Abstract
Avian coccidiosis arises from co-infection involving multiple Eimeria species, which could give rise to substantial economic losses in the global poultry industry. As a result, multivalent anticoccidial vaccines containing common Eimeria antigens offer considerable promise for controlling co-infection in clinical practice. In our previous study, Elongation factor 2 (EF2) was deemed as an immunogenic common antigen across various Eimeria species. This current investigation aimed to further assess the immunogenicity and protective efficacy of EF2 in recombinant subunit vaccine format against three Eimeria species. The EF2 gene cloned from Eimeria maxima (E. maxima) cDNA was designated as EF2 of E. maxima (EmEF2). The immunogenicity of the recombinant protein EmEF2 (rEmEF2) was assessed through Western blot analysis. The evaluation of the vaccine-induced immune response encompassed the determination of T lymphocyte subset proportions, cytokine mRNA transcription levels, and specific IgY concentrations in rEmEF2-vaccinated chickens using flow cytometry, quantitative real-time PCR (qPCR), and indirect enzyme-linked immunosorbent assay (ELISA). Subsequently, the protective efficacy of rEmEF2 was evaluated through vaccination and challenge experiments. The findings demonstrated that rEmEF2 was effectively recognized by the His-tag monoclonal antibody and E. maxima chicken antiserum. Vaccination with rEmEF2 increased the proportions of CD4+ and CD8+ T lymphocytes, elevated IL-4 and IFN-γ mRNA transcription levels, and enhanced IgY antibody levels compared to the control groups. Moreover, compared to the control groups, vaccination with rEmEF2 led to decreased weight loss, reduced oocyst outputs, and alleviated enteric lesions. Furthermore, in the rEmEF2-immunized groups, challenges with E. maxima and E. acervulina resulted in anticoccidial index (ACI) scores of 166.35 and 185.08, showing moderate-to-excellent protective efficacy. Nevertheless, challenges with E. tenella and mixed Eimeria resulted in ACI scores of 144.01 and 127.94, showing low protective efficacy. In conclusion, EmEF2, a common antigen across Eimeria species, demonstrated the capacity to induce a significant cellular and humoral immune response, as well as partial protection against E. maxima, E. acervulina, and E. tenella. These results highlight EmEF2 as a promising candidate antigen for the development of multivalent vaccines targeting mixed infections by Eimeria species.
1. Introduction
Coccidiosis in chickens, characterized by a hemorrhagic presentation, typically results in high mortality rates, subpar weight gains, and inefficient food conversion ratios. This has a profound impact on chicken production and overall welfare [1,2]. In 2016, the global economic burden caused by chicken coccidiosis arising from multiple Eimeria species was estimated to have surpassed GBP 10.4 billion [3,4]. Avian coccidiosis is prevalent worldwide, with an incidence ranging from 70% to 90% [5], often stemming from mixed infections involving multiple Eimeria species [6]. Currently, anticoccidial drugs have been the primary approach for managing avian coccidiosis [7,8]. However, the continuous occurrence of drug resistance and the presence of drug residues in poultry products are constantly escalating, which has led to the search for new approaches, such as the use of anticoccidial vaccines as a substitute for chemoprophylaxis prevention to control coccidiosis [9,10]. In recent years, vaccination with innovative vaccine types, including subunit vaccines, DNA vaccines, and live vector vaccines, has emerged as a promising strategy [11,12,13,14]. Recent research has demonstrated the successful utilization of various Eimeria antigens to develop neotype anticoccidial vaccines that offer effective protection [11,14,15,16,17,18,19,20]. However, the majority of these antigens are derived from single Eimeria species and do not fulfill the clinical requirement for controlling mixed Eimeria infections.
As avian coccidiosis typically arises from mixed infections involving multiple Eimeria species in clinical settings, identifying common proteins shared by various Eimeria species becomes essential for the development of multivalent vaccines against this poultry disease. Several Eimeria common antigens have been reported in this regard. For instance, Talebi discovered a 45 kDa immunogenic protein that was recognized by chicken antiserum against E. maxima and remained conserved among five Eimeria species [21]. A common antigen of all chicken Eimeria species has been clearly identified by Sasai et al., which is present on motile stages and can be recognized by chicken monoclonal antibodies against E. acervulina sporozoites; in addition, the presence of this antigen has been found in two closely related coccidian parasites (Toxoplasma and Neospora) [22]. In our previous studies, we successfully identified five common immunodominant antigens displaying an amino acid sequence similarity of over 93% among three Eimeria species (Eimeria maxima, Eimeria acervulina, and Eimeria tenella). Notably, two of these antigens have been demonstrated to provide effective protection against infections caused by these three Eimeria species, both individually and in combination [23,24,25]. These studies strongly underscore the potential of Eimeria common antigens as prospective candidate components for the development of effective, safe, and stable multivalent vaccines against avian coccidiosis in poultry.
Elongation factor 2 (EF2), a member of the GTP-binding translation elongation factor family, has garnered attention in various research domains [26,27,28,29]. It has been demonstrated to exhibit a high degree of conservation among various apicomplexan protozoa and has been proposed as a potential target for drugs or vaccine candidate antigens against protozoan diseases [30,31]. In the case of Leishmania, Agallou et al. reported remarkable EF2 conservation between strains, including L. infantum, L. donovani, L. major, and L. braziliensis, with amino acid sequence identities ranging from 98% to 100% [32]. Furthermore, EF2 was identified as a T cell-stimulating antigen capable of eliciting protective cellular immune responses against experimental visceral leishmaniosis [32,33,34]. Extensive analysis of 1685 clinically infected Plasmodium samples from 17 countries revealed the high conservation of EF2 in Plasmodium genomic sequences, and inhibiting EF2 at multiple stages of Plasmodium growth resulted in a substantial reduction in the Plasmodium population of up to 98% [35]. In the study of Plasmodium, in order to block the transmission of malaria, Dechering et al. identified drugs targeting EF2 as important candidates [36]. In our previous research, among Eimeria species, EF2 was deemed as a common immunodominant antigen, sharing an astonishing amino acid sequence similarity of 99% among the three Eimeria species studied [24]. However, the protective efficacy of EF2 against infections caused by different Eimeria species remained unknown. In this study, the EF2 gene of Eimeria maxima (EmEF2) was ligated with the prokaryotic expression vector to produce the recombinant protein EmEF2 (rEmEF2). Subsequently, we systematically assessed the cellular and humoral immune responses triggered by EF2 in the form of rEmEF2 in chickens. Finally, we evaluated the protective efficacy of rEmEF2 through vaccination and challenge experiments. These results indicate that EmEF2 may confer partial protection against multiple Eimeria infections and hold promise as a candidate antigen for the development of multivalent vaccines to control avian coccidiosis in practical clinical applications. It may provide new ideas for the development of multivalent vaccines against other pathogens.
2. Materials and Methods
2.1. Animals, Parasites and Antiserum
Hy-Line chickens (1-day-old) were raised under stringent condition in a coccidia-free environment in the Laboratory Animal Center of Nanjing Agricultural University. Meanwhile, chickens had unrestricted access to water and feed without any coccidiostat during the experimental period. Each chicken underwent oral infection with 1 × 104 E. acervulina, and oocysts were collected from feces 4–7 days post-infection. Subsequently, each chicken was orally infected with 1 × 104 E. maxima and 8 × 103 E. tenella, and oocysts were collected from feces 5–8 days post-infection. Oocysts were collected from feces using the saturated saline floatation method. Sporulated oocysts of E. maxima, E. acervulina and E. tenella were stored at 4 °C in 2.5% potassium dichromate. To ensure the viability of the parasites, sporulated oocysts were propagated in chickens seven days prior to the challenge trials. All animal procedures and experiments were subject to rigorous ethical scrutiny and received approval from the Committee on Experimental Animal Welfare and Ethics of Nanjing Agricultural University (Approval number: PTA 2020001). The non-infected chicken serum and E. maxima chicken antiserum were provided by our lab [16] and used for subsequent Western blot analysis.
2.2. Cloning of EmEF2 and Recombinant Plasmid Construction of pET-32a-EmEF2
Total RNA extraction from 1 × 108 E. maxima was carried out utilizing a Total RNA Extraction Kit (Omega Bio-Tek, Norcross, GA, USA). Then, using the HiScript III Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme Biotech, Nanjing, China), the extracted total RNA from E. maxima served as the starting material for cDNA synthesis. The reverse transcription steps were as follows: 1 µg of total RNA and 4 µL of 4 × gDNA wiper Mix were added to a RNase-free EP tube, followed by RNase-free water to 16 µL and 42 °C for 2 min. We then added 4 µL of 5 × HiScript III qRT SuperMix to the tube at 37 °C for 15 min, then 85 °C for 5 s. The cDNA was used for the subsequent experiments. Restriction enzyme-anchored primers were designed and guided by the sequence of E. maxima EF2 (EmEF2) available in GenBank (No. 25335462). The forward primer was anchored with EcoR I (Takara Biotechnology, Dalian, China) and the reverse primer was anchored with Hind III (Takara Biotechnology, Dalian, China) (Table 1). PCR amplification was executed utilizing 2 × Taq Master Mix (Dye Plus) (Vazyme Biotech, Nanjing, China). The PCR program for the amplification of EmEF2 gene was carried out according to the manufacturer’s protocols. In the meantime, changing the extension time based on the length of EmEF2 gene fragment. The program for EmEF2 was as follows: 95 °C, 3 min; 30 cycles (95 °C, 15 s; 60 °C, 30 s; 72 °C, 149 s); and 72 °C, 5 min. The PCR product of EmEF2 was recovered after EcoR I and Hind III digestion in 10 × K Buffer (Takara Biotechnology, Dalian, China) at 37 °C and was subsequently inserted into the pET-32a (Invitrogen Biotechnology, Carlsbad, CA, USA). The constructed pET-32a-EmEF2 was subjected to validation through restriction enzyme digestion and sequence analysis. The complete open reading frame (ORF) of EmEF2 was aligned in the GenBank databases using the Basic Local Alignment Search Tool. Antigenicity analysis of the EmEF2 was performed by Protean of DNAStar software (Version 11.0, DNASTAR Inc., Madison, WI, USA).

Table 1.
Primers of E. maxima EF2 (EmEF2).
2.3. Expression of Recombinant Protein EmEF2 and Western Blot Analysis
pET-32a-EmEF2 was transformed into ampicillin-resistant Escherichia coli (E. coli) BL21 (DE3) (Vazyme Biotech, Nanjing, China) to express the recombinant protein EmEF2 (rEmEF2). The rEmEF2 was purified using a protein affinity chromatography column (His-Trap™ FF, Cytiva, Marlborough, MA, USA). Subsequently, the purification of rEmEF2 was verified through SDS-PAGE analysis. According to the manufacturer’s protocols, the concentration of purified rEmEF2 was measured using the BCA Protein Assay Kit (Beyotime, Shanghai, China). The concentration of rEmEF2 was 100 µg/mL and the rEmEF2 was used for the following Western blot analysis. Then, the rEmEF2 was 400 µg/mL and used in the subsequent determination of the immune responses and experimental assessment of the protective efficacy of rEmEF2. In addition, the rEmEF2 was cryogenically stored by freezing individual tubes at −80 °C.
The presence of the rEmEF2 was determined through a Western blot assay, using E. maxima chicken antiserum, non-infected chicken serum or His-tag monoclonal antibody (Proteintech, Wuhan, China) as primary antibodies, respectively. Here is a concise overview of the procedure: After conducting the SDS-PAGE assay, the purified rEmEF2 was transferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). Subsequently, the PVDF membranes were incubated with E. maxima chicken antiserum (1:100 dilution), non-infected chicken serum (1:100 dilution) or His-tag monoclonal antibody (1:200 dilution) for 1 h at 37 °C, respectively. The non-infected chicken serum was regarded as the negative control. After the PVDF membranes were separately incubated with goat anti-chicken IgY H&L (HRP) (1:20,000 dilution, Abcam, Cambridge, UK) or goat anti-mouse IgG H&L (HRP) (1:10,000 dilution, Abcam, Cambridge, UK) for 45 min at 37 °C. The detection of bound antibodies was achieved by initiating a color development process, utilizing an HRP-DAB substrate chromogenic kit (Tiangen, Beijing, China).
2.4. Determination of the Immune Responses Induced by rEmEF2 in Chickens
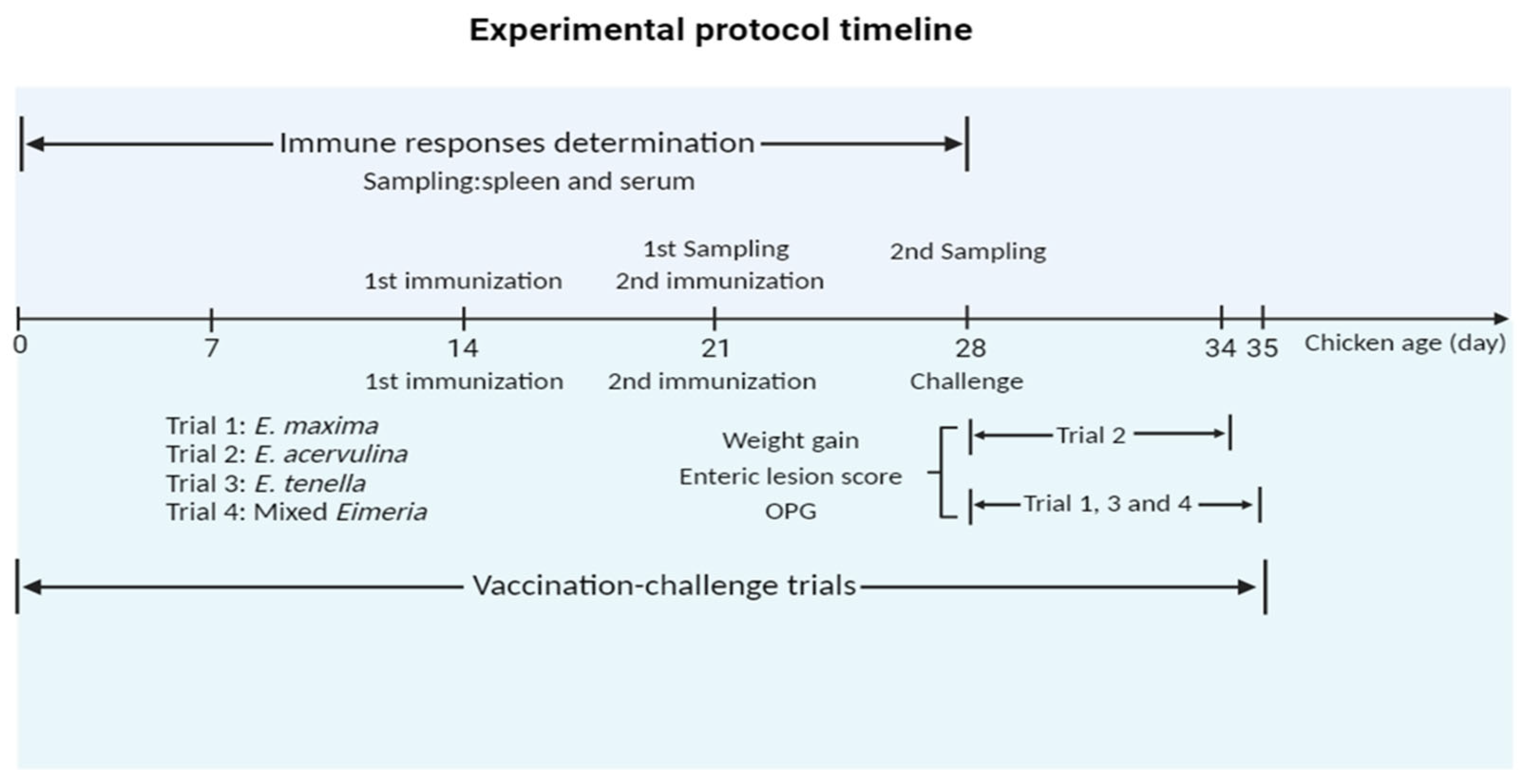
Chickens (14-day-old) were randomly allocated into three groups, and each group comprised six chickens and underwent the primary immunization. Among these groups, two control groups including a PBS control and a pET-32a tag protein control were intramuscularly injected in the leg as follows: PBS and 200 µg of pET-32a tag protein, respectively. In parallel, the experimental group received intramuscular injections of 200 µg of rEmEF2 in the leg. Following the primary immunization, the secondary immunization was administered after a 7-day interval, and the injection dose of the secondary immunization was the same as the primary immunization. The timeline for the determination of immune responses induced by immunized chickens is shown in Figure 1.
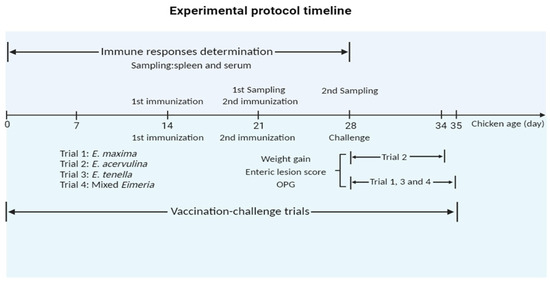
Figure 1.
Experimental protocol timeline. The chickens received the first and second vaccinations at 14 and 21 days of age, respectively. In the timeline for the determination of immune responses induced by immunized chickens, spleen lymphocytes and serum samples were collected from immunized chickens at 21 days old and 28 days old, respectively. In timeline of experimental assessment of protective efficacy of rEmEF2 against three Eimeria, data collection for E. acervulina groups (Trial 2) occurred on the sixth day after the challenge (34 days old), while data collection for other Eimeria species (trial 1, 3 and 4) was conducted on the seventh day (35 days old). The collected data included weight gain, enteric lesion score, and OPG. The figure was created in BioRender.com (https://www.biorender.com/ accessed on 2 December 2023).
On the 7th day following each vaccination, spleen lymphocytes were gathered in three chickens which were stochastically selected in each group. The spleens were meticulously ground in 5 mL of PBS and filtered with cell strainers. The filtrate containing splenocytes was added to the lymphocyte separation solution (TBDscience, Tianjin, China) and centrifuged at 500× g for 20 min, and the lymphocytes located in the middle layer were extracted. The lymphocytes were analyzed using the CD4+ and CD8+ T cell subpopulations. Whereafter, CD3 mouse anti-chicken FITC antibody (Southern Biotechnology Associates, Birmingham, AL, USA), CD4 mouse anti-chicken PE antibody (Southern Biotechnology Associates, Birmingham, AL, USA) and CD8 mouse anti-chicken PE antibody (Southern Biotechnology Associates, Birmingham, AL, USA) were used to detect the T cell subpopulations with a FACS Calibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). In brief, each group’s lymphocytes suspension was adjusted to 1 × 106 cells in 100 µL of PBS. Subsequently, CD3 and CD4 antibody or CD3 and CD8 antibody were used to bind chicken CD3+ CD4+ or CD3+ CD8+, and T cells were incubated for 25 min at 4 °C without light following the manufacturer’s protocols.
To assess the mRNA transcription levels of the IL-4 gene (GenBank No. AJ621735) and IFN-γ gene (GenBank No. Y07922) in immunized chickens, a quantitative real-time PCR (qPCR) assay was conducted, with the GAPDH gene (GenBank No. K01458) serving as internal control. The primer sequences, amplification efficiency (%) and correlation coefficients (r2) of GAPDH, IL-4 and IFN-γ for qPCR are based on previously published articles from our lab [16], and the primer sequences are shown in Table 2. On the 7th day following each vaccination, the total RNA from spleen lymphocytes was extracted from each group of three chickens and subsequently reverse-transcribed to cDNA, following previous protocols. The ChamQTM SYBR qPCR Master Mix kit (Vazyme Biotech, Nanjing, China) was employed as the manufacturer’s instructions by the qPCR assay. The qPCR amplification reaction system contained 10 µL of 2 × ChamQTM SYBR qPCR Master Mix, 0.4 µL of forward and reverse primers, 2 µL of cDNA and 7.2 µL of RNase-free water. The triplicated samples were set in each qPCR assay. The reaction procedure of the qPCR was as follows: 95 °C, 30 s; 40 cycles (95 °C, 10 s; 60 °C, 30 s). The melt curve stage of qPCR was as follows: 95 °C, 15 s; 60 °C, 1 min; 95 °C, 15 s. The qPCR reaction was performed on an ABI prism 7300 Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). The relative quantification of cytokine gene mRNA was determined using the 2−ΔΔCt method for precise quantification based on the methods previously established by Livak and Schmittgen (2001) [37].

Table 2.
Primer sequences of GAPDH, IL-4 and IFN-γ.
Chicken serum samples were collected from three chickens, which were randomly selected on the 7th day following each vaccination in each group. The indirect enzyme-linked immunosorbent assay (ELISA) was conducted to assess the rEmEF2-specific serum IgY antibody levels. In the indirect ELISA procedure, rEmEF2 was diluted to 10 ng/µL, and 200 µL was coated in flat-bottomed 96-well plates (MarxiSorp, Nunc, Waltham, MA, USA) for 16 h at 4 °C. Subsequently, the plates were washed five times with PBST (PBS with 0.05% Tween20) and blocked with 200 µL of PBST containing 5% bovine serum albumin (BSA) (Yifeixue, Nanjing, China) for 2 h at 37 °C. The primary antibody was chicken serum samples (1:50 dilution) for 1 h at 37 °C, while the secondary antibody was goat anti-chicken IgY H&L (HRP) antibody (1:40,000 dilution). Non-infected chicken serum (1:50 dilution) and PBS were used as controls during the analysis. In the end, the color production was detected with 100 µL of 3,3′,5,5′-tetramethylbenzidine (TMB) (Tiangen, Beijing, China) in the dark at RT for 8 min and observed under OD450 absorbance with a microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
2.5. Experimental Assessment of Protective Efficacy of rEmEF2 against Infections by Three Eimeria Species in Chickens
In order to evaluate the protective efficacy of rEmEF2 (Table 3), four immunization challenge trials were carried out. Trial 1, trial 2, trial 3 and trial 4 were performed to evaluate the protective efficacy of rEmEF2 against E. maxima, E. acervulina, E. tenella and mixed Eimeria, respectively. Healthy chickens (14-day-old) were randomly divided into thirteen groups based on similar body weights. Among these, four experimental groups received intramuscular injections of 200 µg of rEmEF2 into the leg, with an injection volume of 0.5 mL. The remaining nine groups served as controls and included a non-immunized non-challenged group, four non-immunized challenged groups and four pET-32a tag protein control groups. The pET-32a tag protein control groups were injected with the same dose of pET-32a tag protein as the experimental groups. The subsequent manipulations were performed as follows. When the chickens reached an age of 21 days, the pET-32a tag protein control groups and the rEmEF2 experimental groups were, respectively, given the same dose as the first immunization for the second immunization. All chickens were orally challenged with 1 × 105 E. maxima (trial 1), 1 × 105 E. acervulina (trial 2), 5 × 104 E. tenella (trial 3), or a mixture of these three Eimeria species (trial 4), except for the non-immunized non-challenged group, at age of 28 days according to the grouping in Table 3. The timeline of the protocol schemes of the trials is shown in Figure 1. The life cycles of various Eimeria species exhibit distinctions, resulting in diverse peak points for fecal oocyst shedding. Specifically, the highest fecal oocyst-shedding point for E. tenella and E. maxima occurs on the seventh day after challenge, whereas E. acervulina reaches its peak on the sixth day post-challenge. The data collection for E. acervulina groups occurred on the sixth day after the challenge, while data collection for other Eimeria species was performed on the seventh day. The collected data included body weight gain, enteric lesion score, oocyst shedding and the anticoccidial index (ACI) to assess the protective efficacy of the rEmEF2. We calculated the body weight gain of each chicken based on the difference in weight between the challenge time and the slaughter time. Based on the methods previously established by Hodgson (1970) [38], oocyst counts were carried out using a McMaster chamber. A numerical scale ranging from normal to severe (0 to 4) was used to score the lesions in the chicken intestine, as per Johnson and Reid (1970) [39]. The oocyst-shedding decrease ratio was calculated using the formula: (mean oocyst amount of the challenged control group-that of the vaccinated groups)/oocyst amount of control group × 100%. ACI was calculated using the formula: (relative rate of weight gain + survival rate) − (lesion index + oocyst index) (McManus et al., 1968) [40]. An ACI of less than 120 was deemed to indicate no protective efficacy, while an ACI ranging from 120 to less than 160 was categorized as indicative of low-level protective efficacy. ACI values falling within the range of 160 to less than 180 were considered to represent moderate protective efficacy, while an ACI of 180 or higher was regarded as indicative of excellent protective efficacy.

Table 3.
Protective efficacy of rEmEF2 vaccines against the challenge infections by E. maxima, E. acervulina, E. tenella and mixed Eimeria.
2.6. Statistical Analysis
The data were analyzed for normal distribution using SPSS software (Version 27.0.1, SPSS Inc., Chicago, IL, USA). Statistical analysis was conducted using GraphPad Prism software (Version 8.0.2, GraphPad Software Inc., San Diego, CA, USA), and the significance of differences between groups was assessed through a Kruskal–Wallis H test. The data were presented in the format of mean ± standard deviation (S.D.). A significance level of p < 0.05 was considered statistically significant, while p > 0.05 indicated a lack of significant difference.
3. Results
3.1. Cloning of EmEF2 and Recombinant Plasmid Construction of pET-32a-EmEF2
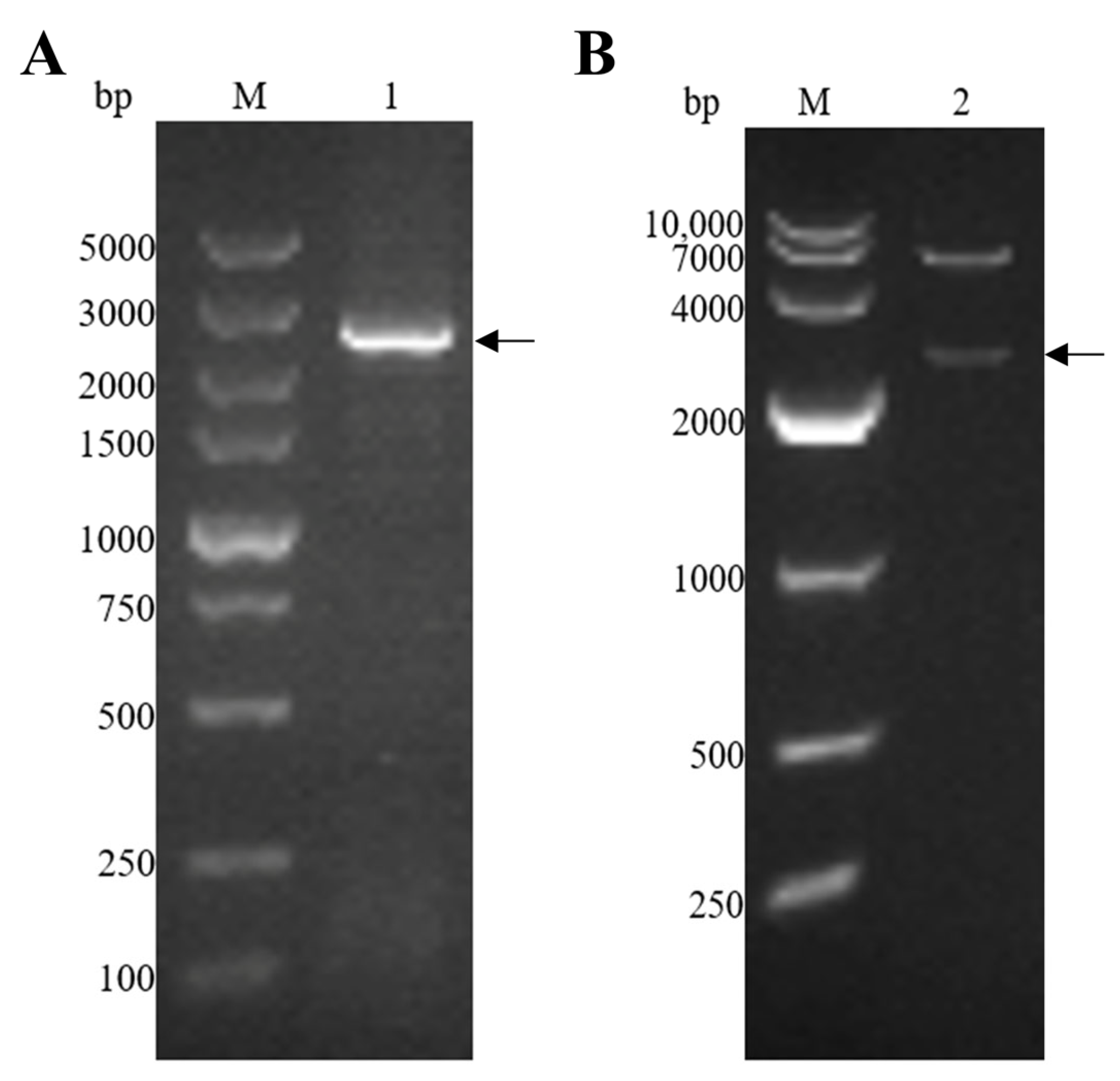
The EmEF2 was successfully amplified using E. maxima cDNA, as previously described. As shown in Figure 2A, the result of the agarose gel electrophoresis showed that the band had a size of 2499 bp, which corresponds to the molecular weight of EmEF2 (Figure 2A, lane 1). Subsequently, the EmEF2 gene was ligated with a prokaryotic expression plasmid: the pET-32a. The recombinant plasmid of pET-32a-EmEF2 was constructed. As shown in Figure 2B, after restriction enzyme digestion (EcoR I and Hind III) of the constructed pET-32a-EmEF2, the results showed that the pET-32a linearized plasmid fragment and the target band of 2499 bp were observed, which were consistent with the expected size of EmEF2 (Figure 2B, lane 2). In addition, the sequence analysis confirmed that EmEF2 shared 100% identity with the sequence of E. maxima EF2 available in GenBank (No. 25335462). DNAStar Protean analysis revealed that EmEF2 is immunogenic.
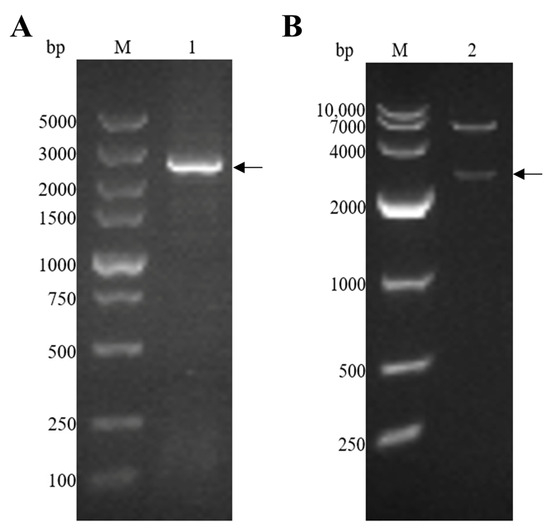
Figure 2.
E. maxima EF2 (EmEF2) gene cloning and recombinant plasmid construction of pET-32a-EmEF2. (A) Cloning of EmEF2 gene from E. maxima cDNA. M: DNA marker DL5000. Lane 1: Amplification product of EmEF2 (2499bp). (B) Restriction enzyme digestion identification of pET-32a–EmEF2. M: DNA marker DL10000. Lane 2: The pET-32a-EmEF2 was identified by EcoR I and Hind III restriction enzyme digestion, resulting in the pET-32a linearized plasmid fragment and the EmEF2.
3.2. Purification of Recombinant Protein EmEF2 and Western Blot Analysis
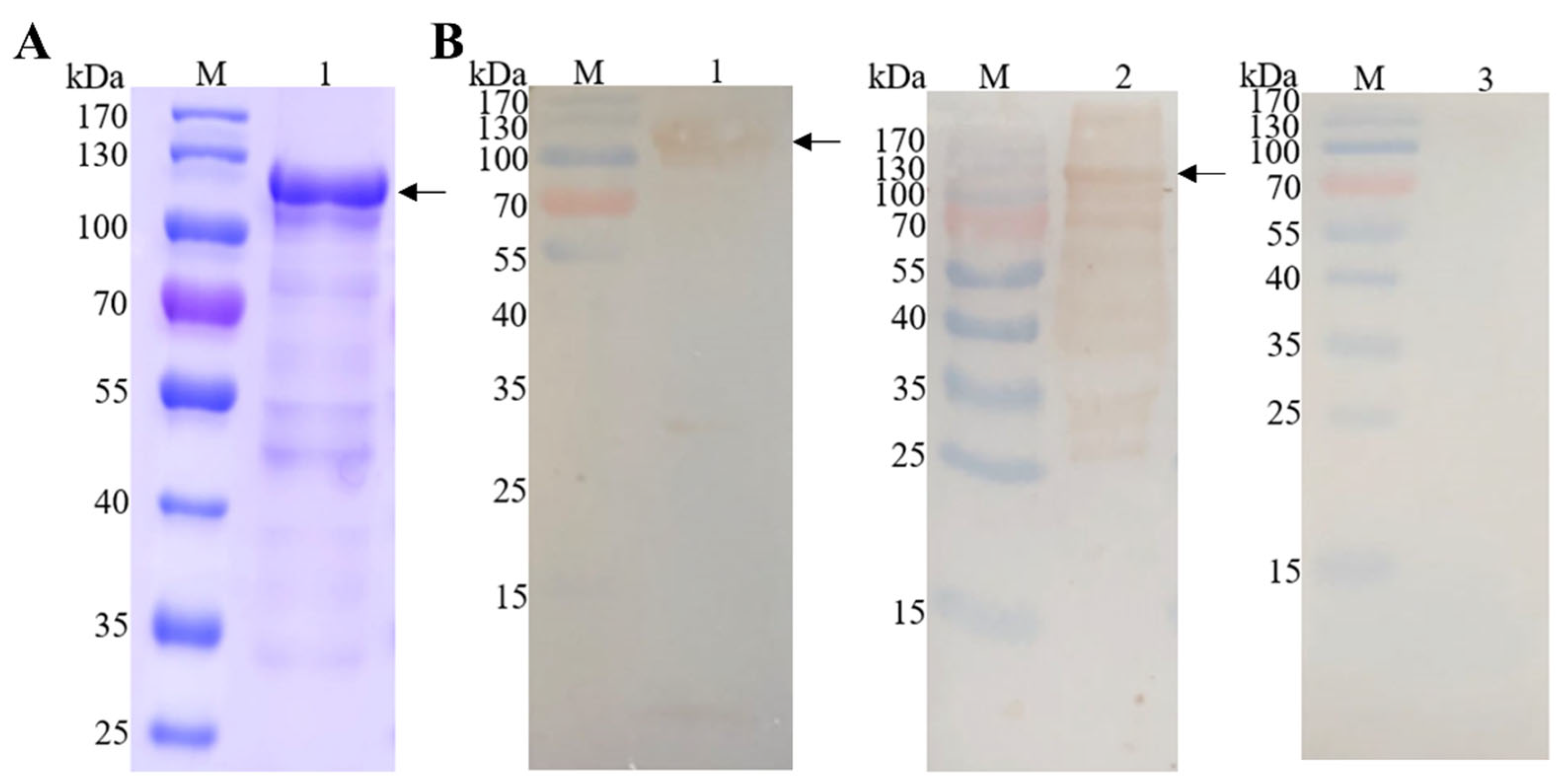
The recombinant protein EmEF2 (rEmEF2) was analyzed through SDS-PAGE and Western blot analysis. As shown in Figure 3A, the rEmEF2 was purified via a protein affinity chromatography column; a band appeared at 110 kDa that was consistent with the expected size of rEmEF2 (Figure 3A, lane 1). Furthermore, Western blot analysis confirmed that the purified rEmEF2 was recognized by the His-tag monoclonal antibody (Figure 3B, lane 1) and E. maxima chicken antiserum (Figure 3B, lane 2). Meanwhile, the rEmEF2 was not recognized by the negative chicken serum (Figure 3B, lane 3).
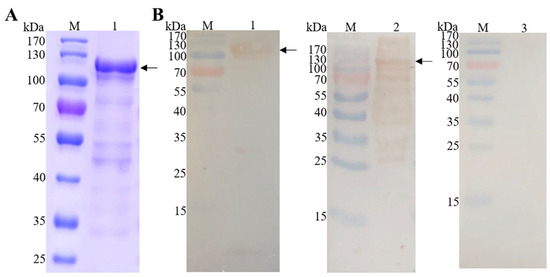
Figure 3.
Purification of recombinant protein EmEF2 (rEmEF2) and Western blot analysis. (A) SDS-PAGE analysis of rEmEF2 purification (110 kDa). M: protein mid-molecular-weight marker. Lane 1: The rEmEF2 was purified via protein affinity chromatography column (110 kDa). (B) Western blot analysis of rEmEF2 (110 kDa). M: protein mid-molecular-weight marker. Lane 1: The purified rEmEF2 was recognized by the His-tag monoclonal antibody as primary antibody and goat anti-mouse IgG H&L (HRP) as secondary antibody. Lane 2: The purified rEmEF2 was recognized by the E. maxima chicken antiserum as primary antibody and goat anti-chicken IgY H&L (HRP) as secondary antibody. Lane 3: The non-infected chicken serum was primary antibody and served as negative control, while the secondary antibody was goat anti-chicken IgY H&L (HRP).
3.3. The Evaluation of Immune Responses Induced by rEmEF2 in Chickens
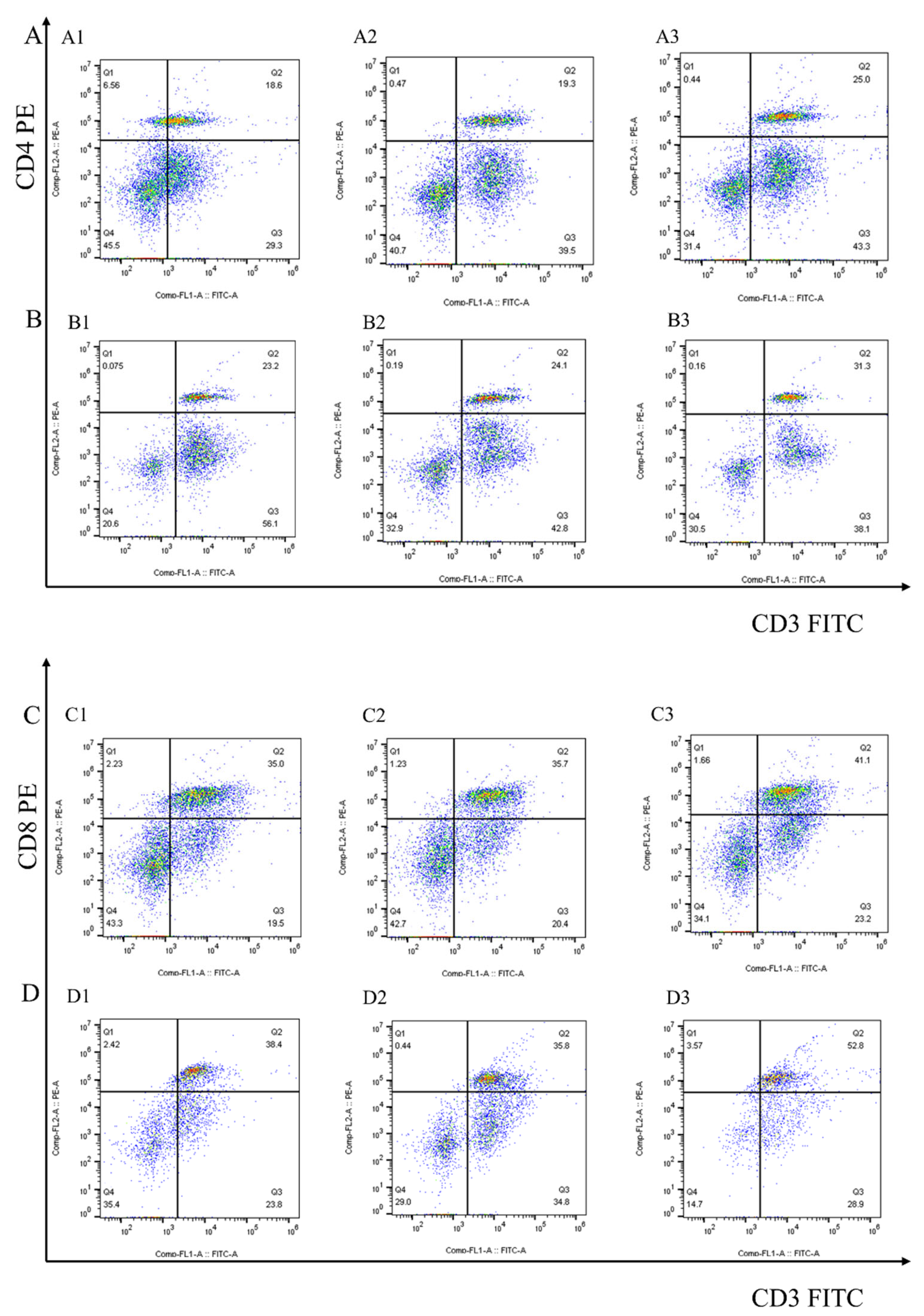
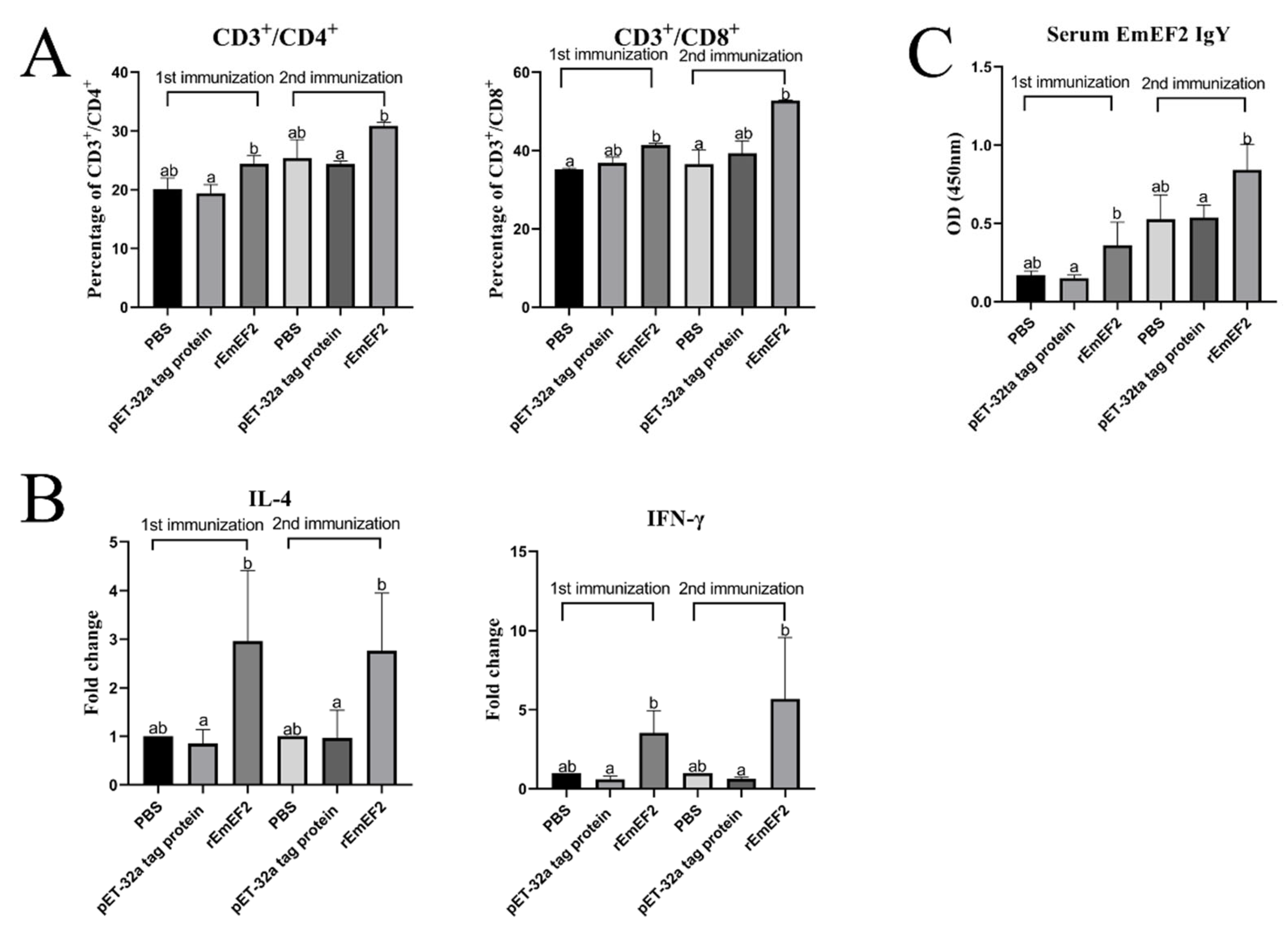
After 7 days of the first and second immunization with rEmEF2, flow cytometry was used to evaluate the proportion of CD4+ and CD8+ T lymphocytes in immunized chickens. The results are shown in Figure 4 and Figure 5A. Remarkably, the proportion of CD4+ T lymphocytes increased after immunization with rEmEF2 (Figure 5A), when compared to the pET-32a tag protein control group (p < 0.05). The proportion of CD8+ T lymphocytes increased after immunization with rEmEF2 (Figure 5A), when compared to the PBS control group (p < 0.05). Notably, among the PBS control group and the pET-32a tag protein control group, these control groups showed no statistically significant differences in the proportions of CD4+ and CD8+ T lymphocytes (p > 0.05).
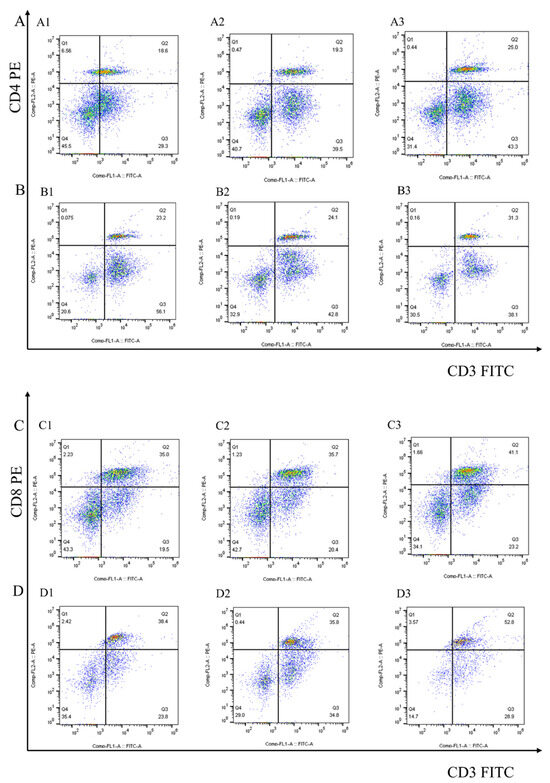
Figure 4.
The proportion of the T cell subpopulation in chickens immunized with PBS, pET-32a tag protein and rEmEF2 was determined by flow cytometry 7 days after the first and second immunization. (A) Detection of CD3+CD4+ T lymphocytes in immunized chickens 7 days after the first immunization. (B) Detection of CD3+CD4+ T lymphocytes in immunized chickens 7 days after the second immunization. (C) Detection of CD3+CD8+ T lymphocytes in immunized chickens 7 days after the first immunization. (D) Detection of CD3+CD8+ T lymphocytes in immunized chickens 7 days after the second immunization. 1: PBS control group. 2: pET-32a tag protein control group. 3: rEmEF2 group.
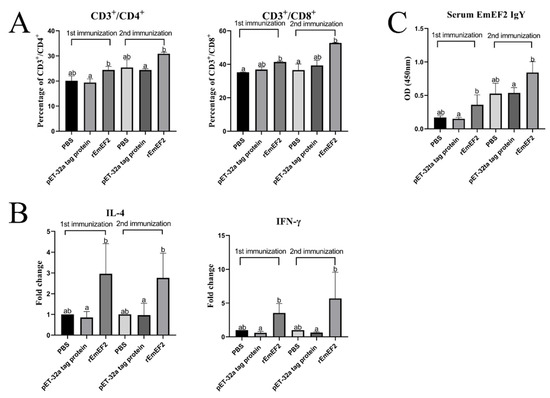
Figure 5.
The immune responses induced in immunized chickens 7 days after the first and second immunization. Chickens received the first and second immunizations when aged 14 and 28 days old, respectively. Spleen lymphocytes and serum samples were collected from chickens in each group 7 days after each immunization. The collected spleen lymphocytes were analyzed for changes in T cell subpopulation and cytokine mRNA transcription. Serum samples were analyzed for changes in rEmEF2-specific serum IgY. Significant difference (p < 0.05) between data was annotated with different letters. No significant difference (p > 0.05) between data was annotated with the same letter. (A) Percentage of T cell subpopulation in the spleen of chickens immunized with PBS, pET-32a tag protein and rEmEF2. Left: change in CD4+ T lymphocytes. Right: change in CD8+ T lymphocytes. (B) Change in the mRNA transcription level of cytokine genes in spleen lymphocytes of chickens immunized with PBS, pET-32a tag protein and rEmEF2. Left: change in IL-4. Right: change in IFN-γ. (C) Change in rEmEF2-specific serum IgY induced by immunized chicken serum samples 7 days after the first and second immunization.
After 7 days of both the first and second immunization with the rEmEF2, the mRNA transcription levels of IL-4 and IFN-γ in immunized chickens were assessed by qPCR analysis. As shown in Figure 5B, the qPCR results indicated that the mRNA transcription levels of cytokine IL-4 and IFN-γ were increased in the rEmEF2 immunization group compared to the pET-32a tag protein control group (p < 0.05). Notably, there were no statistically significant differences in the mRNA transcription levels of cytokines among the control groups (p > 0.05).
The rEmEF2-specific IgY levels in the serum of chickens immunized with rEmEF2 were detected using indirect ELISA assays. As exhibited in Figure 5C, the rEmEF2-specific IgY antibody levels in the rEmEF2-immunized group were higher than the pET-32a tag protein control group seven days after the first and second immunization (p < 0.05). Notably, there was no significant difference in IgY antibody levels between the control groups (p > 0.05).
3.4. Protective Efficacy of rEmEF2 Vaccines against E. maxima, E. acervulina, E. tenella and Mixed Eimeria
The protective efficacy of rEmEF2 was assessed based on parameters such as relative weight gain, oocyst decreased ratio, lesion score and ACI. The results of all data are shown in Table 3. Chickens immunized with rEmEF2 were then orally challenged with E. maxima, E. acervulina, E. tenella and mixed Eimeria, resulting in relative body weight gains, which were 87.06%, 97.58%, 71.87% and 57.56% (Table 3), respectively. Notably, the weight gain of the experimental groups (immunized with rEmEF2) was significantly higher than that of the control groups (non-immunized challenged and pET-32a tag protein control) (p < 0.05). These findings clearly indicate that immunization with rEmEF2 significantly mitigated the weight gain loss caused by Eimeria infection.
The reduction in mean enteric lesion scores and Oocysts Per Gram (OPG) in the rEmEF2-immunized groups were found to be significantly lower compared to the non-immunized challenged groups, as well as the pET-32a tag protein control groups (p < 0.05). Chickens immunized with rEmEF2 were then orally challenged with E. maxima, E. acervulina, E. tenella and mixed Eimeria, resulting in decreased oocyst ratios, which were 70.84%, 83.47%, 94.56% and 92.86% (Table 3), respectively. These findings show that immunization with rEmEF2 alleviated the enteric lesions and reduced oocyst output in the Eimeria-infected chickens.
In the case of rEmEF2-immunized groups, challenges with E. maxima and E. acervulina resulted in ACI scores of 166.35 and 185.08 (Table 3), signifying moderate-to-excellent protective efficacy. However, challenges involving E. tenella and mixed Eimeria species yielded ACI scores of 144.01 and 127.94, indicating a low level of protective efficacy.
4. Discussion
Eimeria species are an obligate intracellular parasitic protozoan and can cause avian coccidiosis clinically, which seriously affects the health, efficiency and sustainable development of the domestic poultry industry and brings tremendous and irreversible loss worldwide [1,41,42]. The data report on avian coccidiosis shows that the prevalence range of Eimeria spp. is widespread and the infection rate is high [41]. Recent reports estimate that avian coccidiosis in poultry is responsible for a global cost of around GBP 10.4 billion [3,4,43]. Anticoccidial drugs and live vaccines have been used strictly in clinical practice because of their various drawbacks, such as drug resistance and residues, the high cost of producing live vaccines, the inconvenient transportation of live vaccines and so on. In existing research on the prevention of avian coccidiosis, neotype vaccines have been revealed as a potential and prospective strategy against Eimeria species that are short of the shortcomings of anticoccidial drugs and traditional vaccines [44,45]. Nevertheless, the search for feasible vaccine candidate antigens has always been a formidable and hard task in the development of subunit, DNA and live vector vaccines. In Eimeria species, various antigens have been identified as antigen candidates for subunit vaccines and DNA vaccines, inducing immune responses and providing immune protection [16,18,19,25]. In this study, an Eimeria common antigen of EF2 was used to construct neotype anticoccidial vaccines: recombinant subunit vaccine (rEmEF2). The vaccination challenge trial showed that the vaccines constructed in the study could provide partial protection against infection by single or mixed Eimeria species. The result indicates that EmEF2, a common antigen, is an effective candidate antigen for the substantial development of neotype vaccines against mixed infection by Eimeria species.
The majority of clinical cases of chicken coccidiosis result from mixed infections [2,46,47,48]. Consequently, commercially available traditional live anticoccidial vaccines are typically multivalent. The same holds true for neotype anticoccidial vaccines, as monovalent vaccines may not adequately address the clinical requirements [5,49]. In recent times, a few strategies have been employed to develop multivalent vaccines for chicken coccidiosis. One approach involves the use of a cocktail of antigens derived from different Eimeria species, which has demonstrated promising immune protection [50,51]. Another strategy entails the design of multiepitope DNA vaccines comprising multiple genes sourced from various chicken Eimeria species, displaying notable protective efficacy against multiple Eimeria strains [45,52]. In this specific study, we chose the common Eimeria antigen EF2 as a candidate antigen and observed that vaccination with EmEF2 provided partial protection against infection by three Eimeria species when administered recombinant subunit vaccine (rEmEF2). These findings present an additional avenue for the development of effective and safe multivalent anticoccidial vaccines.
In the present study, we investigated the immune responses elicited by vaccination with the common Eimeria antigen EF2. Cellular immunity has been recognized as a dominant player in the defense against Eimeria infection, with T cells and their secreted cytokines playing pivotal roles [53,54,55]. Notably, CD4+ T cells and CD8+ T cells have demonstrated significant involvement in combating avian coccidia infections [4]. The IFN-γ as a Th1 cytokine exerts crucial early anticoccidial effects [56,57,58]. In this study, the proportion of CD4+ and CD8+ T lymphocytes showed an increase after the first and second immunization with the rEmEF2, indicating that T cells play a positive role in avian coccidiosis infection. Specifically, IFN-γ mRNA transcript levels increased seven days after the first and second immunization with rEmEF2. These results suggest that EmEF2 positively contributes to resistance against coccidiosis infection, with the observed changes in T lymphocytes and cytokine levels indicative of a robust cell-mediated immune response. In this study, the mRNA transcription level of IL-4 was also increased by vaccination with the rEmEF2. These results align with previous reports [59]. The role of humoral immune responses has been debated in Eimeria infection. Recent studies have demonstrated that maternal or passive immunization can provide protective antibodies, impeding the growth and development of Eimeria species and safeguarding chick offspring [60,61]. In our study, vaccination with rEmEF2 led to an increase in IgY antibody levels following both the initial and booster immunization. These results lend support to the notion that IgY contributes to the anticoccidial immune response. Overall, EmEF2 administered as a recombinant subunit vaccine induced significant cellular and humoral immune responses, underscoring its pivotal role in immune protection.
EF2 has been shown to offer critical protection against various parasites. For example, in Leishmania challenge experiments, EF2 induced elevated levels of IL-12 and IFN-γ, mediated Th1 immune responses, and significantly increased IgG2 antibody levels, resulting in 65% protection in hamsters [33]. Additionally, the localization of Eimeria tenella EF2 (EtEF2) in second-generation merozoites has been determined, with increased expression levels, leading to the inhibition of partial invasion-related proteins following diclazuril treatment [59,62]. These results underscore the significant prospects and potential of EF2 as a vaccine candidate antigen given its high homology across species and its demonstrated capacity to confer significant immune protection.
In this study, we used the whole protein of EmEF2 to construct an anticoccidial vaccine. However, developing peptide vaccines could be considered for controlling chicken coccidiosis. Some researchers have proposed that peptide-based vaccines are composed of immunogenic epitopes of various antigens to generate highly specific immune responses [63,64,65,66,67]. Since cellular immunity plays a crucial role against avian coccidiosis, selecting peptides of T cell epitopes as vaccine candidates for development may effectively control the incidence of avian coccidiosis [68,69,70,71,72]. Therefore, T cell epitopes of several common antigens of chicken coccidia identified in our previous study, including EF2, can be selected for the construction of a multivalent epitope vaccine, which can be targeted to increase the level of cellular immunity induced by the vaccine and thus improve its protective efficacy. We will be conducting such studies in the future.
Immunization with EmEF2 in chickens effectively ameliorated enteric lesions, reduced weight loss, and diminished oocyst output in chickens afflicted with single or mixed Eimeria species infections. Nevertheless, the scope for enhancing their protective efficacy remains. For example, the incorporation of cytokines such as IL-2 or IFN-γ as adjuvants [73,74,75], or their direct inclusion into prokaryotic recombinant plasmids, coupled with potential adjustments to the dosage of EmEF2 vaccines, may serve to augment the immunoprotective potency of EmEF2 vaccines.
Since the intramuscular injection route could provide consistency in systemic immune responses [76], it is commonly employed in studies involving new-generation anticoccidial vaccines. Nasri et al. reported intramuscular injection as the most prevalent administration route for new-generation anticoccidial vaccines (n = 43 studies) [77]. In their report, they took a meta-analysis of the immunization routes used in immunization challenge trials which evaluated the protective efficacy of new-generation anticoccidial vaccine candidates against Eimeria infection in chickens; they found that out of 63 studies, 43 utilized intramuscular injection. Notably, 25 studies employed intramuscular injection for delivering subunit vaccines, indicating its frequent use as a route for administering anticoccidial subunit vaccines in chickens. Therefore, we also employed the intramuscular route in this study, resulting in effective immunoprotection. However, intramuscular injection inevitably causes stress to immune animals, leading to potential deviations in immune responses. Non-injection immunization (e.g., via eye and nasal drops) [78,79,80] is more suitable for large-scale clinical practice than intramuscular immunization. In future studies, we will consider non-injectable vaccination routes to immunize chickens.
5. Conclusions
In conclusion, EmEF2 is highly immunogenic, elicits immune responses and is able to provide partial protection against both single and mixed Eimeria species infections, suggesting that EmEF2 is a promising candidate antigen and offers a hopeful prospect for the development of anticoccidial vaccines to prevent Eimeria infection.
Author Contributions
Conceptualization, X.S.; methodology, X.S., R.Y., L.X., M.L. and X.L.; validation, W.D.; formal analysis, W.D. and X.S.; data curation, W.D.; writing—original draft preparation, Y.M. and W.D.; writing—review and editing, Y.M., M.L. and X.S.; project administration, X.S., R.Y., L.X., M.L. and X.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Grant No. 32273034, 31972705) and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Institutional Review Board Statement
All animal experiments were carried out following the review and approval of the Committee on Experimental Animal Welfare and Ethics of Nanjing Agricultural University (Approval number: PTA 2020001).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available within the article.
Acknowledgments
We gratefully thank Xianglin Pu and Chen Chen for their assistance in test data processing and conducting experiments.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Blake, D.P.; Marugan-Hernandez, V.; Tomley, F.M. Spotlight on avian pathology: Eimeria and the disease coccidiosis. Avian Pathol. 2021, 20, 209–213. [Google Scholar] [CrossRef] [PubMed]
- El-Shall, N.A.; Abd El-Hack, M.E.; Albaqami, N.M.; Khafaga, A.F.; Taha, A.E.; Swelum, A.A.; El-Saadony, M.T.; Salem, H.M.; El-Tahan, A.M.; AbuQamar, S.F.; et al. Phytochemical control of poultry coccidiosis: A review. Poult. Sci. 2022, 101, 101542. [Google Scholar] [CrossRef] [PubMed]
- Blake, D.P.; Knox, J.; Dehaeck, B.; Huntington, B.; Rathinam, T.; Ravipati, V.; Ayoade, S.; Gilbert, W.; Adebambo, A.O.; Jatau, I.D.; et al. Re-calculating the cost of coccidiosis in chickens. Vet. Res. 2020, 51, 115. [Google Scholar] [CrossRef] [PubMed]
- Lillehoj, H.S. Role of T lymphocytes and cytokines in coccidiosis. Int. J. Parasitol. 1998, 28, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Zaheer, T.; Abbas, R.Z.; Imran, M.; Abbas, A.; Butt, A.; Aslam, S.; Ahmad, J. Vaccines against chicken coccidiosis with particular reference to previous decade: Progress, challenges, and opportunities. Parasitol. Res. 2022, 121, 2749–2763. [Google Scholar] [CrossRef] [PubMed]
- Lan, L.H.; Sun, B.B.; Zuo, B.X.; Chen, X.Q.; Du, A.F. Prevalence and drug resistance of avian Eimeria species in broiler chicken farms of Zhejiang province, China. Poult. Sci. 2017, 96, 2104–2109. [Google Scholar] [CrossRef]
- Li, J.; Yang, X.; Jia, Z.; Ma, C.; Pan, X.; Ma, D. Activation of ChTLR15/ChNF-kappaB-ChNLRP3/ChIL-1beta signaling transduction pathway mediated inflammatory responses to E. tenella infection. Vet. Res. 2021, 52, 15. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, R.; Ma, L.; Wang, Y.; Pan, B.; Cai, J.; Wang, M. Eimeria tenella: Expression profiling of toll-like receptors and associated cytokines in the cecum of infected day-old and three-week old SPF chickens. Exp. Parasitol. 2012, 130, 442–448. [Google Scholar] [CrossRef]
- Blake, D.P.; Tomley, F.M. Securing poultry production from the ever-present Eimeria challenge. Trends Parasitol. 2014, 30, 12–19. [Google Scholar] [CrossRef]
- da Silva Giacomini, L.; Fernandes, F.D.; Guerra, R.R.; de Avila Botton, S.; Sangioni, L.A.; Vogel, F.S.F. Production performance and economic analysis of broiler chickens after vaccination with a live attenuated vaccine against avian coccidiosis. Parasitol. Res. 2023, 122, 1677–1683. [Google Scholar] [CrossRef]
- Ma, C.; Li, G.; Chen, W.; Jia, Z.; Yang, X.; Pan, X.; Ma, D. Eimeria tenella: IMP1 protein delivered by Lactococcus lactis induces immune responses against homologous challenge in chickens. Vet. Parasitol. 2021, 289, 109320. [Google Scholar] [CrossRef] [PubMed]
- Venkatas, J.; Adeleke, M.A. A review of Eimeria antigen identification for the development of novel anticoccidial vaccines. Parasitol. Res. 2019, 118, 1701–1710. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, A.N. Progress in recombinant vaccine development against coccidiosis. A review and prospects into the next millennium. Int. J. Parasitol. 1998, 28, 1121–1130. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Ma, C.; Wang, D.; Li, G.; Ma, D. Immune response and protective efficacy of recombinant Enterococcus faecalis displaying dendritic cell-targeting peptide fused with Eimeria tenella 3-1E protein. Poult. Sci. 2020, 99, 2967–2975. [Google Scholar] [CrossRef] [PubMed]
- Appledorn, D.M.; Aldhamen, Y.A.; Depas, W.; Seregin, S.S.; Liu, C.J.; Schuldt, N.; Quach, D.; Quiroga, D.; Godbehere, S.; Zlatkin, I.; et al. A new adenovirus based vaccine vector expressing an Eimeria tenella derived TLR agonist improves cellular immune responses to an antigenic target. PLoS ONE 2010, 5, e9579. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Tian, D.; Su, J.; Liu, X.; Shah, M.A.A.; Li, X.; Xu, L.; Yan, R.; Song, X. Protective Efficacy of Rhomboid-Like Protein 3 as a Candidate Antigen Against Eimeria maxima in Chickens. Front. Microbiol. 2021, 12, 614229. [Google Scholar] [CrossRef] [PubMed]
- Geng, T.; Luo, L.; Wang, Y.; Shen, B.; Fang, R.; Hu, M.; Zhao, J.; Zhou, Y. Evaluation of immunoprotective effects of recombinant proteins and DNA vaccines derived from Eimeria tenella surface antigen 6 and 15 in vivo. Parasitol. Res. 2022, 121, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Pastor-Fernandez, I.; Kim, S.; Marugan-Hernandez, V.; Soutter, F.; Tomley, F.M.; Blake, D.P. Vaccination with transgenic Eimeria tenella expressing Eimeria maxima AMA1 and IMP1 confers partial protection against high-level E. maxima challenge in a broiler model of coccidiosis. Parasites Vectors 2020, 13, 343. [Google Scholar] [CrossRef]
- Wang, M.; Tian, D.; Xu, L.; Lu, M.; Yan, R.; Li, X.; Song, X. Protective efficacy induced by Eimeria maxima rhomboid-like protein 1 against homologous infection. Front. Vet. Sci. 2022, 9, 1049551. [Google Scholar] [CrossRef]
- Yang, G.; Li, J.; Zhang, X.; Zhao, Q.; Liu, Q.; Gong, P. Eimeria tenella: Construction of a recombinant fowlpox virus expressing rhomboid gene and its protective efficacy against homologous infection. Exp. Parasitol. 2008, 119, 30–36. [Google Scholar] [CrossRef]
- Talebi, A. Protein profiles of five avian Eimeria species. Avian Pathol. 1995, 24, 731–735. [Google Scholar] [CrossRef]
- Sasai, K.; Lillehoj, H.S.; Hemphill, A.; Matsuda, H.; Hanioka, Y.; Fukata, T.; Baba, E.; Arakawa, A. A chicken anti-conoid monoclonal antibody identifies a common epitope which is present on motile stages of Eimeria, Neospora, and Toxoplasma. J. Parasitol. 1998, 84, 654–656. [Google Scholar] [CrossRef]
- Liu, J.; Liu, L.; Li, L.; Tian, D.; Li, W.; Xu, L.; Yan, R.; Li, X.; Song, X. Protective immunity induced by Eimeria common antigen 14-3-3 against Eimeria tenella, Eimeria acervulina and Eimeria maxima. BMC Vet. Res. 2018, 14, 337. [Google Scholar] [CrossRef]
- Liu, L.; Huang, X.; Liu, J.; Li, W.; Ji, Y.; Tian, D.; Tian, L.; Yang, X.; Xu, L.; Yan, R.; et al. Identification of common immunodominant antigens of Eimeria tenella, Eimeria acervulina and Eimeria maxima by immunoproteomic analysis. Oncotarget 2017, 8, 34935–34945. [Google Scholar] [CrossRef]
- Tian, L.; Li, W.; Huang, X.; Tian, D.; Liu, J.; Yang, X.; Liu, L.; Yan, R.; Xu, L.; Li, X.; et al. Protective Efficacy of Coccidial Common Antigen Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) against Challenge with Three Eimeria Species. Front. Microbiol. 2017, 8, 1245. [Google Scholar] [CrossRef]
- Flygaard, R.K.; Malacrida, B.; Kiely, P.; Jenner, L.B. Purification and characterization of native human elongation factor 2. Protein Expr. Purif. 2019, 158, 15–19. [Google Scholar] [CrossRef]
- Kaul, G.; Pattan, G.; Rafeequi, T. Eukaryotic elongation factor-2 (eEF2): Its regulation and peptide chain elongation. Cell Biochem. Funct. 2011, 29, 227–234. [Google Scholar] [CrossRef]
- Ryazanov, A.G. Elongation factor-2 kinase and its newly discovered relatives. FEBS Lett. 2002, 514, 26–29. [Google Scholar] [CrossRef]
- Zhu, H.; Yang, X.; Liu, J.; Zhou, L.; Zhang, C.; Xu, L.; Qin, Q.; Zhan, L.; Lu, J.; Cheng, H.; et al. Eukaryotic elongation factor 2 kinase confers tolerance to stress conditions in cancer cells. Cell Stress Chaperones 2015, 20, 217–220. [Google Scholar] [CrossRef]
- Atkinson, G.C. The evolutionary and functional diversity of classical and lesser-known cytoplasmic and organellar translational GTPases across the tree of life. BMC Genom. 2015, 16, 78. [Google Scholar] [CrossRef]
- Susorov, D.; Zakharov, N.; Shuvalova, E.; Ivanov, A.; Egorova, T.; Shuvalov, A.; Shatsky, I.N.; Alkalaeva, E. Eukaryotic translation elongation factor 2 (eEF2) catalyzes reverse translocation of the eukaryotic ribosome. J. Biol. Chem. 2018, 293, 5220–5229. [Google Scholar] [CrossRef]
- Agallou, M.; Pantazi, E.; Tsiftsaki, E.; Toubanaki, D.K.; Gaitanaki, C.; Smirlis, D.; Karagouni, E. Induction of protective cellular immune responses against experimental visceral leishmaniasis mediated by dendritic cells pulsed with the N-terminal domain of Leishmania infantum elongation factor-2 and CpG oligodeoxynucleotides. Mol. Immunol. 2018, 103, 7–20. [Google Scholar] [CrossRef]
- Kushawaha, P.K.; Gupta, R.; Sundar, S.; Sahasrabuddhe, A.A.; Dube, A. Elongation factor-2, a Th1 stimulatory protein of Leishmania donovani, generates strong IFN-gamma and IL-12 response in cured Leishmania-infected patients/hamsters and protects hamsters against Leishmania challenge. J. Immunol. 2011, 187, 6417–6427. [Google Scholar] [CrossRef]
- Probst, P.; Stromberg, E.; Ghalib, H.W.; Mozel, M.; Badaro, R.; Reed, S.G.; Webb, J.R. Identification and characterization of T cell-stimulating antigens from Leishmania by CD4 T cell expression cloning. J. Immunol. 2001, 166, 498–505. [Google Scholar] [CrossRef]
- Baragana, B.; Hallyburton, I.; Lee, M.C.; Norcross, N.R.; Grimaldi, R.; Otto, T.D.; Proto, W.R.; Blagborough, A.M.; Meister, S.; Wirjanata, G.; et al. A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature 2015, 522, 315–320. [Google Scholar] [CrossRef]
- Dechering, K.J.; Duerr, H.P.; Koolen, K.M.J.; Gemert, G.V.; Bousema, T.; Burrows, J.; Leroy, D.; Sauerwein, R.W. Modelling mosquito infection at natural parasite densities identifies drugs targeting EF2, PI4K or ATP4 as key candidates for interrupting malaria transmission. Sci. Rep. 2017, 7, 17680. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hodgson, J.N. Coccidiosis: Oocyst counting technique for coccidiostat evaluation. Exp. Parasitol. 1970, 28, 99–102. [Google Scholar] [CrossRef]
- Johnson, J.; Reid, W.M. Anticoccidial drugs: Lesion scoring techniques in battery and floor-pen experiments with chickens. Exp. Parasitol. 1970, 28, 30–36. [Google Scholar] [CrossRef]
- McManus, E.C.; Campbell, W.C.; Cuckler, A.C. Development of resistance to quinoline coccidiostats under field and laboratory conditions. J. Parasitol. 1968, 54, 1190–1193. [Google Scholar] [CrossRef]
- Britez, J.D.; Rodriguez, A.E.; Di Ciaccio, L.; Marugan-Hernandez, V.; Tomazic, M.L. What Do We Know about Surface Proteins of Chicken Parasites Eimeria? Life 2023, 13, 1295. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, L.; Wang, Z.; Zhu, P.; Chen, Y.; Yu, C.; Chen, S.; Xie, Y. Impacts of Eimeria coinfection on growth performance, intestinal health and immune responses of broiler chickens. Vet. Parasitol. 2023, 322, 110019. [Google Scholar] [CrossRef]
- Tomal, F.; Sadrin, G.; Gaboriaud, P.; Guitton, E.; Sedano, L.; Lallier, N.; Rossignol, C.; Larcher, T.; Rouille, E.; Ledevin, M.; et al. The caecal microbiota promotes the acute inflammatory response and the loss of the intestinal barrier integrity during severe Eimeria tenella infection. Front. Cell. Infect. Microbiol. 2023, 13, 1250080. [Google Scholar] [CrossRef]
- Dhama, K.; Mahendran, M.; Gupta, P.K.; Rai, A. DNA vaccines and their applications in veterinary practice: Current perspectives. Vet. Res. Commun. 2008, 32, 341–356. [Google Scholar] [CrossRef]
- Yu, Z.; Chen, S.; Huang, J.; Ding, W.; Chen, Y.; Su, J.; Yan, R.; Xu, L.; Song, X.; Li, X. A multiepitope vaccine encoding four Eimeria epitopes with PLGA nanospheres: A novel vaccine candidate against coccidiosis in laying chickens. Vet. Res. 2022, 53, 27. [Google Scholar] [CrossRef]
- Blake, D.P.; Worthing, K.; Jenkins, M.C. Exploring Eimeria Genomes to Understand Population Biology: Recent Progress and Future Opportunities. Genes 2020, 11, 1103. [Google Scholar] [CrossRef]
- Brown Jordan, A.; Blake, D.; Beard, J.; Beharry, A.; Serrette, L.; Soleyn, A.; Sookhoo, J.; Blake, L.; Brown, G.; Oura, C. Molecular Identification of Eimeria Species in Broiler Chickens in Trinidad, West Indies. Vet. Sci. 2018, 5, 12. [Google Scholar] [CrossRef]
- Jatau, I.D.; Lawal, I.A.; Kwaga, J.K.; Tomley, F.M.; Blake, D.P.; Nok, A.J. Three operational taxonomic units of Eimeria are common in Nigerian chickens and may undermine effective molecular diagnosis of coccidiosis. BMC Vet. Res. 2016, 12, 86. [Google Scholar] [CrossRef]
- Song, X.; Ren, Z.; Yan, R.; Xu, L.; Li, X. Induction of protective immunity against Eimeria tenella, Eimeria necatrix, Eimeria maxima and Eimeria acervulina infections using multivalent epitope DNA vaccines. Vaccine 2015, 33, 2764–2770. [Google Scholar] [CrossRef]
- Innes, E.A.; Vermeulen, A.N. Vaccination as a control strategy against the coccidial parasites Eimeria, Toxoplasma and Neospora. Parasitology 2006, 133, 145–168. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Wang, C.; Ding, J.; Zhao, C.; Xia, Y.; Hu, Y.; Zhang, L.; Zhou, Y.; Zhao, J.; Fang, R. Evaluation of immunoprotective effects of recombinant protein and DNA vaccine based on Eimeria tenella surface antigen 16 and 22 in vivo. Parasitol. Res. 2021, 120, 1861–1871. [Google Scholar] [CrossRef]
- Song, X.; Xu, L.; Yan, R.; Huang, X.; Li, X. Construction of Eimeria tenella multi-epitope DNA vaccines and their protective efficacies against experimental infection. Vet. Immunol. Immunopathol. 2015, 166, 79–87. [Google Scholar] [CrossRef]
- Chapman, H.D. Milestones in avian coccidiosis research: A review. Poult. Sci. 2014, 93, 501–511. [Google Scholar] [CrossRef]
- Yun, C.H.; Lillehoj, H.S.; Lillehoj, E.P. Intestinal immune responses to coccidiosis. Dev. Comp. Immunol. 2000, 24, 303–324. [Google Scholar] [CrossRef]
- Lillehoj, H.S.; Choi, K.D. Recombinant chicken interferon-gamma-mediated inhibition of Eimeria tenella development in vitro and reduction of oocyst production and body weight loss following Eimeria acervulina challenge infection. Avian Dis. 1998, 42, 307–314. [Google Scholar] [CrossRef]
- Bremner, A.; Kim, S.; Morris, K.M.; Nolan, M.J.; Borowska, D.; Wu, Z.; Tomley, F.; Blake, D.P.; Hawken, R.; Kaiser, P.; et al. Kinetics of the Cellular and Transcriptomic Response to Eimeria maxima in Relatively Resistant and Susceptible Chicken Lines. Front. Immunol. 2021, 12, 653085. [Google Scholar] [CrossRef]
- Kogut, M.H.; Lange, C. Interferon-gamma-mediated inhibition of the development of Eimeria tenella in cultured cells. J. Parasitol. 1989, 75, 313–317. [Google Scholar] [CrossRef]
- Laidlaw, B.J.; Craft, J.E.; Kaech, S.M. The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat. Rev. Immunol. 2016, 16, 102–111. [Google Scholar] [CrossRef]
- Zhou, B.H.; Jia, L.S.; Guo, H.W.; Ding, H.Y.; Yang, J.Y.; Wang, H.W. Eukaryotic elongation factor 2 is involved in the anticoccidial action of diclazuril in the second-generation merozoites of Eimeria tenella. Vet. Parasitol. 2019, 276, 108991. [Google Scholar] [CrossRef] [PubMed]
- Constantinoiu, C.C.; Molloy, J.B.; Jorgensen, W.K.; Coleman, G.T. Antibody response against endogenous stages of an attenuated strain of Eimeria tenella. Vet. Parasitol. 2008, 154, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Wallach, M. Role of antibody in immunity and control of chicken coccidiosis. Trends Parasitol. 2010, 26, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.H.; Wang, H.W.; Wang, X.Y.; Zhang, L.F.; Zhang, K.Y.; Xue, F.Q. Eimeria tenella: Effects of diclazuril treatment on microneme genes expression in second-generation merozoites and pathological changes of caeca in parasitized chickens. Exp. Parasitol. 2010, 125, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Francis, M.J. Recent Advances in Vaccine Technologies. Vet. Clin. N. Am. Small Anim. Pract. 2018, 48, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Nelde, A.; Rammensee, H.G.; Walz, J.S. The Peptide Vaccine of the Future. Mol. Cell. Proteom. 2021, 20, 100022. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, C.L.; Shrimali, P.C.; Clapacs, Z.E.; Files, M.A.; Rudra, J.S. Peptide-based supramolecular vaccine systems. Acta Biomater. 2021, 133, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, N.; Skwarczynski, M.; Toth, I. The influence of component structural arrangement on peptide vaccine immunogenicity. Biotechnol. Adv. 2022, 60, 108029. [Google Scholar] [CrossRef] [PubMed]
- Mahmoodi, S.; Amirzakaria, J.Z.; Ghasemian, A. In silico design and validation of a novel multi-epitope vaccine candidate against structural proteins of Chikungunya virus using comprehensive immunoinformatics analyses. PLoS ONE 2023, 18, e0285177. [Google Scholar] [CrossRef]
- Blake, D.P.; Pastor-Fernandez, I.; Nolan, M.J.; Tomley, F.M. Recombinant anticoccidial vaccines—A cup half full? Infect. Genet. Evol. 2017, 55, 358–365. [Google Scholar] [CrossRef]
- Yang, X.; Song, X.; Liu, J.; Chen, Q.; An, T.; Liu, Q. Protection of hatchlings against coccidiosis by maternal antibodies to four recombinant proteins of Eimeria tenella, Eimeria acervulina and Eimeria maxima. Vet. Parasitol. 2022, 312, 109813. [Google Scholar] [CrossRef]
- Juarez-Estrada, M.A.; Tellez-Isaias, G.; Graham, D.M.; Laverty, L.; Gayosso-Vazquez, A.; Alonso-Morales, R.A. Identification of Eimeria tenella sporozoite immunodominant mimotopes by random phage-display peptide libraries-a proof of concept study. Front. Vet. Sci. 2023, 10, 1223436. [Google Scholar] [CrossRef]
- Ding, J.; Qian, W.; Liu, Q.; Liu, Q. Multi-epitope recombinant vaccine induces immunoprotection against mixed infection of Eimeria spp. Parasitol. Res. 2012, 110, 2297–2306. [Google Scholar] [CrossRef] [PubMed]
- Talebi, A.; Mulcahy, G. Eimeria tenella: B-cell epitope mapping following primary and secondary infections. Exp. Parasitol. 2006, 113, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Gao, M.; Li, J.; Ma, C.; Li, G. Construction of novel cytokine by fusion of chicken IL-2 signal peptide to mature chicken IL-15 and comparison of the adjuvant effects by DNA immunization against Eimeria challenge. Vet. Immunol. Immunopathol. 2013, 156, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Park, I.; Wickramasuriya, S.S.; Arous, J.B.; Koziol, M.E.; Lillehoj, H.S. Co-administration of chicken IL-7 or NK-lysin peptide 2 enhances the efficacy of Eimeria elongation factor-1alpha vaccination against Eimeria maxima infection in broiler chickens. Poult. Sci. 2022, 101, 102013. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, L.; Li, J.; Zheng, J.; Cai, N.; Gong, P.; Li, S.; Li, H.; Zhang, X. A novel recombinant BCG vaccine encoding eimeria tenella rhomboid and chicken IL-2 induces protective immunity against coccidiosis. Korean J. Parasitol. 2014, 52, 251–256. [Google Scholar] [CrossRef]
- Schunk, M.K.; Macallum, G.E. Applications and optimization of immunization procedures. ILAR J. 2005, 46, 241–257. [Google Scholar] [CrossRef]
- Nasri, T.; Sangmaneedet, S.; Nam, N.H.; Worawong, K.; Taweenan, W.; Sukon, P. Protective efficacy of new-generation anticoccidial vaccine candidates against Eimeria infection in chickens: A meta-analysis of challenge trials. Vet. Parasitol. 2022, 306, 109724. [Google Scholar] [CrossRef]
- Elnaggar, A.; Mahmoud, H.; Saber, S. Quality control procedure for Coccidial vaccines versus different routes of immunization. Vet. World 2022, 15, 2342–2347. [Google Scholar] [CrossRef]
- Gaghan, C.; Adams, D.; Mohammed, J.; Crespo, R.; Livingston, K.; Kulkarni, R.R. Characterization of vaccine-induced immune responses against coccidiosis in broiler chickens. Vaccine 2022, 40, 3893–3902. [Google Scholar] [CrossRef]
- Li, W.C.; Zhang, X.K.; Du, L.; Pan, L.; Gong, P.T.; Li, J.H.; Yang, J.; Li, H.; Zhang, X.C. Eimeria maxima: Efficacy of recombinant Mycobacterium bovis BCG expressing apical membrane antigen1 against homologous infection. Parasitol. Res. 2013, 112, 3825–3833. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).