluation of recombinant E2 protein-based and whole-virus inactivated candidate vaccines against chikungunya virus. Vaccine 2012, 30, 6142–6149. [Google Scholar] [CrossRef]
- Shukla, M.; Chandley, P.; Tapryal, S.; Kumar, N.; Mukherjee, S.P.; Rohatgi, S. Expression, Purification, and Refolding of Chikungunya Virus Full-Length Envelope E2 Protein along with B-Cell and T-Cell Epitope Analyses Using Immuno-Informatics Approaches. ACS Omega 2022, 7, 3491–3513. [Google Scholar] [CrossRef]
- Sharma, P.; Sharma, P.; Ahmad, S.; Kumar, A. Chikungunya Virus Vaccine Development: Through Computational Proteome Exploration for Finding of HLA and cTAP Binding Novel Epitopes as Vaccine Candidates. Int. J. Pept. Res. Ther. 2022, 28, 50. [Google Scholar] [CrossRef]
- Nair, S.R.; Abraham, R.; Sreekumar, E. Generation of a Live-Attenuated Strain of Chikungunya Virus from an Indian Isolate for Vaccine Development. Vaccines 2022, 10, 1939. [Google Scholar] [CrossRef]
- Slathia, P.S. DNA Vaccine Design for Chikungunya Virus Based On the Conserved Epitopes Derived from Structural Protein. In Proceedings of the International Conference on Bioinformatics, Computational Biology and Biomedical Informatics, Northbrook, IL, USA, 7–10 August 2022; pp. 849–850. [Google Scholar]
- Tiwari, M.; Parida, M.; Santhosh, S.; Khan, M.; Dash, P.K.; Rao, P.L. Assessment of immunogenic potential of Vero adapted formalin inactivated vaccine derived from novel ECSA genotype of Chikungunya virus. Vaccine 2009, 27, 2513–2522. [Google Scholar] [CrossRef]
- Saraswat, S.; Athmaram, T.; Parida, M.; Agarwal, A.; Saha, A.; Dash, P.K. Expression and characterization of yeast derived chikungunya virus like particles (CHIK-VLPs) and its evaluation as a potential vaccine candidate. PLoS Negl. Trop. Dis. 2016, 10, e0004782. [Google Scholar] [CrossRef]
- Khan, M.; Dhanwani, R.; Rao, P.V.L.; Parida, M. Subunit vaccine formulations based on recombinant envelope proteins of Chikungunya virus elicit balanced Th1/Th2 response and virus-neutralizing antibodies in mice. Virus Res. 2012, 167, 236–246. [Google Scholar] [CrossRef]
- Sumathy, K.; Ella, K.M. Genetic diversity of chikungunya virus, India 2006–2010: Evolutionary dynamics and serotype analyses. J. Med. Virol. 2012, 84, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Sheela, P.J.; Sumathy, K. Serological correlates of immune protection conferred by Chikungunya virus infection. Acta Virol. 2013, 57, 471–473. [Google Scholar] [CrossRef]
- Ella, M.K.; Sumathy, K.; Pydigummala, J.S.; Hegde, N.R. Vaccine for Chikungunya Virus Infection. U.S. Patent 8,865,184 B2, 21 October 2014. [Google Scholar]
- Caglioti, C.; Lalle, E.; Castilletti, C.; Carletti, F.; Capobianchi, M.R.; Bordi, L. Chikungunya virus infection: An overview. New Microbiol. 2013, 36, 211–227. [Google Scholar]
- Mavale, M.; Parashar, D.; Sudeep, A.; Gokhale, M.; Ghodke, Y.; Geevarghese, G.; Arankalle, V.; Mishra, A.C. Venereal transmission of chikungunya virus by Aedes aegypti mosquitoes (Diptera: Culicidae). Am. J. Trop. Med. Hyg. 2010, 83, 1242–1244. [Google Scholar] [CrossRef] [PubMed]
- Mourya, D.; Yadav, P.; Mishra, A. Effect of temperature stress on immature stages and susceptibility of Aedes aegypti mosquitoes to chikungunya virus. Am. J. Trop. Med. Hyg. 2004, 70, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Mourya, D.; Gokhale, M.; Malunjkar, A.; Bhat, H.; Banerjee, K. Inheritance of oral susceptibility of Aedes aegypti to chikungunya virus. Am. J. Trop. Med. Hyg. 1994, 51, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Mourya, D.; Singh, D.K.; Yadav, P.; Gokhale, M.; Barde, P.; Narayan, N.; Thakare, J.; Mishra, A.; Shouche, Y. Role of gregarine parasite Ascogregarina culicis (Apicomplexa: Lecudinidae) in the maintenance of Chikungunya virus in vector mosquito. J. Eukaryot. Microbiol. 2003, 50, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.; Gokhale, M.; Barde, P.; Singh, D.K.; Mishra, A.; Mourya, D. Experimental transmission of Chikungunya virus by Anopheles stephensi mosquitoes. Acta Virol. 2003, 47, 45–47. [Google Scholar] [PubMed]
- Mourya, D.; Ranadive, S.; Gokhale, M.; Barde, P.; Padbidri, V.; Banerjee, K. Putative chikungunya virus-specific receptor proteins on the midgut brush border membrane of Aedes aegypti mosquito. Indian J. Med. Res. 1998, 107, 10–14. [Google Scholar]
- Mourya, D.; Hemingway, J.; Leake, C. Post-inoculation changes in enzyme activity of Aedes aegypti infected with Chikungunya virus. Acta Virol. 1995, 39, 31–35. [Google Scholar]
- Mourya, D.T.; Gokhale, M.D.; Majumdar, T.D.; Yadav, P.D.; Kumar, V.; Mavale, M.S. Experimental Zika virus infection in Aedes aegypti: Susceptibility, transmission & co-infection with dengue & chikungunya viruses. Indian J. Med. Res. 2018, 147, 88. [Google Scholar]
- Ghosh, A.; Alladi, P.; Narayanappa, G.; Vasanthapuram, R.; Desai, A. The time course analysis of morphological changes induced by Chikungunya virus replication in mammalian and mosquito cells. Acta Virol. 2018, 62, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.; Desai, A.; Krishna, S.S.; Vasanthapuram, R. Molecular mimicry between chikungunya virus and host components: A possible mechanism for the arthritic manifestations. PLoS Negl. Trop. Dis. 2017, 11, e0005238. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Mullapudi, T.; Bomanna, S.; Tyagi, B.; Ravi, V.; Desai, A. Understanding the mechanism of Chikungunya virus vector competence in three species of mosquitoes. Med. Vet. Entomol. 2019, 33, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Desai, A.; Ravi, V.; Narayanappa, G.; Tyagi, B.K. Chikungunya virus interacts with heat shock cognate 70 protein to facilitate its entry into mosquito cell line. Intervirology 2017, 60, 247–262. [Google Scholar] [CrossRef]
- Sirisena, P.; Kumar, A.; Sunil, S. Evaluation of Aedes aegypti (Diptera: Culicidae) life table attributes upon chikungunya virus replication reveals impact on egg-laying pathways. J. Med. Entomol. 2018, 55, 1580–1587. [Google Scholar] [CrossRef]
| No | Institute | Principal Investigator | Department/Area of Interest | Funding | Location |
|---|---|---|---|---|---|
| 1. | AIIMS | Rakesh Lodha | Department of Pediatrics | New Delhi | |
| 2. | BBIL | K. Sumathy | BBIL | Hyderabad | |
| 3. | CRME | Brij Kishore Tyagi | Vector-borne Diseases | DBT, CSIR | Madurai |
| 4. | CSTM | Anusri Tripathi | Molecular Biology & Molecular Genetics | Kolkata | |
| 5. | DRDE | Manmohan Parida | Division of Virology | DRDO | Gwalior |
| 6. | DRDE | Paban Kumar Dash | Division of Virology | DRDO | Gwalior |
| 7. | DRDE | P.V. Lakshmana Rao | Division of Virology | DRDO | Gwalior |
| 8. | DRDE | S.R. Santhosh | Division of Virology | DRDO | Gwalior |
| 9. | ICGEB | Sujatha Sunil | Vector Borne Diseases | CSIR, DBT, DST, ICGEB, Ministry of Ayush | New Delhi |
| 10. | ICMR | Shyamalendu Chatterjee | Epidemiology | ICMR | Kolkata |
| 11. | ICMR- RMRC | Paluru Vijayachari | Molecular Epidemiology and Clinical Microbiology | DBT, ICMR-RMRC | Port Blair |
| 12. | ICMR-VCRC | Muniaraj Mayilsamy | Parasitology | ICMR | Madurai |
| 13. | ICMR-VCRC | Narendran Pradeep Kumar | Vector Biology & Control | ICMR | Kottayam |
| 14. | IIM | Dileep Mavalankar | Center for Management of Health Services Public Systems Group | Ahmedabad | |
| 15. | IITD | Ashok Kumar Patel | Virology | DST, DBT | New Delhi |
| 16. | IITR | Shailly Tomar | Antiviral Research, Molecular and Structural Virology | DBT, DST, DRDO, ICMR | Roorkee |
| 17. | ILS | Soma Chattopadhyay | Infectious Disease Biology | UGC, DBT, DST, ICMR | Bhubaneswar |
| 18. | JH | Pratima Ray | Department of Biotechnology | DBT | New Delhi |
| 19. | JMI | Shama Parveen | Evolution of Dengue and Chikungunya Viruses | UGC, CCRUM, DST, CSIR, ICMR | New Delhi |
| 20. | Narayana Nethralaya Post Graduate Institute of Ophthalmology | Padmamalini Mahendradas | Ophthalmology | Bangalore | |
| 21. | NIE | Vidya Ramachandran | Impacts of CHIKV Outbreaks in Chennai | ICMR-NIE | Chennai |
| 22. | NIE | Manoj V. Murhekar | ICMR | Chennai | |
| 23. | NIMHANS | Vasanthapuram Ravi | Department of Neurovirology | Bangalore | |
| 24. | NIRRH | Itta Krishna Chaaithanya | Vector Biology and Clinical Virology | ICMR, DBT | Mumbai |
| 25. | NIV | Vidya A. Arankalle | Virology and Vaccinology | ICMR, NIV | Pune |
| 26. | NIV | Kalichamy Alagarasu | Dengue/Chikungunya Group | SERB | Pune |
| 27. | NIV | Akhilesh Chandra Mishra | Epidemiology and Control Planning of Vector-borne Diseases | ICMR | Pune |
| 28. | NIV | Devendra Tarachand Mourya | Molecular Virology and Epidemiology | NIV | Pune |
| 29. | NIV | Sarah S. Cherian | Bioinformatics and Computational Biology | NIV | Pune |
| 30. | NIV | Deepti Parashar | Molecular Virology | ICMR | Pune |
| 31. | RGCB | Easwaran Sreekumar | Molecular Virology Laboratory | DBT | Thiruvananthapuram |
| 32. | RMRC | Prafulla Dutta | Arbovirology Group | ICMR | Dibrugarh |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varikkodan, M.M.; Kunnathodi, F.; Azmi, S.; Wu, T.-Y. An Overview of Indian Biomedical Research on the Chikungunya Virus with Particular Reference to Its Vaccine, an Unmet Medical Need. Vaccines 2023, 11, 1102. https://doi.org/10.3390/vaccines11061102
Varikkodan MM, Kunnathodi F, Azmi S, Wu T-Y. An Overview of Indian Biomedical Research on the Chikungunya Virus with Particular Reference to Its Vaccine, an Unmet Medical Need. Vaccines. 2023; 11(6):1102. https://doi.org/10.3390/vaccines11061102
Chicago/Turabian StyleVarikkodan, Muhammed Muhsin, Faisal Kunnathodi, Sarfuddin Azmi, and Tzong-Yuan Wu. 2023. "An Overview of Indian Biomedical Research on the Chikungunya Virus with Particular Reference to Its Vaccine, an Unmet Medical Need" Vaccines 11, no. 6: 1102. https://doi.org/10.3390/vaccines11061102
APA StyleVarikkodan, M. M., Kunnathodi, F., Azmi, S., & Wu, T.-Y. (2023). An Overview of Indian Biomedical Research on the Chikungunya Virus with Particular Reference to Its Vaccine, an Unmet Medical Need. Vaccines, 11(6), 1102. https://doi.org/10.3390/vaccines11061102








