Analytical Performance of a Multiplexed Microarray Assay for Rapid Identification and Quantification of a Multivalent mRNA Vaccine
Abstract
:1. Introduction
2. Materials and Methods
2.1. mRNA Constructs
2.2. Oligonucleotide Sequence Design
2.3. Microarray Printing and Initial Down-Selection
2.4. Assay Procedure for mRNA Encapsulated in LNPs
2.5. Encapsulated mRNA Reactivity and Specificity
2.6. Monovalent versus Multivalent Comparison of mRNAs Encapsulated in LNPs
2.7. Limits of Quantification for mRNAs Encapsulated in LNPs
2.8. Precision and Accuracy Testing for mRNAs Encapsulated in LNPs
2.9. Assay Procedure for Unencapsulated mRNA
2.10. Reactivity and Specificity for Unencapsulated mRNA
2.11. Precision and Accuracy Testing for Unencapsulated mRNA
3. Results
3.1. VaxArray mRNA Assay Overview
3.2. Assay Is Highly Reactive and Specific for mRNA Encapsulated in LNPs and Exhibits Similar Mono- and Quadrivalent Signal Responses
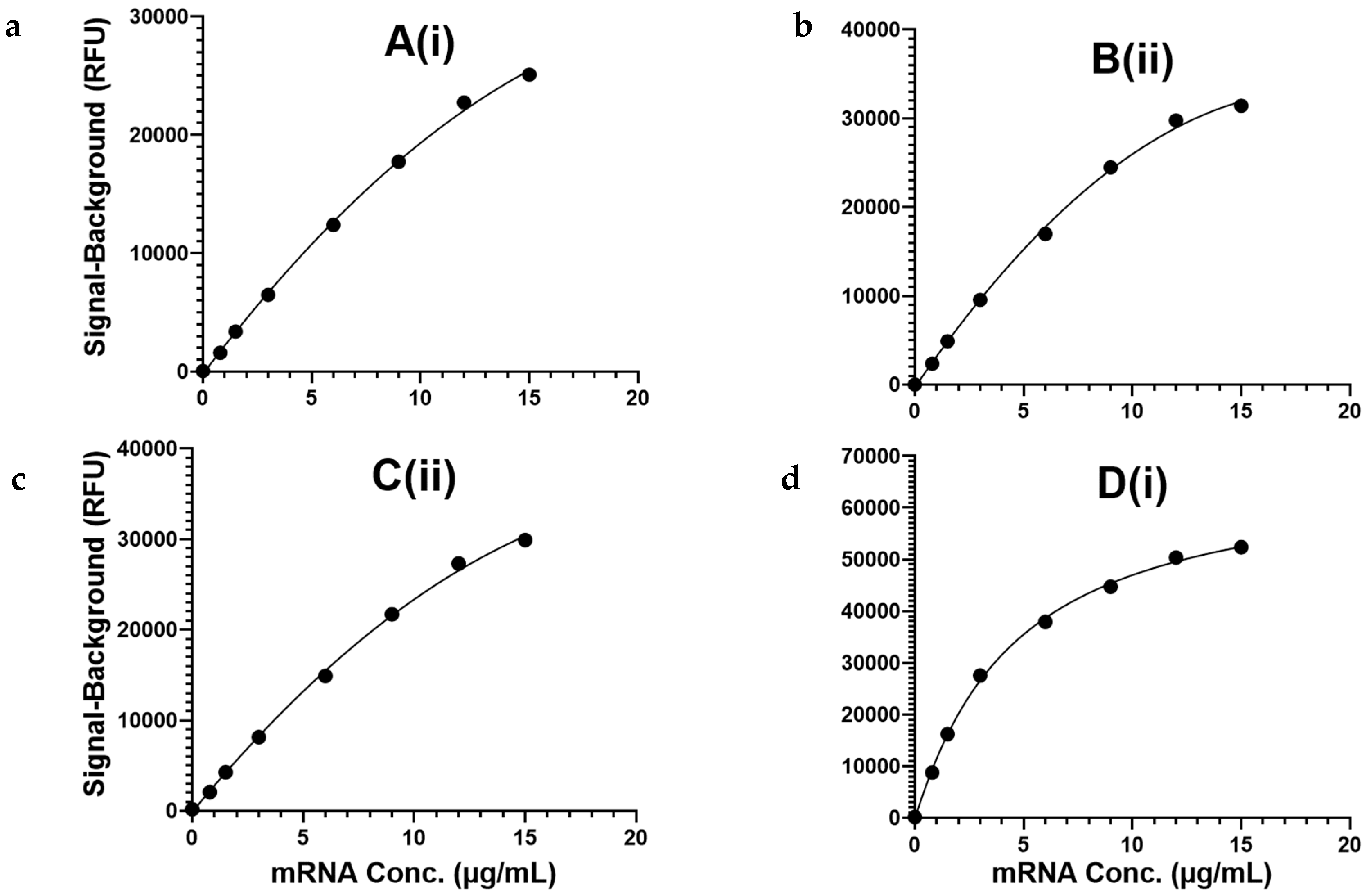
3.3. Assay Exhibits Analytical Sensitivity < 1 µg/mL and a ≥100-Fold Quantification Range for mRNA Encapsulated in LNPs
3.4. Assay Exhibits Precision of ≤10% RSD and Accuracy of 100% ± 10% Expected for Encapsulated mRNA
3.5. Assay Can Be Used Upstream to Identify and Quantify Naked mRNA
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Macchia, A.; Ferrante, D.; Angeleri, P.; Biscayart, C.; Mariani, J.; Esteban, S.; Tablado, M.R.; de Quirós, F.G.B. Evaluation of a COVID-19 Vaccine Campaign and SARS-CoV-2 Infection and Mortality Among Adults Aged 60 Years and Older in a Middle-Income Country. JAMA Netw. Open 2021, 4, e2130800. [Google Scholar] [CrossRef]
- Castelli, J.M.; Rearte, A.; Olszevicki, S.; Voto, C.; Del Valle Juarez, M.; Pesce, M.; Iovane, A.N.; Paz, M.; Chaparro, M.E.; Buyayisqui, M.P.; et al. Effectiveness of mRNA-1273, BNT162b2, and BBIBP-CorV vaccines against infection and mortality in children in Argentina, during predominance of delta and omicron covid-19 variants: Test negative, case-control study. BMJ 2022, 379, e073070. [Google Scholar] [CrossRef]
- Rahmani, K.; Shavaleh, R.; Forouhi, M.; Disfani, H.F.; Kamandi, M.; Oskooi, R.K.; Foogerdi, M.; Soltani, M.; Rahchamani, M.; Mohaddespour, M.; et al. The effectiveness of COVID-19 vaccines in reducing the incidence, hospitalization, and mortality from COVID-19: A systematic review and meta-analysis. Front. Public Health 2022, 10, 873596. [Google Scholar] [CrossRef]
- Mallapaty, S.; Callaway, E.; Kozlov, M.; Ledford, H.; Pickrell, J.; Van Noorden, R. How COVID vaccines shaped 2021 in eight powerful charts. Nature 2021, 600, 580–583. [Google Scholar] [CrossRef]
- Chavda, V.P.; Yao, Q.; Vora, L.K.; Apostolopoulos, V.; Patel, C.A.; Bezbaruah, R.; Patel, A.B.; Chen, Z.-S. Fast-track development of vaccines for SARS-CoV-2: The shots that saved the world. Front. Immunol. 2022, 13, 961198. [Google Scholar] [CrossRef]
- Lewis, L.M.; Badkar, A.V.; Cirelli, D.; Combs, R.; Lerch, T.F. The Race to Develop the Pfizer-BioNTech COVID-19 Vaccine: From the Pharmaceutical Scientists’ Perspective. J. Pharm. Sci. 2023, 112, 640–647. [Google Scholar] [CrossRef]
- Thorn, C.R.; Sharma, D.; Combs, R.; Bhujbal, S.; Romine, J.; Zheng, X.; Sunasara, K.; Badkar, A. The journey of a lifetime—Development of Pfizer’s COVID-19 vaccine. Curr. Opin. Biotechnol. 2022, 78, 102803. [Google Scholar] [CrossRef]
- Kim, Y.C.; Dema, B.; Reyes-Sandoval, A. COVID-19 vaccines: Breaking record times to first-in-human trials. NPJ Vaccines 2020, 5, 34. [Google Scholar] [CrossRef]
- Barbier, A.J.; Jiang, A.Y.; Zhang, P.; Wooster, R.; Anderson, D.G. The clinical progress of mRNA vaccines and immunotherapies. Nat. Biotechnol. 2022, 40, 840–854. [Google Scholar] [CrossRef]
- Ball, P. The lightning-fast quest for COVID vaccines—And what it means for other diseases. Nature 2021, 589, 16–18. [Google Scholar] [CrossRef]
- Lorentzen, C.L.; Haanen, J.B.; Met, Ö.; Svane, I.M. Clinical advances and ongoing trials on mRNA vaccines for cancer treatment. Lancet Oncol. 2022, 23, e450–e458. [Google Scholar] [CrossRef]
- Immuowatch RNA-Based Drug Products, 7th ed.; Mab Designs: Bristol, CT, USA, 2023; Available online: https://www.mabdesign.fr/wp-content/uploads/2023/06/Immunowatch-Edition-7-RNA-based-drug-products.pdf (accessed on 3 October 2024).
- Arevalo, C.P.; Bolton, M.J.; Le Sage, V.; Ye, N.; Furey, C.; Muramatsu, H.; Alameh, M.G.; Pardi, N.; Drapeau, E.M.; Parkhouse, K.; et al. A multivalent nucleoside-modified mRNA vaccine against all known influenza virus subtypes. Science 2022, 378, 899–904. [Google Scholar] [CrossRef]
- Big mRNA players focus on flu vaccines. Nat. Biotechnol. 2022, 40, 1706. [CrossRef]
- 5 Years of Age and Older Bivalent Pfizer-BioNTech COVID-19 Vaccine Standing Orders for Administering Vaccine. Available online: https://www.cdc.gov/vaccines/covid-19/info-by-product/pfizer/downloads/Pfizer-StandingOrders-5yrs-Older-508.pdf (accessed on 3 October 2024).
- Dolgin, E. mRNA flu shots move into trials. Nat. Rev. Drug Discov. 2021, 20, 801–803. [Google Scholar] [CrossRef]
- Arthur, R. Moderna Takes mRNA Influenza Candidate into Phase 3 Trials. In Biopharma Reporter; William Reed Ltd.: Broadfield Park, West Sussex, UK; Available online: https://www.biopharma-reporter.com/Article/2022/06/07/moderna-takes-mrna-influenza-candidate-into-phase-3-trial (accessed on 3 October 2024).
- Rosa, S.S.; Prazeres, D.M.F.; Azevedo, A.M.; Marques, M.P.C. mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine 2021, 39, 2190–2200. [Google Scholar] [CrossRef]
- Jackson, N.A.C.; Kester, K.E.; Casimiro, D.; Gurunathan, S.; DeRosa, F. The promise of mRNA vaccines: A biotech and industrial perspective. NPJ Vaccines 2020, 5, 11. [Google Scholar] [CrossRef]
- Sanyal, G.; Särnefält, A.; Kumar, A. Considerations for bioanalytical characterization and batch release of COVID-19 vaccines. NPJ Vaccines 2021, 6, 53. [Google Scholar] [CrossRef]
- Sanyal, G. Development of functionally relevant potency assays for monovalent and multivalent vaccines delivered by evolving technologies. NPJ Vaccines 2022, 7, 50. [Google Scholar] [CrossRef]
- Poveda, C.; Biter, A.B.; Bottazzi, M.E.; Strych, U. Establishing Preferred Product Characterization for the Evaluation of RNA Vaccine Antigens. Vaccines 2019, 7, 131. [Google Scholar] [CrossRef]
- Analytical Procedures for mRNA Vaccine Quality (Draft Guidelines), 2nd ed.; USP-NF: North Bethesda, MD, USA, 2023; Available online: https://www.usp.org/sites/default/files/usp/document/our-work/biologics/documents/vaccine-mrna-guidelines-2.pdf (accessed on 10 May 2023).
- Gao, R.Y.; Riley, C.M.; Toth, E.; Blair, R.H.; Gerold, M.N.; McCormick, C.; Taylor, A.W.; Hu, T.; Rowlen, K.L.; Dawson, E.D. Rapid Identity and Quantity CQA Test for Multivalent mRNA Drug Product Formulations. Vaccines 2022, 10, 1704. [Google Scholar] [CrossRef]
- Freyn, A.W.; Ramos da Silva, J.; Rosado, V.C.; Bliss, C.M.; Pine, M.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; de Souza Ferreira, L.C.; Weissman, D.; et al. A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol. Ther. 2020, 28, 1569–1584. [Google Scholar] [CrossRef]
- Ma, H.; Bell, K.N.; Loker, R.N. qPCR and qRT-PCR analysis: Regulatory points to consider when conducting biodistribution and vector shedding studies. Mol. Ther. Methods Clin. Dev. 2021, 20, 152–168. [Google Scholar] [CrossRef]
- Koetsier, G.; Cantor, E. Technical Note: A Practical Guide to Analyzing Nucleic Acid Concentration and Purity with Microvolume Spectrophotometers; New England BioLabs, Inc.: Ipswich, MA, USA, 2019; Available online: https://www.neb.com/-/media/nebus/files/application-notes/technote_mvs_analysis_of_nucleic_acid_concentration_and_purity.pdf (accessed on 3 October 2024).
- Baldwin, J.; Piplani, S.; Sakala, I.G.; Honda-Okubo, Y.; Li, L.; Petrovsky, N. Rapid development of analytical methods for evaluating pandemic vaccines: A COVID-19 perspective. Bioanalysis 2021, 13, 1805–1826. [Google Scholar] [CrossRef]
- ModernaTX, Inc. A Safety, Tolerability, and Immunogenicity Study of mRNA-1345 and mRNA-1365 in Participants Aged 5 Months to 24 Months. Available online: https://clinicaltrials.gov/ct2/show/NCT05743881 (accessed on 3 October 2024).
- ModernaTX, Inc. A Safety, Reactogenicity, and Immunogenicity Study of mRNA-1045 (Influenza and Respiratory Syncytial Virus [RSV]) or mRNA-1230 (Influenza, RSV, and Severe Acute Respiratory Syndrome Coronavirus 2 [SARS-CoV-2]) Vaccine in Adults 50 to 75 Years Old. Available online: https://clinicaltrials.gov/ct2/show/NCT05585632 (accessed on 10 May 2023).


| Capture Oligo | Signal to Background Ratio (S/B) ± σ (n = 8 Replicates) | |||
|---|---|---|---|---|
| mRNA A | mRNA B | mRNA C | mRNA D | |
| A(i) | 11 ± 1 | 1 ± 0 | 1 ± 0 | 1 ± 0 |
| B(ii) | 1 ± 0 | 13 ± 1 | 1 ± 0 | 1 ± 0 |
| C(ii) | 1 ± 0 | 1 ± 0 | 14 ± 0 | 1 ± 0 |
| D(i) | 1 ± 0 | 1 ± 0 | 1 ± 0 | 22 ± 1 |
| Capture Oligo | Lower Limit of Quantification (LLOQ) (µg/mL) | Upper Limit of Quantification (ULOQ) (µg/mL) | Quantification Range (ULOQ/LLOQ) |
|---|---|---|---|
| A(i) | 0.1 | ≥20 | ≥200 |
| B(ii) | ≤0.05 | ≥20 | ≥400 |
| C(ii) | ≤0.05 | ≥20 | ≥400 |
| D(i) | ≤0.05 | 5 | ≥100 |
| Capture Oligo | Precision (%RSD) | Accuracy (% Expected) | ||||||
|---|---|---|---|---|---|---|---|---|
| Slide 1 (n = 8) | Slide 2 (n = 8) | Slide 3 (n = 8) | All Slides (n = 24) | Slide 1 (n = 8) | Slide 2 (n = 8) | Slide 3 (n = 8) | All Slides (n = 24) | |
| A(i) | 5% | 6% | 6% | 8% | 91% | 93% | 103% | 96% |
| B(ii) | 4% | 3% | 3% | 5% | 94% | 100% | 102% | 99% |
| C(ii) | 4% | 5% | 4% | 6% | 96% | 93% | 102% | 97% |
| D(i) | 9% | 6% | 6% | 10% | 90% | 100% | 106% | 99% |
| Capture Oligo | Signal to Background Ratio (S/B) | |||
|---|---|---|---|---|
| mRNA A | mRNA B | mRNA C | mRNA D | |
| A(i) | 14 | 1 | 1 | 1 |
| B(ii) | 1 | 13 | 1 | 1 |
| C(ii) | 1 | 1 | 10 | 1 |
| D(i) | 1 | 1 | 1 | 16 |
| Capture Oligo | Precision (%RSD) | Accuracy (% Expected) | ||||||
|---|---|---|---|---|---|---|---|---|
| Slide 1 (n = 8) | Slide 2 (n = 8) | Slide 3 (n = 8) | All Slides (n = 24) | Slide 1 (n = 8) | Slide 2 (n = 8) | Slide 3 (n = 8) | All Slides (n = 24) | |
| A(i) | 4% | 6% | 10% | 8% | 97% | 95% | 105% | 99% |
| B(ii) | 7% | 7% | 8% | 7% | 99% | 102% | 98% | 100% |
| C(ii) | 5% | 7% | 8% | 8% | 98% | 97% | 107% | 101% |
| D(i) | 9% | 14% | 14% | 16% | 93% | 81% | 105% | 93% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gerold, M.N.; Toth, E.; Blair, R.H.; Gao, R.Y.; Nadkarni, D.V.; Barua, S.; Woods, J.; Rowlen, K.L.; Dawson, E.D. Analytical Performance of a Multiplexed Microarray Assay for Rapid Identification and Quantification of a Multivalent mRNA Vaccine. Vaccines 2024, 12, 1144. https://doi.org/10.3390/vaccines12101144
Gerold MN, Toth E, Blair RH, Gao RY, Nadkarni DV, Barua S, Woods J, Rowlen KL, Dawson ED. Analytical Performance of a Multiplexed Microarray Assay for Rapid Identification and Quantification of a Multivalent mRNA Vaccine. Vaccines. 2024; 12(10):1144. https://doi.org/10.3390/vaccines12101144
Chicago/Turabian StyleGerold, Megan N., Evan Toth, Rebecca H. Blair, Rachel Y. Gao, Durgesh V. Nadkarni, Sutapa Barua, Joshua Woods, Kathy L. Rowlen, and Erica D. Dawson. 2024. "Analytical Performance of a Multiplexed Microarray Assay for Rapid Identification and Quantification of a Multivalent mRNA Vaccine" Vaccines 12, no. 10: 1144. https://doi.org/10.3390/vaccines12101144






