Immunogenicity and Cross-Protective Efficacy Induced by an Inactivated Recombinant Avian Influenza A/H5N1 (Clade 2.3.4.4b) Vaccine against Co-Circulating Influenza A/H5Nx Viruses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells, Specific Pathogen-Free (SPF) Embryonated Eggs and Chicks, and Viruses
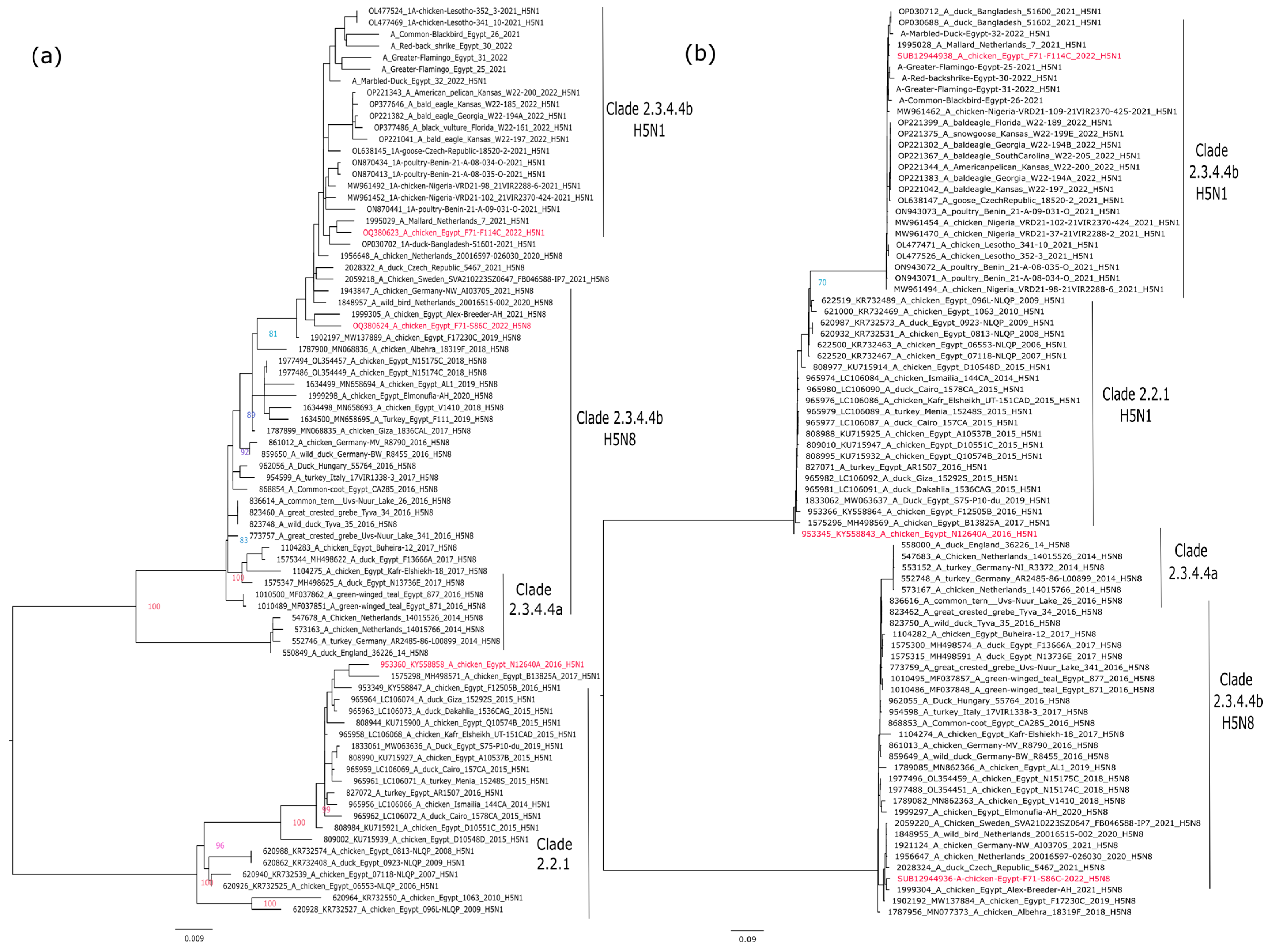
2.2. Phylogenetic Analysis
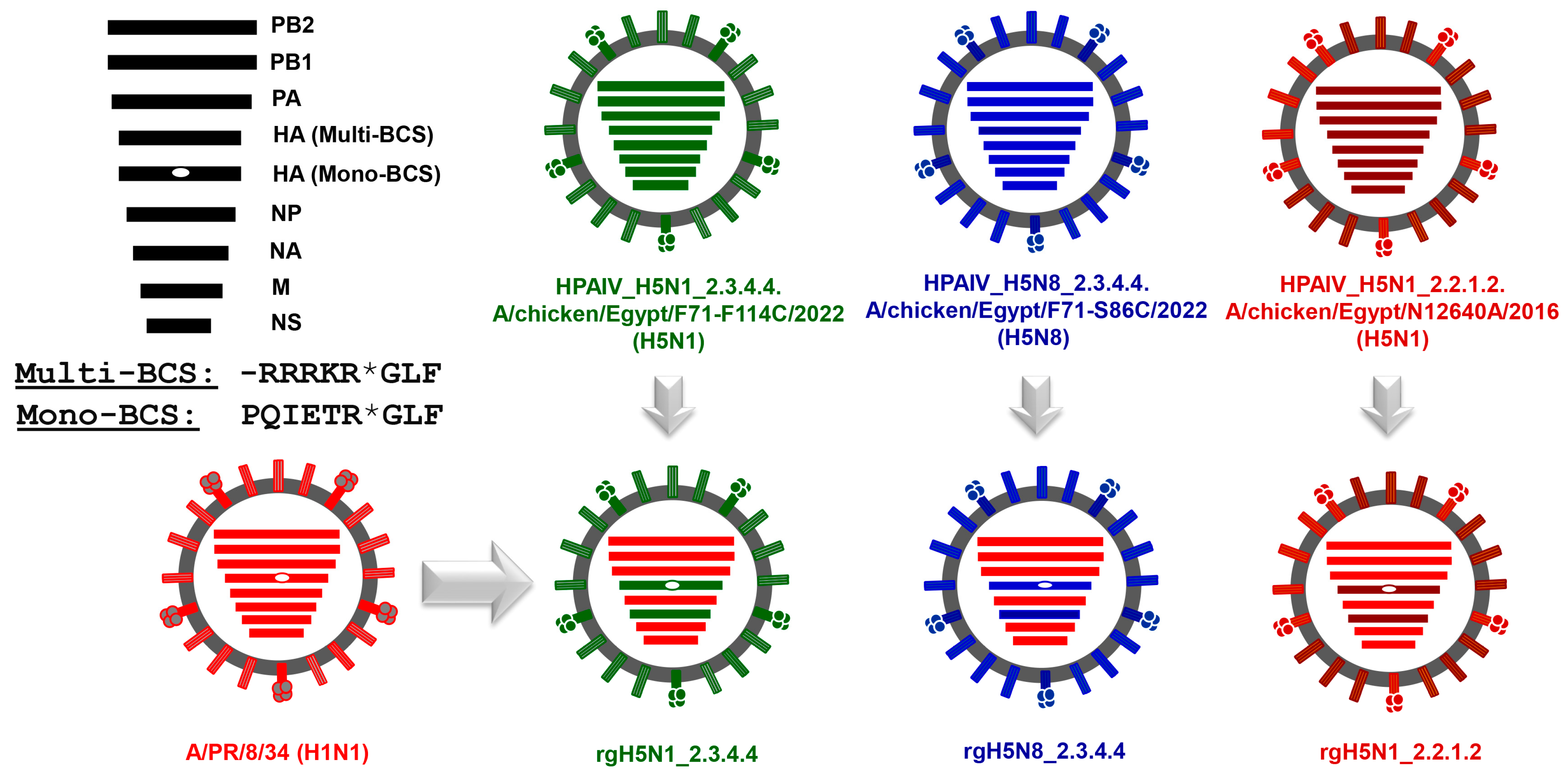
2.3. Construction of Reverse Genetics Plasmids
2.4. Rescue and Propagation of PR8-Based Candidate Vaccines against HPAI H5N1_2.3.4.4, HPAI H5N8_2.3.4.4, and HPAI H5N1_2.2.1.2 Viruses
2.5. Immunization of SPF Chickens and Seroconversion Determination
2.6. Challenge Infection and Viruses Shedding
2.7. Biosafety and Ethical Approval
2.8. Statistical Analyses
3. Results
3.1. Phylogenetic Analysis of the Candidate Vaccine Strains
3.2. Generation of the Candidate H5-Type Vaccine Strains
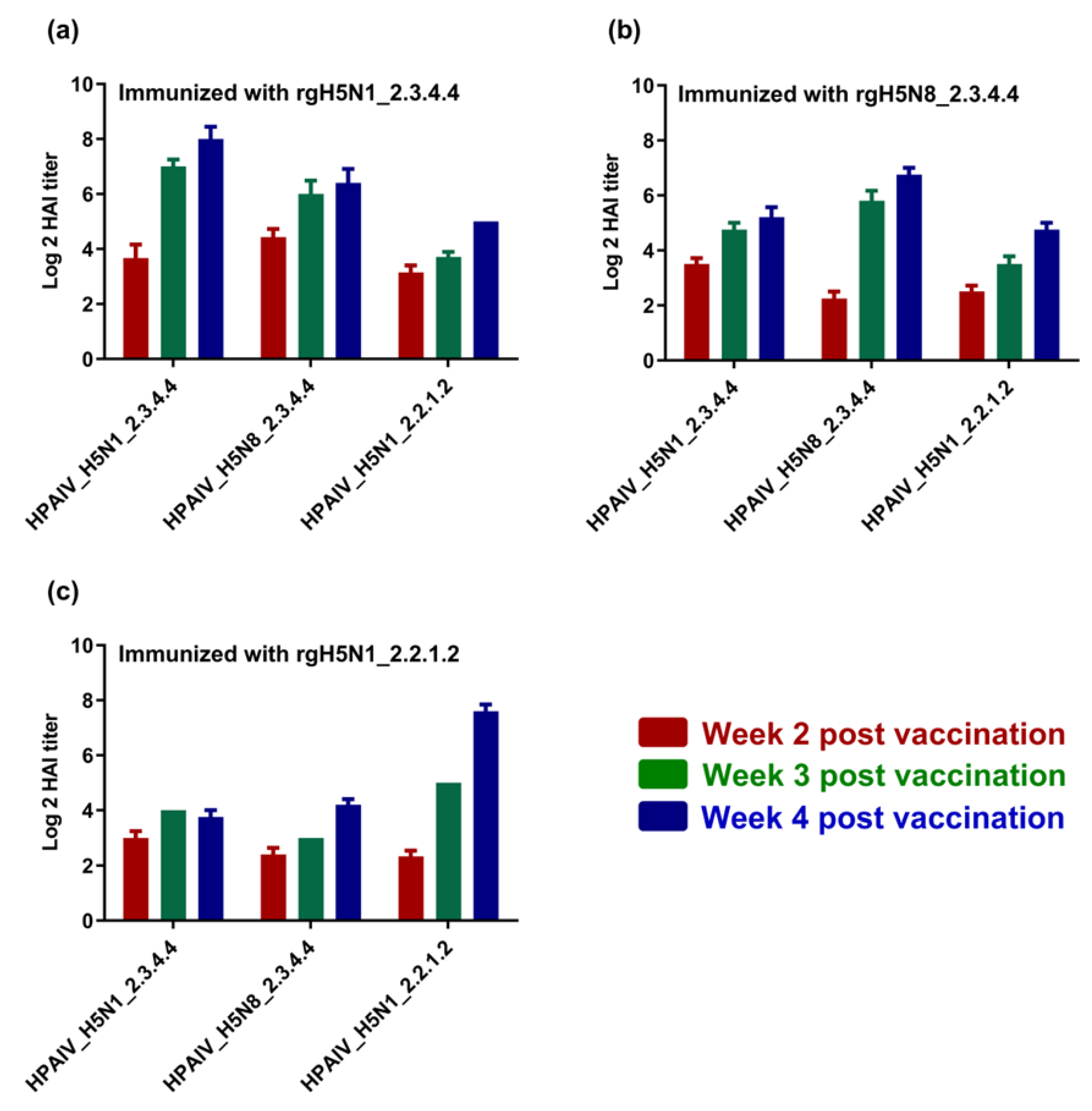
3.3. Immunogenicity of the Formulated H5-Type Vaccine and Cross-Reactivity against Homologous and Heterologous HPAI H5-Type Strains
3.4. Protectiveness of the Formulated Vaccines against HPAI H5-Type Viruses and Survival of Immunized Chickens following Homologous and Heterologous Challenge Infection
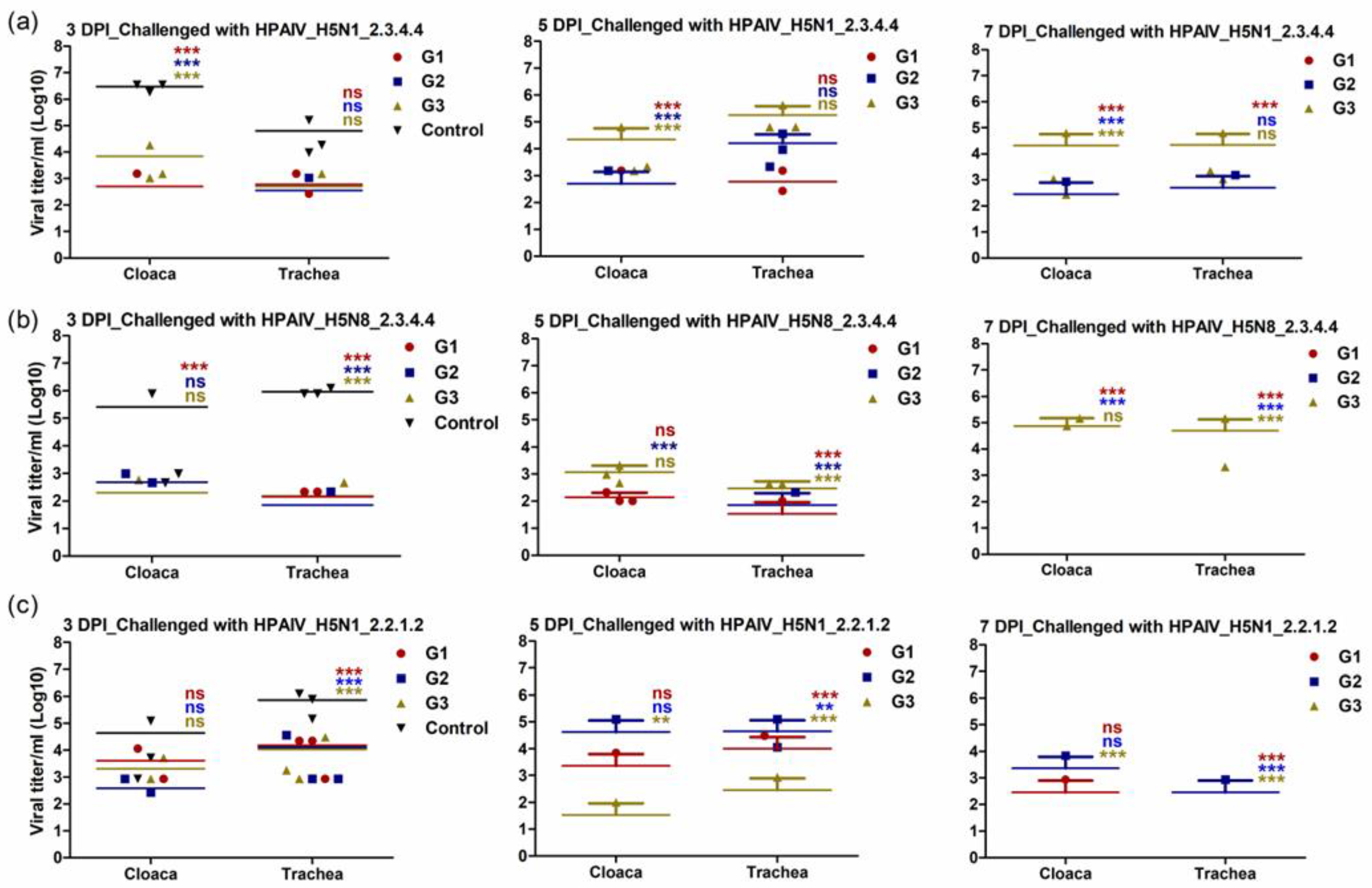
3.5. Immunized Chickens with rgH5N1_2.3.4.4 Showed Limited Shedding following Challenge Infection with Homologous and Heterologous HPAI H5Nx Strains
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tian, J.; Bai, X.; Li, M.; Zeng, X.; Xu, J.; Li, P.; Wang, M.; Song, X.; Zhao, Z.; Tian, G.; et al. Highly Pathogenic Avian Influenza Virus (H5N1) Clade 2.3.4.4b Introduced by Wild Birds, China, 2021. Emerg. Infect. Dis. 2023, 29, 1367–1375. [Google Scholar] [CrossRef]
- Youk, S.; Torchetti, M.K.; Lantz, K.; Lenoch, J.B.; Killian, M.L.; Leyson, C.; Bevins, S.N.; Dilione, K.; Ip, H.S.; Stallknecht, D.E.; et al. H5N1 highly pathogenic avian influenza clade 2.3.4.4b in wild and domestic birds: Introductions into the United States and reassortments, December 2021–April 2022. Virology 2023, 587, 109860. [Google Scholar] [CrossRef]
- Ariyama, N.; Pardo-Roa, C.; Muñoz, G.; Aguayo, C.; Ávila, C.; Mathieu, C.; Almonacid, L.; Medina, R.; Brito, B.; Johow, M.; et al. Highly Pathogenic Avian Influenza A(H5N1) Clade 2.3.4.4b Virus in Wild Birds, Chile. Emerg. Infect. Dis. J. 2023, 29. [Google Scholar]
- Ruiz-Saenz, J.; Martinez-Gutierrez, M.; Pujol, F.H. Multiple introductions of highly pathogenic avian influenza H5N1 clade 2.3.4.4b into South America. Travel Med. Infect. Dis. 2023, 53, 102591. [Google Scholar] [CrossRef]
- Mosaad, Z.; Elhusseiny, M.H.; Zanaty, A.; Fathy, M.M.; Hagag, N.M.; Mady, W.H.; Said, D.; Elsayed, M.M.; Erfan, A.M.; Rabie, N.; et al. Emergence of Highly Pathogenic Avian Influenza A Virus (H5N1) of Clade 2.3.4.4b in Egypt, 2021–2022. Pathogens 2023, 12, 90. [Google Scholar] [CrossRef]
- Guyonnet, V.; Peters, A.R. Are current avian influenza vaccines a solution for smallholder poultry farmers? Gates Open Res. 2020, 4, 122. [Google Scholar] [CrossRef]
- Dey, P.; Ahuja, A.; Panwar, J.; Choudhary, P.; Rani, S.; Kaur, M.; Sharma, A.; Kaur, J.; Yadav, A.K.; Sood, V.; et al. Immune Control of Avian Influenza Virus Infection and Its Vaccine Development. Vaccines 2023, 11, 593. [Google Scholar] [CrossRef]
- El-Shall, N.A.; Awad, A.M.; Sedeik, M.E. Examination of the protective efficacy of two avian influenza H5 vaccines against clade 2.3.4.4b H5N8 highly pathogenic avian influenza virus in commercial broilers. Res. Vet. Sci. 2021, 140, 125–133. [Google Scholar] [CrossRef]
- Hagag, N.M.; Yehia, N.; El-Husseiny, M.H.; Adel, A.; Shalaby, A.G.; Rabie, N.; Samy, M.; Mohamed, M.; El-Oksh, A.S.A.; Selim, A.; et al. Molecular Epidemiology and Evolutionary Analysis of Avian Influenza A(H5) Viruses Circulating in Egypt, 2019–2021. Viruses 2022, 14, 1758. [Google Scholar] [CrossRef]
- Kim, S.H. Challenge for One Health: Co-Circulation of Zoonotic H5N1 and H9N2 Avian Influenza Viruses in Egypt. Viruses 2018, 10, 121. [Google Scholar] [CrossRef]
- Fasanmi, O.G.; Odetokun, I.A.; Balogun, F.A.; Fasina, F.O. Public health concerns of highly pathogenic avian influenza H5N1 endemicity in Africa. Vet. World 2017, 10, 1194–1204. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, A.; Kanrai, P.; Petersen, H.; Ibrahim, S.; Rautenschlein, S.; Pleschka, S. Efficient Generation of Recombinant Influenza A Viruses Employing a New Approach to Overcome the Genetic Instability of HA Segments. PLoS ONE 2015, 10, e0116917. [Google Scholar] [CrossRef] [PubMed]
- Subbarao, K.; Chen, H.; Swayne, D.; Mingay, L.; Fodor, E.; Brownlee, G.; Xu, X.; Lu, X.; Katz, J.; Cox, N.; et al. Evaluation of a genetically modified reassortant H5N1 influenza A virus vaccine candidate generated by plasmid-based reverse genetics. Virology 2003, 305, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Bu, Z. Development and application of avian influenza vaccines in China. Curr. Top. Microbiol. Immunol. 2009, 333, 153–162. [Google Scholar]
- Krammer, F.; Schultz-Cherry, S. We need to keep an eye on avian influenza. Nat. Rev. Immunol. 2023, 23, 267–268. [Google Scholar] [CrossRef]
- Brauer, R.; Chen, P. Influenza virus propagation in embryonated chicken eggs. J. Vis. Exp. JoVE 2015, 97, e52421. [Google Scholar]
- Karakus, U.; Crameri, M.; Lanz, C.; Yángüez, E. Propagation and Titration of Influenza Viruses. In Influenza Virus: Methods and Protocols; Yamauchi, Y., Ed.; Springer: New York, NY, USA, 2018; pp. 59–88. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Mostafa, A.; Kanrai, P.; Ziebuhr, J.; Pleschka, S. Improved dual promotor-driven reverse genetics system for influenza viruses. J. Virol. Methods 2013, 193, 603–610. [Google Scholar] [CrossRef]
- Mostafa, A.; Mahmoud, S.H.; Shehata, M.; Müller, C.; Kandeil, A.; El-Shesheny, R.; Nooh, H.Z.; Kayali, G.; Ali, M.A.; Pleschka, S. PA from a Recent H9N2 (G1-like) Avian Influenza a Virus (AIV) Strain Carrying Lysine 367 Confers Altered Replication Efficiency and Pathogenicity to Contemporaneous H5N1 in Mammalian Systems. Viruses 2020, 12, 1046. [Google Scholar] [CrossRef]
- Ping, J.; Lopes, T.J.S.; Nidom, C.A.; Ghedin, E.; Macken, C.A.; Fitch, A.; Imai, M.; Maher, E.A.; Neumann, G.; Kawaoka, Y. Development of high-yield influenza A virus vaccine viruses. Nat. Commun. 2015, 6, 8148. [Google Scholar] [CrossRef] [PubMed]
- Claes, F.; Morzaria, S.P.; Donis, R.O. Emergence and dissemination of clade 2.3.4.4 H5Nx influenza viruses-how is the Asian HPAI H5 lineage maintained. Curr. Opin. Virol. 2016, 16, 158–163. [Google Scholar] [CrossRef]
- Gutiérrez, R.A.; Naughtin, M.J.; Horm, S.V.; San, S.; Buchy, P. A(H5N1) Virus Evolution in South East Asia. Viruses 2009, 1, 335–361. [Google Scholar] [CrossRef] [PubMed]
- Vijaykrishna, D.; Bahl, J.; Riley, S.; Duan, L.; Zhang, J.X.; Chen, H.; Peiris, J.S.; Smith, G.J.; Guan, Y. Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses. PLoS Pathog. 2008, 4, e1000161. [Google Scholar] [CrossRef] [PubMed]
- WHO/OIE/FAO. Continued evolution of highly pathogenic avian influenza A (H5N1): Updated nomenclature. Influenza Other Respir. Viruses 2012, 6, 1–5. [Google Scholar] [CrossRef]
- Li, Y.; Li, M.; Li, Y.; Tian, J.; Bai, X.; Yang, C.; Shi, J.; Ai, R.; Chen, W.; Zhang, W.; et al. Outbreaks of Highly Pathogenic Avian Influenza (H5N6) Virus Subclade 2.3.4.4h in Swans, Xinjiang, Western China, 2020. Emerg. Infect. Dis. 2020, 26, 2956–2960. [Google Scholar] [CrossRef]
- Cui, P.; Shi, J.; Wang, C.; Zhang, Y.; Xing, X.; Kong, H.; Yan, C.; Zeng, X.; Liu, L.; Tian, G.; et al. Global dissemination of H5N1 influenza viruses bearing the clade 2.3.4.4b HA gene and biologic analysis of the ones detected in China. Emerg. Microbes Infect. 2022, 11, 1693–1704. [Google Scholar] [CrossRef]
- Cui, P.; Zeng, X.; Li, X.; Li, Y.; Shi, J.; Zhao, C.; Qu, Z.; Wang, Y.; Guo, J.; Gu, W.; et al. Genetic and biological characteristics of the globally circulating H5N8 avian influenza viruses and the protective efficacy offered by the poultry vaccine currently used in China. Sci. China. Life Sci. 2022, 65, 795–808. [Google Scholar] [CrossRef]
- Baek, Y.G.; Lee, Y.N.; Lee, D.H.; Shin, J.I.; Lee, J.H.; Chung, D.H.; Lee, E.K.; Heo, G.B.; Sagong, M.; Kye, S.J.; et al. Multiple Reassortants of H5N8 Clade 2.3.4.4b Highly Pathogenic Avian Influenza Viruses Detected in South Korea during the Winter of 2020–2021. Viruses 2021, 13, 490. [Google Scholar] [CrossRef]
- Adlhoch, C.; Fusaro, A.; Gonzales, J.L.; Kuiken, T.; Marangon, S.; Niqueux, É.; Staubach, C.; Terregino, C.; Aznar, I.; Guajardo, I.M.; et al. Avian influenza overview February–May 2021. EFSA J. Eur. Food Saf. Auth. 2021, 19, e06951. [Google Scholar]
- Adlhoch, C.; Fusaro, A.; Gonzales, J.L.; Kuiken, T.; Marangon, S.; Niqueux, É.; Staubach, C.; Terregino, C.; Aznar, I.; Muñoz Guajardo, I.; et al. Avian influenza overview September–December 2021. EFSA J. Eur. Food Saf. Auth. 2021, 19, e07108. [Google Scholar]
- Salaheldin, A.H.; El-Hamid, H.S.; Elbestawy, A.R.; Veits, J.; Hafez, H.M.; Mettenleiter, T.C.; Abdelwhab, E.M. Multiple Introductions of Influenza A(H5N8) Virus into Poultry, Egypt, 2017. Emerg. Infect. Dis. 2018, 24, 943–946. [Google Scholar] [CrossRef]
- Eggert, D.; Swayne, D.E. Single vaccination provides limited protection to ducks and geese against H5N1 high pathogenicity avian influenza virus. Avian Dis 2010, 54, 1224–1229. [Google Scholar] [CrossRef] [PubMed]
- Spackman, E.; Swayne, D.E.; Pantin-Jackwood, M.J.; Wan, X.-F.; Torchetti, M.K.; Hassan, M.; Suarez, D.L.; Sá e Silva, M. Variation in protection of four divergent avian influenza virus vaccine seed strains against eight clade 2.2.1 and 2.2.1.1. Egyptian H5N1 high pathogenicity variants in poultry. Influenza Other Respir. Viruses 2014, 8, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.S.; Webby, R.J. Traditional and new influenza vaccines. Clin. Microbiol. Rev. 2013, 26, 476–492. [Google Scholar] [CrossRef] [PubMed]
- Saeed El-Naggar, M.; Mohammed Ibrahim, H.; Mohammed Salem, H.; Marouf, S.A. A Novel Locally Prepared Inactivated Bivalent Mycoplasma Vaccine for Chicken Flocks in Egypt. Adv. Anim. Vet. Sci. 2021, 10, 55–61. [Google Scholar] [CrossRef]
- Ibrahim, M.; Zakaria, S.; Bazid, A.I.; Kilany, W.H.; Zain El-Abideen, M.A.; Ali, A. A single dose of inactivated oil-emulsion bivalent H5N8/H5N1 vaccine protects chickens against the lethal challenge of both highly pathogenic avian influenza viruses. Comp. Immunol. Microbiol. Infect. Dis. 2021, 74, 101601. [Google Scholar] [CrossRef]
- Cate, T.R.; Rayford, Y.; Niño, D.; Winokur, P.; Brady, R.; Belshe, R.; Chen, W.; Atmar, R.L.; Couch, R.B. A high dosage influenza vaccine induced significantly more neuraminidase antibody than standard vaccine among elderly subjects. Vaccine 2010, 28, 2076–2079. [Google Scholar] [CrossRef]
- Kamel, M.N.; Mahmoud, S.H.; Moatasim, Y.; El Taweel, A.; Shehata, M.; Shehata, M.R.; AbdElSalam, E.T.; Ali, M.A.; Mostafa, A. Immunogenicity and effectiveness of a bivalent influenza A/H1N2 vaccine strain against seasonal human influenza A viruses in mice. J. Genet. Eng. Biotechnol. 2022, 20, 155. [Google Scholar] [CrossRef]

| Challenge-Infection Strain | Group/Vaccine | Days Post-Infection | Number of Deaths | PROTECTION (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||||
| Challenge with HPAIV_H5N1_2.3.4.4 | G1/rgH5N1_2.3.4.4 | 0 | 100% | |||||||||
| G2/rgH5N8_2.3.4.4 | 0 | 100% | ||||||||||
| G3/rgH5N1_2.2.1.2 | 1 | 1 | 2 | 60% | ||||||||
| Control | 2 | 3 | 5 | 0% | ||||||||
| Challenge with HPAIV_H5N8_2.3.4.4 | G1/rgH5N1_2.3.4.4 | 1 | 1 | 80% | ||||||||
| G2/rgH5N8_2.3.4.4 | 0 | 100% | ||||||||||
| G3/rgH5N1_2.2.1.2 | 1 | 1 | 1 | 3 | 40% | |||||||
| Control | 2 | 3 | 5 | 0% | ||||||||
| Challenge with HPAIV_H5N1_2.2.1.2 | G1/rgH5N1_2.3.4.4 | 1 | 1 | 2 | 60% | |||||||
| G2/rgH5N8_2.3.4.4 | 3 | 3 | 40% | |||||||||
| G3/rgH5N1_2.2.1.2 | 1 | 1 | 80% | |||||||||
| Control | 2 | 3 | 5 | 0% | ||||||||
| Amino Acid Residue * | H5N1_2.3.4.4b | H5N8_2.3.4.4b | H5N1_2.2.1.2 |
|---|---|---|---|
| 69 | E | E | E |
| 71 | I | I | L |
| 83 | A | A | I |
| 95 | L | L | F |
| 133 | A | A | A |
| 140 | A | A | R |
| 162 | I | I | K |
| 183 | A | A | A |
| 189 | N | N | K |
| 194 | T | T | N |
| 270 | E | E | E |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahmoud, S.H.; Khalil, A.A.; Abo Shama, N.M.; El Sayed, M.F.; Soliman, R.A.; Hagag, N.M.; Yehia, N.; Naguib, M.M.; Arafa, A.-S.; Ali, M.A.; et al. Immunogenicity and Cross-Protective Efficacy Induced by an Inactivated Recombinant Avian Influenza A/H5N1 (Clade 2.3.4.4b) Vaccine against Co-Circulating Influenza A/H5Nx Viruses. Vaccines 2023, 11, 1397. https://doi.org/10.3390/vaccines11091397
Mahmoud SH, Khalil AA, Abo Shama NM, El Sayed MF, Soliman RA, Hagag NM, Yehia N, Naguib MM, Arafa A-S, Ali MA, et al. Immunogenicity and Cross-Protective Efficacy Induced by an Inactivated Recombinant Avian Influenza A/H5N1 (Clade 2.3.4.4b) Vaccine against Co-Circulating Influenza A/H5Nx Viruses. Vaccines. 2023; 11(9):1397. https://doi.org/10.3390/vaccines11091397
Chicago/Turabian StyleMahmoud, Sara H., Ahmed A. Khalil, Noura M. Abo Shama, Marwa F. El Sayed, Reem A. Soliman, Naglaa M. Hagag, Nahed Yehia, Mahmoud M. Naguib, Abdel-Sattar Arafa, Mohamed A. Ali, and et al. 2023. "Immunogenicity and Cross-Protective Efficacy Induced by an Inactivated Recombinant Avian Influenza A/H5N1 (Clade 2.3.4.4b) Vaccine against Co-Circulating Influenza A/H5Nx Viruses" Vaccines 11, no. 9: 1397. https://doi.org/10.3390/vaccines11091397
APA StyleMahmoud, S. H., Khalil, A. A., Abo Shama, N. M., El Sayed, M. F., Soliman, R. A., Hagag, N. M., Yehia, N., Naguib, M. M., Arafa, A.-S., Ali, M. A., El-Safty, M. M., & Mostafa, A. (2023). Immunogenicity and Cross-Protective Efficacy Induced by an Inactivated Recombinant Avian Influenza A/H5N1 (Clade 2.3.4.4b) Vaccine against Co-Circulating Influenza A/H5Nx Viruses. Vaccines, 11(9), 1397. https://doi.org/10.3390/vaccines11091397







