Host Genetic Variation Impacts SARS-CoV-2 Vaccination Response in the Diversity Outbred Mouse Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus Stocks
2.2. Mice
2.3. Genotyping
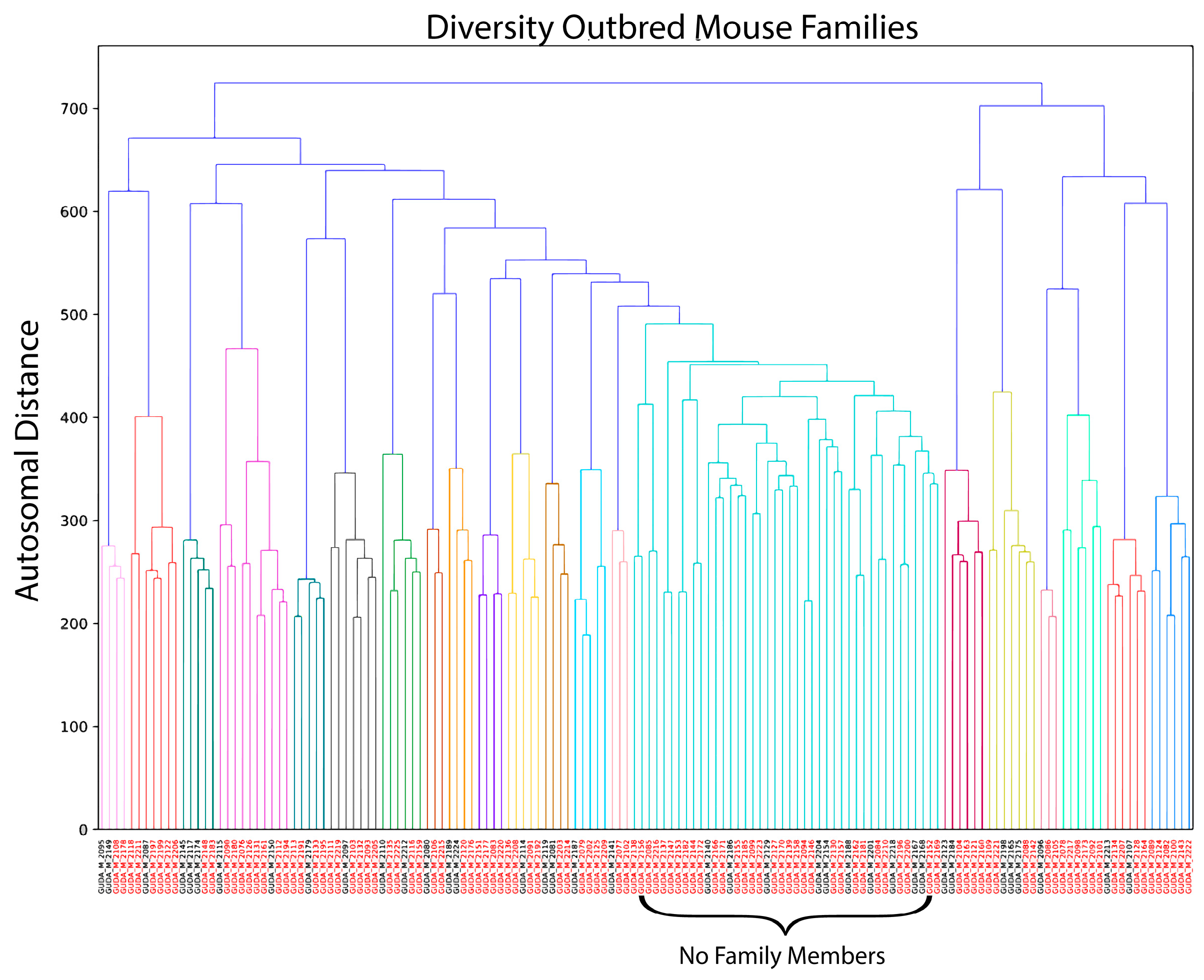
2.4. Genetic Clustering
2.5. Vaccination
2.6. Virus Challenge
2.7. Neutralizing Antibody
2.8. Viral Plaque Assays
2.9. Statistical Analysis
3. Results
3.1. Case–Control Design in an Outbred Mouse Population
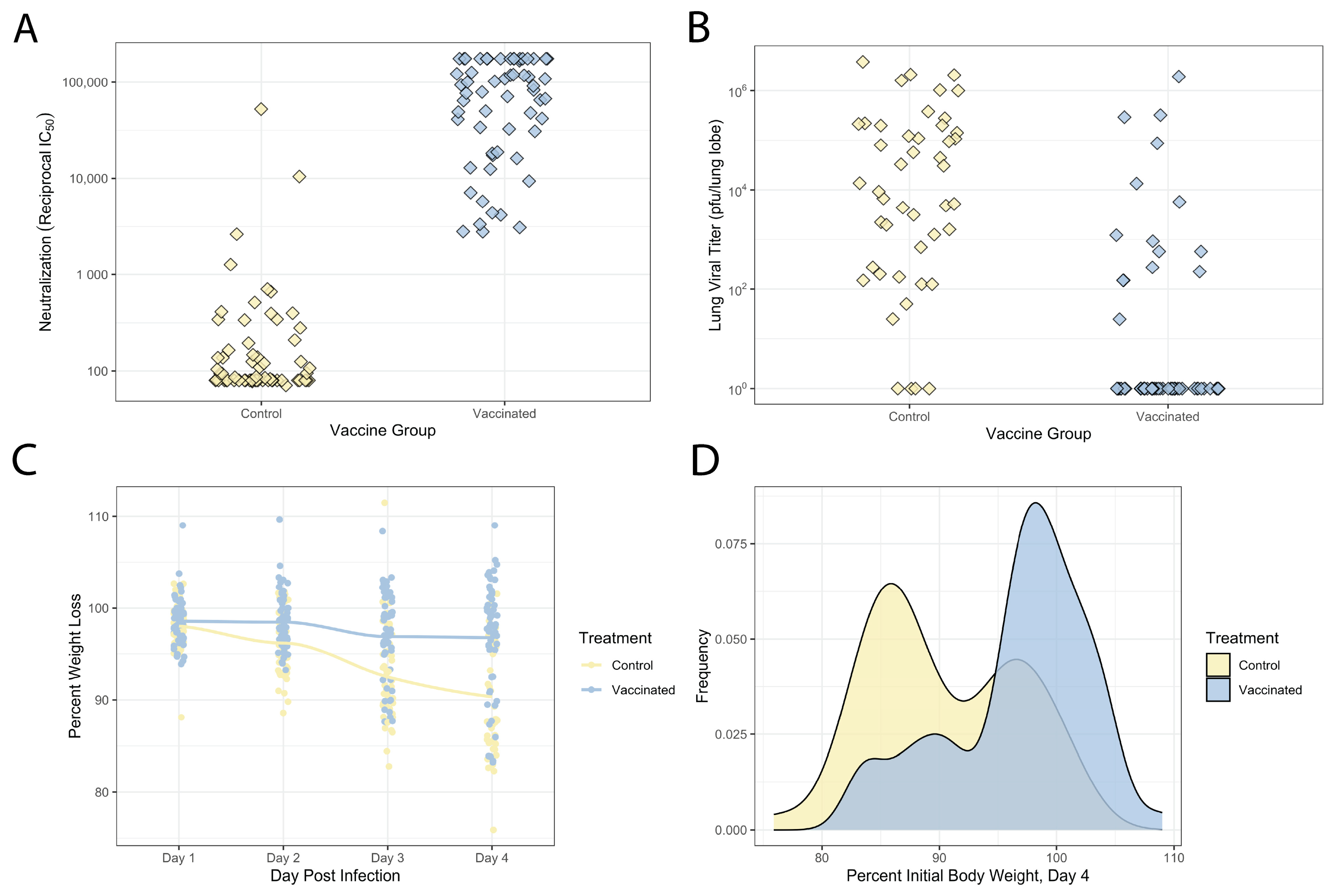
3.2. Variable Vaccination and Disease Response across This DO Population
3.3. Genetic Diversity Contributes to Vaccination Outcomes
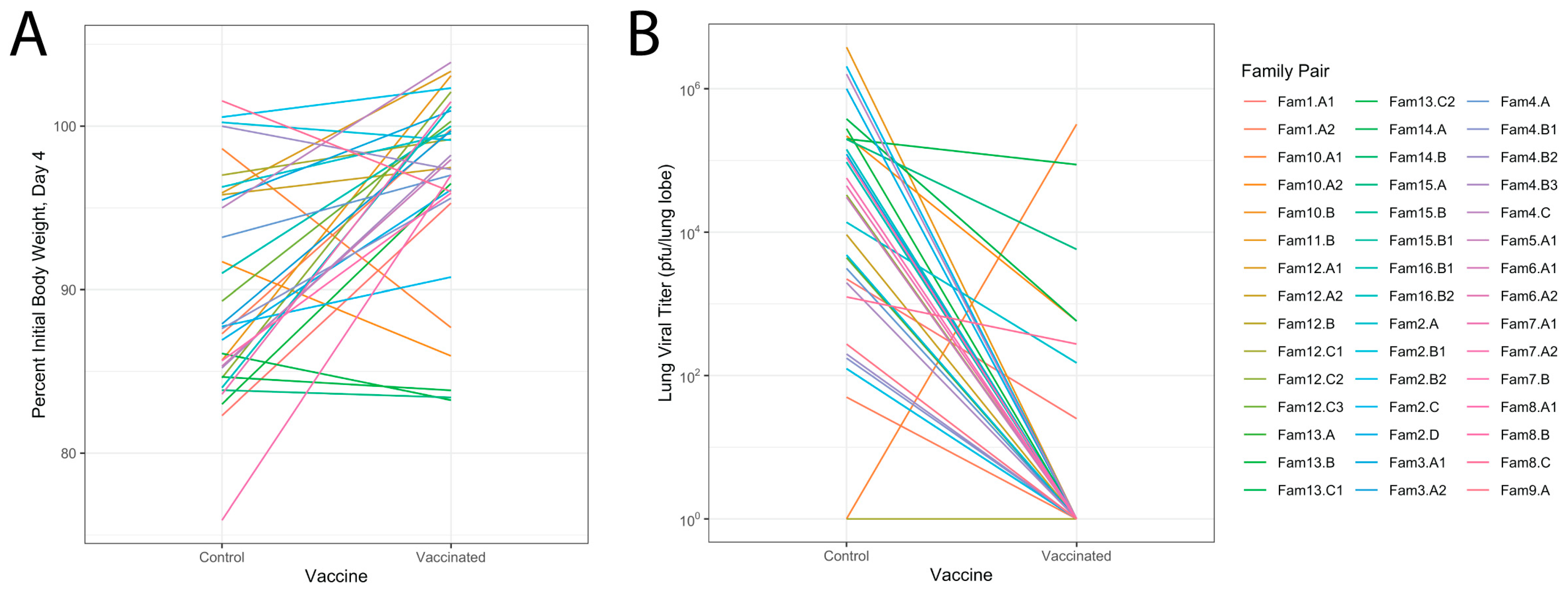
3.4. Relationship of Infectious Responses among Vaccine Treatments and Families
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- COVID-19 Map. Available online: https://coronavirus.jhu.edu/map.html (accessed on 18 December 2023).
- Yeyati, E.L.; Filippini, F. Social and Economic Impact of COVID-19. Brookings Institution. Available online: https://www.brookings.edu/articles/social-and-economic-impact-of-covid-19/ (accessed on 18 December 2023).
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Heath, P.T.; Galiza, E.P.; Baxter, D.N.; Boffito, M.; Browne, D.; Burns, F.; Chadwick, D.R.; Clark, R.; Cosgrove, C.; Galloway, J.; et al. Safety and Efficacy of NVX-CoV2373 COVID-19 Vaccine. N. Engl. J. Med. 2021, 385, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Hu, Y.; Xu, M.; Chen, Z.; Yang, W.; Jiang, Z.; Li, M.; Jin, H.; Cui, G.; Chen, P.; et al. Safety, Tolerability, and Immunogenicity of an Inactivated SARS-CoV-2 Vaccine (CoronaVac) in Healthy Adults Aged 60 Years and Older: A Randomised, Double-Blind, Placebo-Controlled, Phase 1/2 Clinical Trial. Lancet Infect. Diseases 2021, 21, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Nasreen, S.; Chung, H.; He, S.; Brown, K.A.; Gubbay, J.B.; Buchan, S.A.; Fell, D.B.; Austin, P.C.; Schwartz, K.L.; Sundaram, M.E.; et al. Effectiveness of COVID-19 Vaccines against Symptomatic SARS-CoV-2 Infection and Severe Outcomes with Variants of Concern in Ontario. Nat. Microbiol. 2022, 7, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Andrews, N.; Stowe, J.; Kirsebom, F.; Toffa, S.; Rickeard, T.; Gallagher, E.; Gower, C.; Kall, M.; Groves, N.; O’Connell, A.-M.; et al. COVID-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N. Engl. J. Med. 2022, 386, 1532–1546. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Huo, J.; Zhou, D.; Zahradník, J.; Supasa, P.; Liu, C.; Duyvesteyn, H.M.E.; Ginn, H.M.; Mentzer, A.J.; Tuekprakhon, A.; et al. SARS-CoV-2 Omicron-B.1.1.529 Leads to Widespread Escape from Neutralizing Antibody Responses. Cell 2022, 185, 467–484.e15. [Google Scholar] [CrossRef]
- Willett, B.J.; Grove, J.; MacLean, O.A.; Wilkie, C.; De Lorenzo, G.; Furnon, W.; Cantoni, D.; Scott, S.; Logan, N.; Ashraf, S.; et al. SARS-CoV-2 Omicron Is an Immune Escape Variant with an Altered Cell Entry Pathway. Nat. Microbiol. 2022, 7, 1161–1179. [Google Scholar] [CrossRef]
- Davis, C.; Logan, N.; Tyson, G.; Orton, R.; Harvey, W.T.; Perkins, J.S.; Mollett, G.; Blacow, R.M.; COVID-19 Genomics UK (COG-UK) Consortium; Peacock, T.P.; et al. Reduced Neutralisation of the Delta (B.1.617.2) SARS-CoV-2 Variant of Concern Following Vaccination. PLoS Pathog. 2021, 17, e1010022. [Google Scholar] [CrossRef]
- Ovsyannikova, I.G.; Salk, H.M.; Kennedy, R.B.; Haralambieva, I.H.; Zimmermann, M.T.; Grill, D.E.; Oberg, A.L.; Poland, G.A. Gene Signatures Associated with Adaptive Humoral Immunity Following Seasonal Influenza A/H1N1 Vaccination. Genes. Immun. 2016, 17, 371–379. [Google Scholar] [CrossRef]
- Tsang, T.K.; Wang, C.; Tsang, N.N.Y.; Fang, V.J.; Perera, R.A.P.M.; Malik Peiris, J.S.; Leung, G.M.; Cowling, B.J.; Ip, D.K.M. Impact of Host Genetic Polymorphisms on Response to Inactivated Influenza Vaccine in Children. Npj Vaccines 2023, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Poland, G.A.; Ovsyannikova, I.G.; Jacobson, R.M.; Vierkant, R.A.; Jacobsen, S.J.; Pankratz, V.S.; Schaid, D.J. Identification of an Association between HLA Class II Alleles and Low Antibody Levels after Measles Immunization. Vaccine 2001, 20, 430–438. [Google Scholar] [CrossRef]
- Jacobson, R.M.; Poland, G.A.; Vierkant, R.A.; Pankratz, V.S.; Schaid, D.J.; Jacobsen, S.J.; Sauver, J.S.; Moore, S.B. The Association of Class I HLA Alleles and Antibody Levels after a Single Dose of Measles Vaccine. Hum. Immunol. 2003, 64, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Hennig, B.J.; Fielding, K.; Broxholme, J.; Diatta, M.; Mendy, M.; Moore, C.; Pollard, A.J.; Rayco-Solon, P.; Sirugo, G.; van der Sande, M.A.; et al. Host Genetic Factors and Vaccine-Induced Immunity to Hepatitis B Virus Infection. PLoS ONE 2008, 3, e1898. [Google Scholar] [CrossRef] [PubMed]
- Gemmati, D.; Longo, G.; Gallo, I.; Silva, J.A.; Secchiero, P.; Zauli, G.; Hanau, S.; Passaro, A.; Pellegatti, P.; Pizzicotti, S.; et al. Host Genetics Impact on SARS-CoV-2 Vaccine-Induced Immunoglobulin Levels and Dynamics: The Role of TP53, ABO, APOE, ACE2, HLA-A, and CRP Genes. Front. Genet. 2022, 13, 1028081. [Google Scholar] [CrossRef] [PubMed]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schäfer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA Vaccine Design Enabled by Prototype Pathogen Preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Fontela, C.; Widerspick, L.; Albrecht, R.A.; Beer, M.; Carroll, M.W.; de Wit, E.; Diamond, M.S.; Dowling, W.E.; Funnell, S.G.P.; García-Sastre, A.; et al. Advances and Gaps in SARS-CoV-2 Infection Models. PLOS Pathog. 2022, 18, e1010161. [Google Scholar] [CrossRef]
- Zeng, M.; Nourishirazi, E.; Guinet, E.; Nouri-Shirazi, M. The Genetic Background Influences the Cellular and Humoral Immune Responses to Vaccines. Clin. Exp. Immunol. 2016, 186, 190–204. [Google Scholar] [CrossRef]
- Kuipers, K.; van Selm, S.; van Opzeeland, F.; Langereis, J.D.; Verhagen, L.M.; Diavatopoulos, D.A.; de Jonge, M.I. Genetic Background Impacts Vaccine-Induced Reduction of Pneumococcal Colonization. Vaccine 2017, 35, 5235–5241. [Google Scholar] [CrossRef]
- Trammell, R.A.; Liberati, T.A.; Toth, L.A. Host Genetic Background and the Innate Inflammatory Response of Lung to Influenza Virus. Microbes Infect. 2012, 14, 50–58. [Google Scholar] [CrossRef]
- Churchill, G.A.; Gatti, D.M.; Munger, S.C.; Svenson, K.L. The Diversity Outbred Mouse Population. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2012, 23, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Threadgill, D.W.; Miller, D.R.; Churchill, G.A.; de Villena, F.P.-M. The Collaborative Cross: A Recombinant Inbred Mouse Population for the Systems Genetic Era. ILAR J. 2011, 52, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.M.; Proulx, M.K.; Olive, A.J.; Laddy, D.; Mishra, B.B.; Moss, C.; Gutierrez, N.M.; Bellerose, M.M.; Barreira-Silva, P.; Phuah, J.Y.; et al. Tuberculosis Susceptibility and Vaccine Protection Are Independently Controlled by Host Genotype. mBio 2016, 7, e01516-16. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.L.; Rossi, A.P.; Beamer, G.L.; Gatti, D.M.; Kramnik, I.; Elkins, K.L. The Diversity Outbred Mouse Population Is an Improved Animal Model of Vaccination against Tuberculosis That Reflects Heterogeneity of Protection. mSphere 2020. [Google Scholar] [CrossRef]
- Kurtz, S.L.; Mittereder, L.R.; Lehman, C.C.; Khan, H.; Gould, V.A.; Elkins, K.L. Intravenous BCG Vaccination of Diversity Outbred Mice Results in Moderately Enhanced Protection against Challenge with Mycobacterium Tuberculosis Compared to Intradermal Vaccination. Infect. Immun. 2023, 91, e00168-23. [Google Scholar] [CrossRef] [PubMed]
- McHugh, K.J.; Mandalapu, S.; Kolls, J.K.; Ross, T.M.; Alcorn, J.F. A Novel Outbred Mouse Model of 2009 Pandemic Influenza and Bacterial Co-Infection Severity. PLoS ONE 2013, 8, e82865. [Google Scholar] [CrossRef]
- Dinnon, K.H.; Leist, S.R.; Schäfer, A.; Edwards, C.E.; Martinez, D.R.; Montgomery, S.A.; West, A.; Yount, B.L.; Hou, Y.J.; Adams, L.E.; et al. A Mouse-Adapted Model of SARS-CoV-2 to Test COVID-19 Countermeasures. Nature 2020, 586, 560–566. [Google Scholar] [CrossRef]
- Leist, S.R.; Dinnon, K.H.; Schäfer, A.; Tse, L.V.; Okuda, K.; Hou, Y.J.; West, A.; Edwards, C.E.; Sanders, W.; Fritch, E.J.; et al. A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell 2020, 183, 1070–1085.e12. [Google Scholar] [CrossRef]
- Sigmon, J.S.; Blanchard, M.W.; Baric, R.S.; Bell, T.A.; Brennan, J.; Brockmann, G.A.; Burks, A.W.; Calabrese, J.M.; Caron, K.M.; Cheney, R.E.; et al. Content and Performance of the MiniMUGA Genotyping Array: A New Tool to Improve Rigor and Reproducibility in Mouse Research. Genetics 2020, 216, 905–930. [Google Scholar] [CrossRef]
- Martinez, D.R.; Schaefer, A.; Gobeil, S.; Li, D.; De la Cruz, G.; Parks, R.; Lu, X.; Barr, M.; Manne, K.; Mansouri, K.; et al. A Broadly Neutralizing Antibody Protects against SARS-CoV, Pre-Emergent Bat CoVs, and SARS-CoV-2 Variants in Mice. BioRxiv Prepr. Serv. Biol. 2021. [Google Scholar] [CrossRef]
- Gralinski, L.E.; Ferris, M.T.; Aylor, D.L.; Whitmore, A.C.; Green, R.; Frieman, M.B.; Deming, D.; Menachery, V.D.; Miller, D.R.; Buus, R.J.; et al. Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross. PLoS Genet. 2015, 11, e1005504. [Google Scholar] [CrossRef] [PubMed]
- Gralinski, L.E.; Menachery, V.D.; Morgan, A.P.; Totura, A.L.; Beall, A.; Kocher, J.; Plante, J.; Harrison-Shostak, D.C.; Schäfer, A.; Pardo-Manuel de Villena, F.; et al. Allelic Variation in the Toll-Like Receptor Adaptor Protein Ticam2 Contributes to SARS-Coronavirus Pathogenesis in Mice. G3 GenesGenomesGenetics 2017, 7, 1653–1663. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.B.; Swarts, J.L.; Leist, S.R.; Schäfer, A.; Menachery, V.D.; Gralinski, L.E.; Jeng, S.; Miller, D.R.; Mooney, M.A.; McWeeney, S.K.; et al. Baseline T Cell Immune Phenotypes Predict Virologic and Disease Control upon SARS-CoV Infection in Collaborative Cross Mice. PLoS Pathog. 2021, 17, e1009287. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, A.; Leist, S.R.; Gralinski, L.E.; Martinez, D.R.; Winkler, E.S.; Okuda, K.; Hawkins, P.E.; Gully, K.L.; Graham, R.L.; Scobey, D.T.; et al. A Multitrait Locus Regulates Sarbecovirus Pathogenesis. mBio 2022, 13, e0145422. [Google Scholar] [CrossRef] [PubMed]
- Velavan, T.P.; Pallerla, S.R.; Rüter, J.; Augustin, Y.; Kremsner, P.G.; Krishna, S.; Meyer, C.G. Host Genetic Factors Determining COVID-19 Susceptibility and Severity. EBioMedicine 2021, 72, 103629. [Google Scholar] [CrossRef]
- Shelton, J.F.; Shastri, A.J.; Ye, C.; Weldon, C.H.; Filshtein-Sonmez, T.; Coker, D.; Symons, A.; Esparza-Gordillo, J.; 23andMe COVID-19 Team; Aslibekyan, S.; et al. Trans-Ancestry Analysis Reveals Genetic and Nongenetic Associations with COVID-19 Susceptibility and Severity. Nat. Genet. 2021, 53, 801–808. [Google Scholar] [CrossRef]
- Pairo-Castineira, E.; Clohisey, S.; Klaric, L.; Bretherick, A.D.; Rawlik, K.; Pasko, D.; Walker, S.; Parkinson, N.; Fourman, M.H.; Russell, C.D.; et al. Genetic Mechanisms of Critical Illness in COVID-19. Nature 2021, 591, 92–98. [Google Scholar] [CrossRef]
- Li, P.; Ke, Y.; Shen, W.; Shi, S.; Wang, Y.; Lin, K.; Guo, X.; Wang, C.; Zhang, Y.; Zhao, Z. Targeted Screening of Genetic Associations with COVID-19 Susceptibility and Severity. Front. Genet. 2022, 13. [Google Scholar] [CrossRef]
- Schaid, D.J.; Haralambieva, I.H.; Larrabee, B.R.; Ovsyannikova, I.G.; Kennedy, R.B.; Poland, G.A. Heritability of Vaccine-Induced Measles Neutralizing Antibody Titers. Vaccine 2017, 35, 1390–1394. [Google Scholar] [CrossRef]
- Ovsyannikova, I.G.; Kennedy, R.B.; O’Byrne, M.; Jacobson, R.M.; Pankratz, V.S.; Poland, G.A. Genome-Wide Association Study of Antibody Response to Smallpox Vaccine. Vaccine 2012, 30, 4182–4189. [Google Scholar] [CrossRef]
- Vogel, A.B.; Kanevsky, I.; Che, Y.; Swanson, K.A.; Muik, A.; Vormehr, M.; Kranz, L.M.; Walzer, K.C.; Hein, S.; Güler, A.; et al. BNT162b Vaccines Protect Rhesus Macaques from SARS-CoV-2. Nature 2021, 592, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Scheaffer, S.M.; Lee, D.; Whitener, B.; Ying, B.; Wu, K.; Liang, C.-Y.; Jani, H.; Martin, P.; Amato, N.J.; Avena, L.E.; et al. Bivalent SARS-CoV-2 mRNA Vaccines Increase Breadth of Neutralization and Protect against the BA.5 Omicron Variant in Mice. Nat. Med. 2023, 29, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, U.B.; Namachivayam, M.; Jeewon, R.; Huang, J.-D.; Durairajan, S.S.K. Animal Models for SARS-CoV-2 and SARS-CoV-1 Pathogenesis, Transmission and Therapeutic Evaluation. World J. Virol. 2022, 11, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Shao, W.; Chen, X.; Zhang, B.; Wang, G.; Zhang, W. Real-World Effectiveness of COVID-19 Vaccines: A Literature Review and Meta-Analysis. Int. J. Infect. Dis. 2022, 114, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Feikin, D.R.; Higdon, M.M.; Abu-Raddad, L.J.; Andrews, N.; Araos, R.; Goldberg, Y.; Groome, M.J.; Huppert, A.; O’Brien, K.L.; Smith, P.G.; et al. Duration of Effectiveness of Vaccines against SARS-CoV-2 Infection and COVID-19 Disease: Results of a Systematic Review and Meta-Regression. Lancet Lond. Engl. 2022, 399, 924–944. [Google Scholar] [CrossRef] [PubMed]
- Stowe, J.; Andrews, N.; Kirsebom, F.; Ramsay, M.; Bernal, J.L. Effectiveness of COVID-19 Vaccines against Omicron and Delta Hospitalisation, a Test Negative Case-Control Study. Nat. Commun. 2022, 13, 5736. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.S.; Collier, A.Y.; Yu, J.; Liu, J.; Chandrashekar, A.; McMahan, K.; Jacob-Dolan, C.; He, X.; Roy, V.; Hauser, B.M.; et al. Durability of Heterologous and Homologous COVID-19 Vaccine Boosts. JAMA Netw. Open 2022, 5, e2226335. [Google Scholar] [CrossRef]
- Atmar, R.L.; Lyke, K.E.; Deming, M.E.; Jackson, L.A.; Branche, A.R.; El Sahly, H.M.; Rostad, C.A.; Martin, J.M.; Johnston, C.; Rupp, R.E.; et al. Homologous and Heterologous COVID-19 Booster Vaccinations. N. Engl. J. Med. 2022, 386, 1046–1057. [Google Scholar] [CrossRef]
- Wu, K.; Choi, A.; Koch, M.; Elbashir, S.; Ma, L.; Lee, D.; Woods, A.; Henry, C.; Palandjian, C.; Hill, A.; et al. Variant SARS-CoV-2 mRNA Vaccines Confer Broad Neutralization as Primary or Booster Series in Mice. Vaccine 2021, 39, 7394–7400. [Google Scholar] [CrossRef]
- Liu, J.; Budylowski, P.; Samson, R.; Griffin, B.D.; Babuadze, G.; Rathod, B.; Colwill, K.; Abioye, J.A.; Schwartz, J.A.; Law, R.; et al. Preclinical Evaluation of a SARS-CoV-2 mRNA Vaccine PTX-COVID19-B. Sci. Adv. 2022, 8, eabj9815. [Google Scholar] [CrossRef]
- Tian, J.-H.; Patel, N.; Haupt, R.; Zhou, H.; Weston, S.; Hammond, H.; Logue, J.; Portnoff, A.D.; Norton, J.; Guebre-Xabier, M.; et al. SARS-CoV-2 Spike Glycoprotein Vaccine Candidate NVX-CoV2373 Immunogenicity in Baboons and Protection in Mice. Nat. Commun. 2021, 12, 372. [Google Scholar] [CrossRef]
- Wørzner, K.; Sheward, D.J.; Schmidt, S.T.; Hanke, L.; Zimmermann, J.; McInerney, G.; Hedestam, G.B.K.; Murrell, B.; Christensen, D.; Pedersen, G.K. Adjuvanted SARS-CoV-2 Spike Protein Elicits Neutralizing Antibodies and CD4 T Cell Responses after a Single Immunization in Mice. EBioMedicine 2021, 63, 103197. [Google Scholar] [CrossRef] [PubMed]
- Seephetdee, C.; Buasri, N.; Bhukhai, K.; Srisanga, K.; Manopwisedjaroen, S.; Lertjintanakit, S.; Phueakphud, N.; Pakiranay, C.; Kangwanrangsan, N.; Srichatrapimuk, S.; et al. Mice Immunized with the Vaccine Candidate HexaPro Spike Produce Neutralizing Antibodies against SARS-CoV-2. Vaccines 2021, 9, 498. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Li, R.; Guo, J.; Li, M.; Ma, L.; Dai, J.; Shi, Y.; Dai, J.; Huang, Y.; Dai, C.; et al. Immunization with a Prefusion SARS-CoV-2 Spike Protein Vaccine (RBMRNA-176) Protects against Viral Challenge in Mice and Nonhuman Primates. Vaccines 2022, 10, 1698. [Google Scholar] [CrossRef] [PubMed]
- Pairo-Castineira, E.; Rawlik, K.; Bretherick, A.D.; Qi, T.; Wu, Y.; Nassiri, I.; McConkey, G.A.; Zechner, M.; Klaric, L.; Griffiths, F.; et al. GWAS and Meta-Analysis Identifies 49 Genetic Variants Underlying Critical COVID-19. Nature 2023, 617, 764–768. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.; Pearson, M.; Das, S.; Sardell, J.; Chocian, K.; Gardner, S. Genetic Risk Factors for Severe and Fatigue Dominant Long COVID and Commonalities with ME/CFS Identified by Combinatorial Analysis. J. Transl. Med. 2023, 21, 775. [Google Scholar] [CrossRef]
- Lammi, V.; Nakanishi, T.; Jones, S.E.; Andrews, S.J.; Karjalainen, J.; Cortés, B.; O’Brien, H.E.; Fulton-Howard, B.E.; Haapaniemi, H.H.; Schmidt, A.; et al. Genome-Wide Association Study of Long COVID 2023. Available online: https://www.medrxiv.org/content/10.1101/2023.06.29.23292056v1 (accessed on 16 November 2023).
- COVID-19 Host Genetics Initiative Mapping the Human Genetic Architecture of COVID-19. Nature 2021, 600, 472–477. [CrossRef]
- Severe COVID-19 GWAS Group; Ellinghaus, D.; Degenhardt, F.; Bujanda, L.; Buti, M.; Albillos, A.; Invernizzi, P.; Fernández, J.; Prati, D.; Baselli, G.; et al. Genomewide Association Study of Severe COVID-19 with Respiratory Failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar] [CrossRef]
- Zecevic, M.; Kotur, N.; Ristivojevic, B.; Gasic, V.; Skodric-Trifunovic, V.; Stjepanovic, M.; Stevanovic, G.; Lavadinovic, L.; Zukic, B.; Pavlovic, S.; et al. Genome-Wide Association Study of COVID-19 Outcomes Reveals Novel Host Genetic Risk Loci in the Serbian Population. Front. Genet. 2022, 13, 911010. [Google Scholar] [CrossRef]
- Mentzer, A.J.; O’Connor, D.; Bibi, S.; Chelysheva, I.; Clutterbuck, E.A.; Demissie, T.; Dinesh, T.; Edwards, N.J.; Felle, S.; Feng, S.; et al. Human Leukocyte Antigen Alleles Associate with COVID-19 Vaccine Immunogenicity and Risk of Breakthrough Infection. Nat. Med. 2023, 29, 147–157. [Google Scholar] [CrossRef]
- Bolze, A.; Neveux, I.; Schiabor Barrett, K.M.; White, S.; Isaksson, M.; Dabe, S.; Lee, W.; Grzymski, J.J.; Washington, N.L.; Cirulli, E.T. HLA-A∗03:01 Is Associated with Increased Risk of Fever, Chills, and Stronger Side Effects from Pfizer-BioNTech COVID-19 Vaccination. Hum. Genet. Genom. Adv. 2022, 3, 100084. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Shi, D.; Shen, W.; Shi, S.; Guo, X.; Li, J.; Xu, S.; Zhang, Y.; Zhao, Z. Pilot Genome-Wide Association Study of Antibody Response to Inactivated SARS-CoV-2 Vaccines. Front. Immunol. 2022, 13, 1054147. [Google Scholar] [CrossRef] [PubMed]
- Robertson, S.J.; Bedard, O.; McNally, K.L.; Shaia, C.; Clancy, C.S.; Lewis, M.; Broeckel, R.M.; Chiramel, A.I.; Shannon, J.G.; Sturdevant, G.L.; et al. Genetically Diverse Mouse Models of SARS-CoV-2 Infection Reproduce Clinical Variation in Type I Interferon and Cytokine Responses in COVID-19. Nat. Commun. 2023, 14, 4481. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.B.; Montefiori, D.C.; McDermott, A.B.; Fong, Y.; Benkeser, D.; Deng, W.; Zhou, H.; Houchens, C.R.; Martins, K.; Jayashankar, L.; et al. Immune Correlates Analysis of the mRNA-1273 COVID-19 Vaccine Efficacy Clinical Trial. Science 2022, 375, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing Antibody Levels Are Highly Predictive of Immune Protection from Symptomatic SARS-CoV-2 Infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef]
- Bergwerk, M.; Gonen, T.; Lustig, Y.; Amit, S.; Lipsitch, M.; Cohen, C.; Mandelboim, M.; Gal Levin, E.; Rubin, C.; Indenbaum, V.; et al. COVID-19 Breakthrough Infections in Vaccinated Health Care Workers. N. Engl. J. Med. 2021, 385, 1629–1630. [Google Scholar] [CrossRef]
- Yamamoto, S.; Matsuda, K.; Maeda, K.; Horii, K.; Okudera, K.; Oshiro, Y.; Inamura, N.; Takeuchi, J.S.; Konishi, M.; Ozeki, M.; et al. Neutralizing Antibodies after Three Doses of the BNT162b2 Vaccine, Breakthrough Infection, and Symptoms during the Omicron-Predominant Wave. Int. J. Infect. Dis. 2023, 128, 347–354. [Google Scholar] [CrossRef]
- Yamamoto, S.; Matsuda, K.; Maeda, K.; Horii, K.; Okudera, K.; Oshiro, Y.; Inamura, N.; Nemoto, T.; Takeuchi, J.S.; Li, Y.; et al. Preinfection Neutralizing Antibodies, Omicron BA.5 Breakthrough Infection, and Long COVID: A Propensity Score-Matched Analysis. J. Infect. Dis. 2023, 228, 1652–1661. [Google Scholar] [CrossRef]
- Servellita, V.; Syed, A.M.; Morris, M.K.; Brazer, N.; Saldhi, P.; Garcia-Knight, M.; Sreekumar, B.; Khalid, M.M.; Ciling, A.; Chen, P.-Y.; et al. Neutralizing Immunity in Vaccine Breakthrough Infections from the SARS-CoV-2 Omicron and Delta Variants. Cell 2022, 185, 1539–1548.e5. [Google Scholar] [CrossRef]
- Lai, R.; Gong, D.N.; Williams, T.; Ogunsola, A.F.; Cavallo, K.; Arlehamn, C.S.L.; Acolatse, S.; Beamer, G.L.; Ferris, M.T.; Sassetti, C.M.; et al. Host Genetic Background Is a Barrier to Broadly Effective Vaccine–Mediated Protection against Tuberculosis. J. Clin. Investig. 2023, 133, e167762. [Google Scholar] [CrossRef]
- Nelde, A.; Bilich, T.; Heitmann, J.S.; Maringer, Y.; Salih, H.R.; Roerden, M.; Lübke, M.; Bauer, J.; Rieth, J.; Wacker, M.; et al. SARS-CoV-2-Derived Peptides Define Heterologous and COVID-19-Induced T Cell Recognition. Nat. Immunol. 2021, 22, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Kaplonek, P.; Fischinger, S.; Cizmeci, D.; Bartsch, Y.C.; Kang, J.; Burke, J.S.; Shin, S.A.; Dayal, D.; Martin, P.; Mann, C.; et al. mRNA-1273 Vaccine-Induced Antibodies Maintain Fc Effector Functions across SARS-CoV-2 Variants of Concern. Immunity 2022, 55, 355–365.e4. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, M.; Penfold, R.S.; Merino, J.; Sudre, C.H.; Molteni, E.; Berry, S.; Canas, L.S.; Graham, M.S.; Klaser, K.; Modat, M.; et al. Risk Factors and Disease Profile of Post-Vaccination SARS-CoV-2 Infection in UK Users of the COVID Symptom Study App: A Prospective, Community-Based, Nested, Case-Control Study. Lancet Infect. Dis. 2022, 22, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Gebo, K.A.; Abraham, A.G.; Habtehyimer, F.; Patel, E.U.; Laeyendecker, O.; Gniadek, T.J.; Fernandez, R.E.; Baker, O.R.; Ram, M.; et al. Dynamics of Inflammatory Responses after SARS-CoV-2 Infection by Vaccination Status in the USA: A Prospective Cohort Study. Lancet Microbe 2023, 4, e692–e703. [Google Scholar] [CrossRef]
- Scola, L.; Ferraro, D.; Sanfilippo, G.L.; De Grazia, S.; Lio, D.; Giammanco, G.M. Age and Cytokine Gene Variants Modulate the Immunogenicity and Protective Effect of SARS-CoV-2 mRNA-Based Vaccination. Vaccines 2023, 11, 413. [Google Scholar] [CrossRef]
- Mosedale, M.; Cai, Y.; Eaddy, J.S.; Kirby, P.J.; Wolenski, F.S.; Dragan, Y.; Valdar, W. Human-Relevant Mechanisms and Risk Factors for TAK-875-Induced Liver Injury Identified via a Gene Pathway-Based Approach in Collaborative Cross Mice. Toxicology 2021, 461, 152902. [Google Scholar] [CrossRef]

| Treatment | IC50 Heritability | IC80 Heritability |
|---|---|---|
| Vaccinated | 80.6% | 74.3% |
| Treatment | Day 4 Percent Weight Loss Heritability | Vaccinated Lung Viral Titer Heritability |
|---|---|---|
| PBS | 68.0% | 41.2% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz Cisneros, M.C.; Anderson, E.J.; Hampton, B.K.; Parotti, B.; Sarkar, S.; Taft-Benz, S.; Bell, T.A.; Blanchard, M.; Dillard, J.A.; Dinnon, K.H., III; et al. Host Genetic Variation Impacts SARS-CoV-2 Vaccination Response in the Diversity Outbred Mouse Population. Vaccines 2024, 12, 103. https://doi.org/10.3390/vaccines12010103
Cruz Cisneros MC, Anderson EJ, Hampton BK, Parotti B, Sarkar S, Taft-Benz S, Bell TA, Blanchard M, Dillard JA, Dinnon KH III, et al. Host Genetic Variation Impacts SARS-CoV-2 Vaccination Response in the Diversity Outbred Mouse Population. Vaccines. 2024; 12(1):103. https://doi.org/10.3390/vaccines12010103
Chicago/Turabian StyleCruz Cisneros, Marta C., Elizabeth J. Anderson, Brea K. Hampton, Breantié Parotti, Sanjay Sarkar, Sharon Taft-Benz, Timothy A. Bell, Matthew Blanchard, Jacob A. Dillard, Kenneth H. Dinnon, III, and et al. 2024. "Host Genetic Variation Impacts SARS-CoV-2 Vaccination Response in the Diversity Outbred Mouse Population" Vaccines 12, no. 1: 103. https://doi.org/10.3390/vaccines12010103
APA StyleCruz Cisneros, M. C., Anderson, E. J., Hampton, B. K., Parotti, B., Sarkar, S., Taft-Benz, S., Bell, T. A., Blanchard, M., Dillard, J. A., Dinnon, K. H., III, Hock, P., Leist, S. R., Madden, E. A., Shaw, G. D., West, A., Baric, R. S., Baxter, V. K., Pardo-Manuel de Villena, F., Heise, M. T., & Ferris, M. T. (2024). Host Genetic Variation Impacts SARS-CoV-2 Vaccination Response in the Diversity Outbred Mouse Population. Vaccines, 12(1), 103. https://doi.org/10.3390/vaccines12010103








