The Impact of Clinical Factors and SARS-CoV-2 Variants on Antibody Production in Vaccinated German Healthcare Professionals Infected Either with the Delta or the Omicron Variant
Abstract
1. Introduction
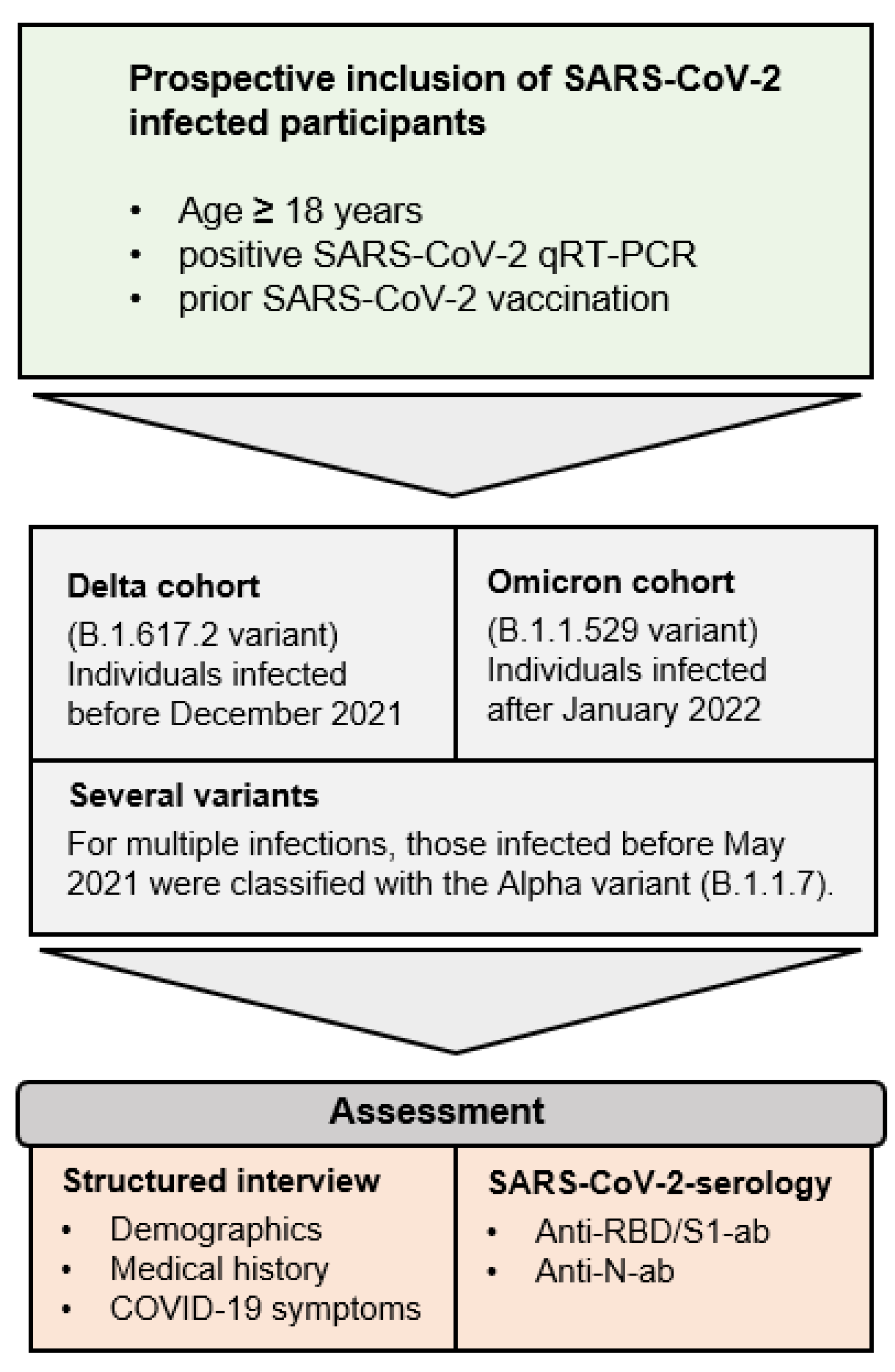
2. Materials and Methods
2.1. Participant Recruitment and Sample Collection
2.2. Anti-SARS-CoV-2 Antibody Detection
2.3. General Data Analysis
3. Results
3.1. Demographics and Vaccination Strategies
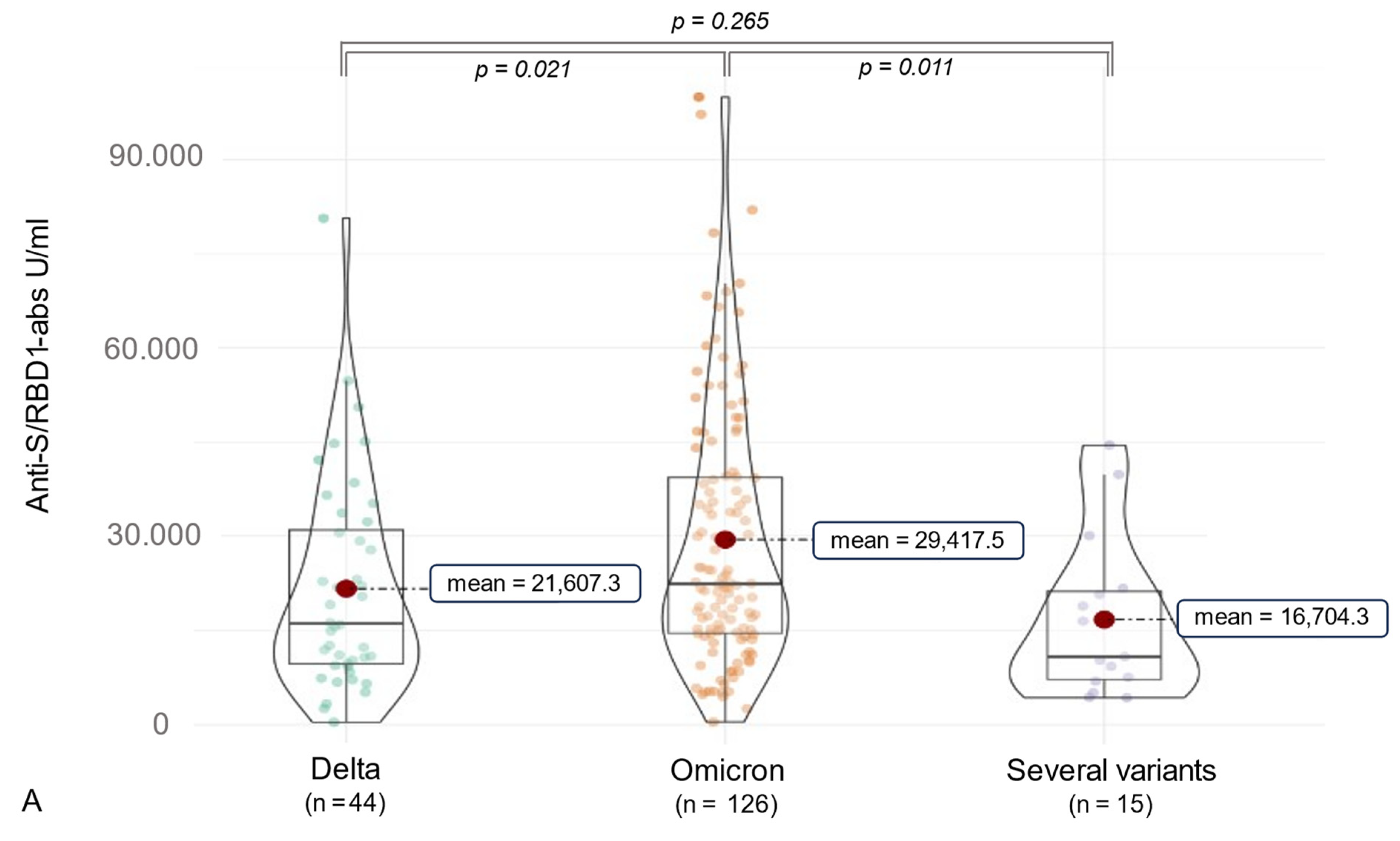
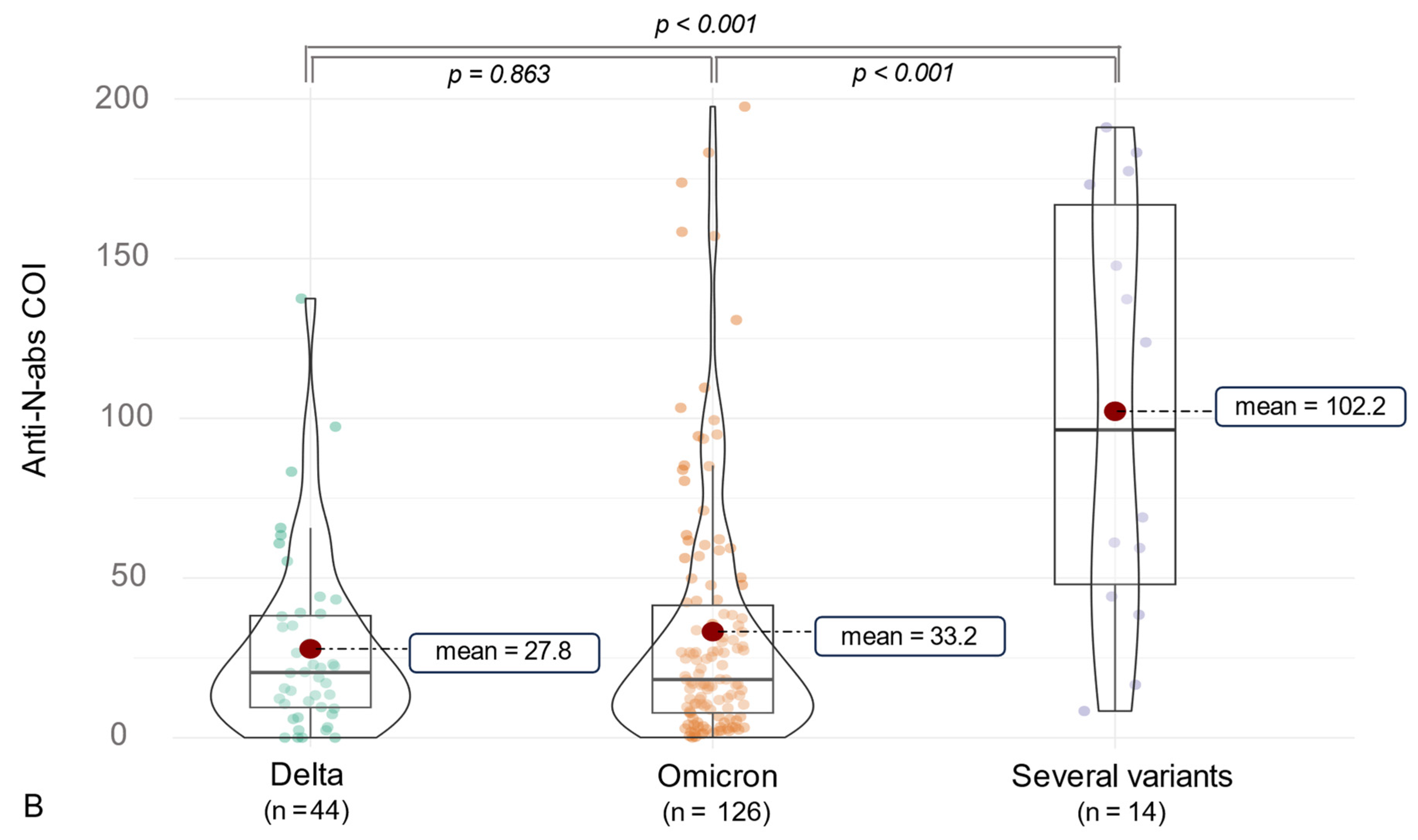
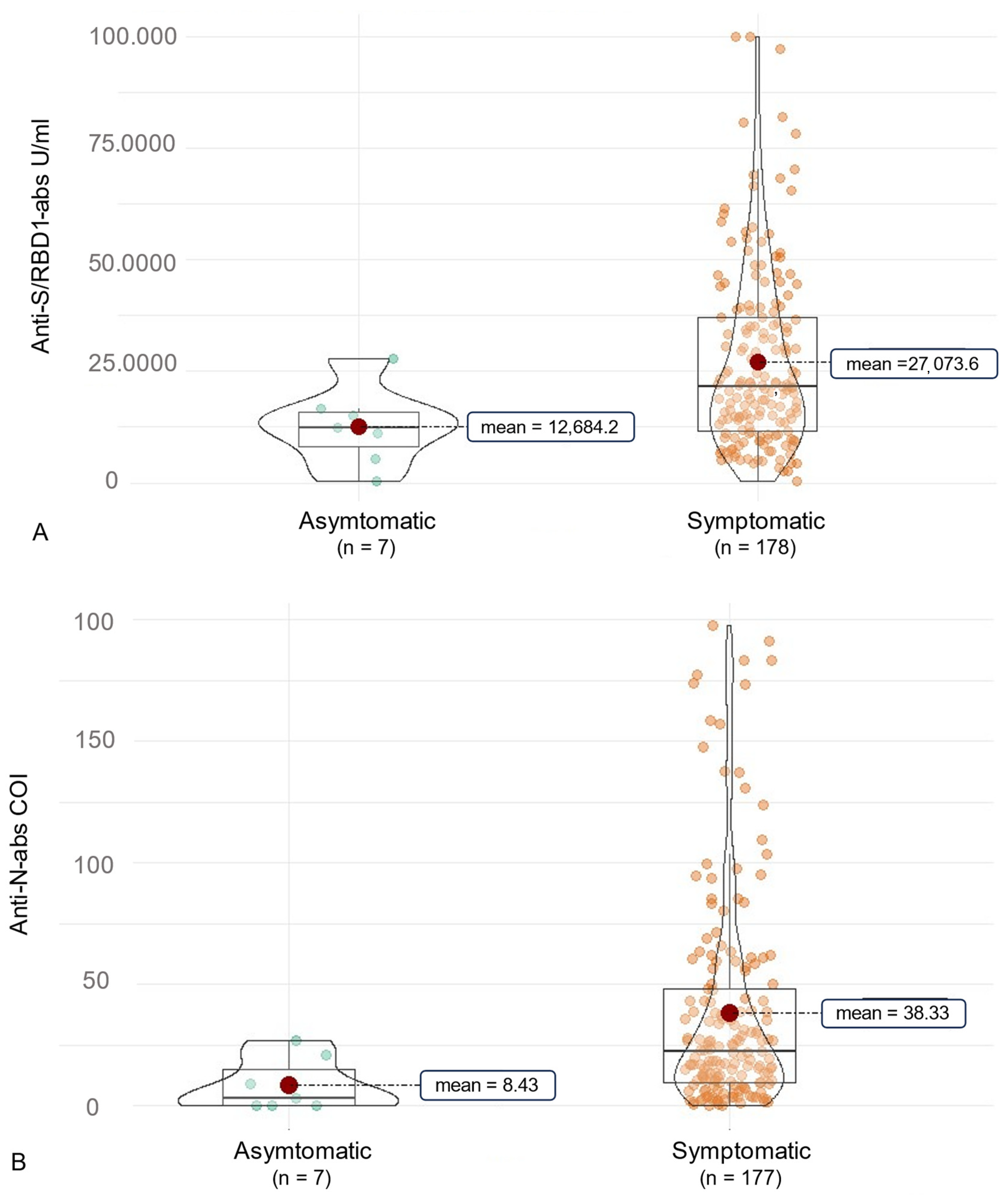
3.2. Anti-SARS-CoV-2 Antibody (ab) Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mathieu, E.; Ritchie, H.; Ortiz-Ospina, E.; Roser, M.; Hasell, J.; Appel, C.; Giattino, C.; Rodés-Guirao, L. A global database of COVID-19 vaccinations. Nat. Hum. Behav. 2021, 5, 947–953. [Google Scholar] [CrossRef]
- Ssentongo, P.; Ssentongo, A.E.; Voleti, N.; Groff, D.; Sun, A.; Ba, D.M.; Nunez, J.; Parent, L.J.; Chinchilli, V.M.; Paules, C.I. SARS-CoV-2 vaccine effectiveness against infection, symptomatic and severe COVID-19: A systematic review and meta-analysis. BMC Infect. Dis. 2022, 22, 439. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.W., Jr.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Zhao, J.; Zhao, S.; Ou, J.; Zhang, J.; Lan, W.; Guan, W.; Wu, X.; Yan, Y.; Zhao, W.; Wu, J.; et al. COVID-19: Coronavirus Vaccine Development Updates. Front. Immunol. 2020, 11, 602256. [Google Scholar] [CrossRef]
- Mistry, P.; Barmania, F.; Mellet, J.; Peta, K.; Strydom, A.; Viljoen, I.M.; James, W.; Gordon, S.; Pepper, M.S. SARS-CoV-2 Variants, Vaccines, and Host Immunity. Front Immunol. 2022, 12, 809244. [Google Scholar] [CrossRef] [PubMed]
- Horn, M.P.; Jonsdottir, H.R.; Brigger, D.; Damonti, L.; Suter-Riniker, F.; Endrich, O.; Froehlich, T.K.; Fiedler, M.; Largiadèr, C.R.; Marschall, J.; et al. Serological testing for SARS-CoV-2 antibodies in clinical practice: A comparative diagnostic accuracy study. Allergy 2022, 77, 2090–2103. [Google Scholar] [CrossRef] [PubMed]
- Gerhards, C.; Thiaucourt, M.; Hetjens, M.; Haselmann, V.; Neumaier, M.; Kittel, M. Heterologous Vector—mRNA Based SARS-CoV-2 Vaccination Strategy Appears Superior to a Homologous Vector—Based Vaccination Scheme in German Healthcare Workers Regarding Humoral SARS-CoV-2 Response Indicating a High Boosting Effect by mRNA Vaccines. Vaccines 2023, 11, 701. [Google Scholar] [CrossRef] [PubMed]
- Dyer, O. COVID-19: Omicron is causing more infections but fewer hospital admissions than delta, South African data show. BMJ 2021, 375, n3104. [Google Scholar] [CrossRef]
- Maslo, C.; Friedland, R.; Toubkin, M.; Laubscher, A.; Akaloo, T.; Kama, B. Characteristics and Outcomes of Hospitalized Patients in South Africa During the COVID-19 Omicron Wave Compared With Previous Waves. JAMA 2022, 327, 583. [Google Scholar] [CrossRef] [PubMed]
- Bouzid, D.; Visseaux, B.; Kassasseya, C.; Daoud, A.; Fémy, F.; Hermand, C.; Truchot, J.; Beaune, S.; Javaud, N.; Peyrony, O.; et al. Comparison of Patients Infected with Delta Versus Omicron COVID-19 Variants Presenting to Paris Emergency Departments. Ann. Intern. Med. 2022, 175, 831–837. [Google Scholar] [CrossRef]
- Suzuki, K.; Ichikawa, T.; Suzuki, S.; Tanino, Y.; Kakinoki, Y. Clinical characteristics of the severe acute respiratory syndrome coronavirus 2 omicron variant compared with the delta variant: A retrospective case-control study of 318 outpatients from a single sight institute in Japan. PeerJ 2022, 10, e13762. [Google Scholar] [CrossRef] [PubMed]
- ISO 15189; Medical laboratories—Requirements for quality and competence Fourth edition 2022-12. ISO: Geneva, Switzerland, 2022.
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Gerhards, C.; Thiaucourt, M.; Kittel, M.; Becker, C.; Ast, V.; Hetjens, M.; Neumaier, M.; Haselmann, V. Longitudinal assessment of anti-SARS-CoV-2 antibody dynamics and clinical features following convalescence from a COVID-19 infection. Int. J. Infect. Dis. 2021, 107, 221–227. [Google Scholar] [CrossRef]
- Lopez Bernal, J.; Andrews, N.; Gower, C.; Stowe, J.; Robertson, C.; Tessier, E.; Simmons, R.; Cottrell, S.; Robertson, R.; O’Doherty, M. Early effectiveness of COVID-19 vaccination with BNT162b2 mRNA vaccine and ChAdOx1 adenovirus vector vaccine on symptomatic disease, hospitalisations and mortality in older adults in England. MedRxiv 2021. [Google Scholar]
- Sales-Moioli, A.I.L.; Galvão-Lima, L.J.; Pinto, T.K.B.; Cardoso, P.H.; Silva, R.D.; Fernandes, F.; Barbalho, I.M.P.; Farias, F.L.O.; Veras, N.V.R.; Souza, G.F.; et al. Effectiveness of COVID-19 Vaccination on Reduction of Hospitalizations and Deaths in Elderly Patients in Rio Grande do Norte, Brazil. Int. J. Environ. Res. Public Health 2022, 19, 13902. [Google Scholar] [CrossRef] [PubMed]
- Vasileiou, E.; Simpson, C.R.; Shi, T.; Kerr, S.; Agrawal, U.; Akbari, A.; Bedston, S.; Beggs, J.; Bradley, D.; Chuter, A.; et al. Interim findings from first-dose mass COVID-19 vaccination roll-out and COVID-19 hospital admissions in Scotland: A national prospective cohort study. Lancet 2021, 397, 1646–1657. [Google Scholar] [CrossRef]
- Porru, S.; Spiteri, G.; Monaco, M.G.L.; Valotti, A.; Carta, A.; Lotti, V.; Diani, E.; Lippi, G.; Gibellini, D.; Verlato, G. Post-Vaccination SARS-CoV-2 Infections among Health Workers at the University Hospital of Verona, Italy: A Retrospective Cohort Survey. Vaccines 2022, 10, 272. [Google Scholar] [CrossRef]
- Tenforde, M.W.; Self, W.H.; Adams, K.; Gaglani, M.; Ginde, A.A.; McNeal, T.; Ghamande, S.; Douin, D.J.; Talbot, H.K.; Casey, J.D.; et al. Association Between mRNA Vaccination and COVID-19 Hospitalization and Disease Severity. JAMA 2021, 326, 2043–2054. [Google Scholar] [CrossRef]
- Nyberg, T.; Ferguson, N.M.; Nash, S.G.; Webster, H.H.; Flaxman, S.; Andrews, N.; Hinsley, W.; Bernal, J.L.; Kall, M.; Bhatt, S.; et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: A cohort study. Lancet 2022, 399, 1303–1312. [Google Scholar] [CrossRef]
- Park, S.; Lim, S.Y.; Kim, J.Y.; Park, H.; Lim, J.S.; Bae, S.; Kim, J.; Jung, J.; Kim, M.J.; Chong, Y.P.; et al. Clinical and Virological Characteristics of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) B.1.617.2 (Delta) Variant: A Prospective Cohort Study. Clin. Infect. Dis. 2022, 75, e27–e34. [Google Scholar] [CrossRef]
- Duong, B.V.; Larpruenrudee, P.; Fang, T.; Hossain, S.I.; Saha, S.C.; Gu, Y.; Islam, M.S. Is the SARS-CoV-2 Omicron Variant Deadlier and More Transmissible Than Delta Variant? Int. J. Environ. Res. Public Health 2022, 19, 4586. [Google Scholar] [CrossRef]
- Mella-Torres, A.; Escobar, A.; Barrera-Avalos, C.; Vargas-Salas, S.; Pirazzoli, M.; Gonzalez, U.; Valdes, D.; Rojas, P.; Luraschi, R.; Vallejos-Vidal, E.; et al. Epidemiological characteristics of Omicron and Delta SARS-CoV-2 variant infection in Santiago, Chile. Front. Public Health 2022, 10, 984433. [Google Scholar] [CrossRef] [PubMed]
- Dobrowolska, K.; Brzdęk, M.; Zarębska-Michaluk, D.; Rzymski, P.; Rogalska, M.; Moniuszko-Malinowska, A.; Szymanek-Pasternak, A.; Jaroszewicz, J.; Dutkiewicz, E.; Kowalska, J.; et al. Differences between the course of SARS-CoV-2 infections in the periods of the Delta and Omicron variants dominance in Poland. Pol. Arch. Intern. Med. 2023, 133, 16403. [Google Scholar] [CrossRef]
- Shrestha, L.B.; Foster, C.; Rawlinson, W.; Tedla, N.; Bull, R.A. Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: Implications for immune escape and transmission. Rev. Med. Virol. 2022, 32, e2381. [Google Scholar] [CrossRef]
- Kim, P.S.; Dimcheff, D.E.; Siler, A.; Schildhouse, R.J.; Chensue, S.W. Effect of monoclonal antibody therapy on the endogenous SARS-CoV-2 antibody response. Clin. Immunol. 2022, 236, 108959. [Google Scholar] [CrossRef]
- Zhao, J.; Alshukairi, A.N.; Baharoon, S.A.; Ahmed, W.A.; Bokhari, A.A.; Nehdi, A.M.; Layqah, L.A.; Alghamdi, M.G.; Al Gethamy, M.M.; Dada, A.M.; et al. Recovery from the Middle East respiratory syndrome is associated with antibody and T cell responses. Sci. Immunol. 2017, 2, eaan5393. [Google Scholar] [CrossRef]
- Zhong, W.; Roberts, A.D.; Woodland, D.L. Antibody-Independent Antiviral Function of Memory CD4+ T Cells In Vivo Requires Regulatory Signals from CD8+ Effector T Cells. J. Immunol. 2001, 167, 1379–1386. [Google Scholar] [CrossRef] [PubMed]
- Tarke, A.; Sidney, J.; Methot, N.; Yu, E.D.; Zhang, Y.; Dan, J.M.; Goodwin, B.; Rubiro, P.; Sutherland, A.; Wang, E.; et al. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep. Med. 2021, 2, 100355. [Google Scholar] [CrossRef] [PubMed]
- Fernandes-Matano, L.; Salas-Lais, A.G.; Grajales-Muñiz, C.; Hernández-Ávila, M.; Garfias-Becerra, Y.O.; Rodríguez-Sepúlveda, M.C.; Segura-Sánchez, C.; Montes-Herrera, D.; Mendoza-Sánchez, D.; Angeles-Martínez, J.; et al. Longevity and Neutralizing Capacity of IgG Antibodies against SARS-CoV-2 Generated by the Application of BNT162b2, AZD1222, Convidecia, Sputnik V, and CoronaVac Vaccines: A Cohort Study in the Mexican Population. Microbiol. Spectr. 2023, 11, e02376-22. [Google Scholar] [CrossRef]
- Meyer, B.; Torriani, G.; Yerly, S.; Mazza, L.; Calame, A.; Arm-Vernez, I.; Zimmer, G.; Agoritsas, T.; Stirnemann, J.; Spechbach, H.; et al. Validation of a commercially available SARS-CoV-2 serological Immunoassay. Clin. Microbiol. Infect. 2020, 26, 1386–1394. [Google Scholar] [CrossRef]
- Bonelli, F.; Sarasini, A.; Zierold, C.; Calleri, M.; Bonetti, A.; Vismara, C.; Blocki, F.A.; Pallavicini, L.; Chinali, A.; Campisi, D.; et al. Clinical and Analytical Performance of an Automated Serological Test That Identifies S1/S2-Neutralizing IgG in COVID-19 Patients Semiquantitatively. J. Clin. Microbiol. 2020, 58, e01224-20. [Google Scholar] [CrossRef] [PubMed]
- da Costa, C.H.S.; de Freitas, C.A.B.; Alves, C.N.; Lameira, J. Assessment of mutations on RBD in the Spike protein of SARS-CoV-2 Alpha, Delta and Omicron variants. Sci. Rep. 2022, 12, 8540. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Bi, Y.; Luo, Y.; Tao, Q.; Gong, S.; Wang, Y.; Xiong, L.; Xia, X.; Zheng, J.C. Cognitive dysfunction of patients infected with SARS-CoV-2 omicron variant in Shanghai, China. Transl. Neurodegener. 2023, 12, 28. [Google Scholar] [CrossRef] [PubMed]


| Variable | All Subjects (n = 185) | |
|---|---|---|
| Demographics | ||
| Sex F/M (%) | 142/43 (76.8/23.2) | |
| Age (mean (SD)) | 36.04 (11.65) | |
| BMI (median [IQR]) | 23.66 [21.72, 27.85] | |
| Smoking (%) | 28 (15.1) | |
| Medical profession (%) | 184 (99.5) | |
| Patient contact | ||
| Working in intensive care unit (%) | 35 (18.9) | |
| Working in a normal care unit (%) | 66 (35.7) | |
| Rare patient contact (%) | 27 (14.6) | |
| No patient contact (%) | 57 (30.8) | |
| Medical history | ||
| Lung disease (%) | 18 (9.7) | |
| Autoimmune disease (%) | 17 (9.2) | |
| Immunosuppression (%) | 3 (33.3) | |
| First vaccination dose | ||
| BioNTech/Pfizer (%) | 120 (64.9) | |
| Moderna (%) | 16 (8.6) | |
| AstraZeneca (%) | 48 (25.9) | |
| Different (%) | 1 (0.5) | |
| COVID-19 before first vaccination (%) | 0/4 (2.16) * | |
| Days between positive qPCR and first vaccination (median [IQR]) | 313.00 [250.00, 373.00] | |
| Second vaccination dose | ||
| BioNTech/Pfizer (%) | 155 (84.2) | |
| Moderna (%) | 16 (8.7) | |
| AstraZeneca (%) | 13 (7.1) | |
| COVID-19 before second vaccination (%) | 1/5 (2.7) * | |
| Days between positive qPCR and second vaccination (mean (SD)) | 256.42 (88.3) | |
| Third vaccination dose | ||
| BioNTech/Pfizer (%) | 156 (92) | |
| Moderna (%) | 29 (18.0) | |
| COVID-19 before third vaccination (%) | 26/30 (16.2) * | |
| Positive qPCR before third vaccination (days) (median [IQR]) | −92.00 [−103.50, −79.50] | |
| Positive qPCR after third vaccination (days) (median [IQR]) | 81.00 [62.00, 105.00] | |
| Influenza vaccination (%) | 76 (41.1) | |
| Indication for SARS-CoV-2 qPCR *1 | ||
| Routine Hospital Instructions (%) | 37 (20.0) | |
| Symptomatic (%) | 105 (56.8) | |
| SARS-CoV-2 contact (%) | 87 (47.0) | |
| COVID-19 outpatient care (%) | 184 (99.5) | |
| Anti-SARS-CoV-2 abs | (n = 185) |
|---|---|
| Anti-N (positive) (%) | 177 (96.2) |
| Anti-N (median [IQR]) | 21.12 [9.07, 45.12] |
| Anti-RBD/S1 (positive) (%) | 185 (100.0) |
| Anti-RBD/S1 (U/mL) (median [IQR]) | 21,247.00 [11,494.00, 36,538.00] |
| Type of SARS-CoV-2 Variant | ||||||
|---|---|---|---|---|---|---|
| Anti-SARS-CoV-2 abs | Alpha and Delta | Delta and Omicron | Alpha and Omicron | Omicron and Omicron | p-Values | BH Correction |
| n | 2 | 10 | 3 | 4 | ||
| Anti-N abs (median [IQR]) | 53.75 [46.14, 61.37] | 99.22 [48.03, 166.85] | 0.519 | 0.566 | ||
| Anti-N abs (median [IQR]) | 53.75 [46.14, 61.37] | 153.50 [138.65, 168.35] | 0.121 | 0.219 | ||
| Anti-N abs (median [IQR]) | 53.75 [46.14, 61.37] | 15.98 [11.91, 16.63] | 0.064 | 0.154 | ||
| Anti-N abs (median [IQR]) | 99.22 [48.03, 166.85] | 153.50 [138.65, 168.35] | 0.390 | 0.468 | ||
| Anti-N abs (median [IQR]) | 99.22 [48.03, 166.85] | 15.98 [11.91, 16.63] | 0.024 | 0.154 | ||
| Anti-N abs (median [IQR]) | 153.50 [138.65, 168.35] | 15.98 [11.91, 16.63] | 0.064 | 0.154 | ||
| Anti-S/RBD abs (median [IQR]) | 4682.00 [4493.50, 4870.50] | 19,780.00 [11,070.75, 27,964.50] | 0.032 | 0.154 | ||
| Anti-S/RBD abs (median [IQR]) | 4682.00 [4493.50, 4870.50] | 10,258.00 [7318.00, 10,535.00] | 0.248 | 0.331 | ||
| Anti-S/RBD abs (median [IQR]) | 4682.00 [4493.50, 4870.50] | 13,251.00 [10,215.50, 28,164.25] | 0.064 | 0.154 | ||
| Anti-S/RBD abs (median [IQR]) | 19,780.00 [11,070.75, 27,964.50] | 10,258.00 [7318.00, 10,535.00] | 0.128 | 0.219 | ||
| Anti-S/RBD abs (median [IQR]) | 19,780.00 [11,070.75, 27,964.50] | 13,251.00 [10,215.50, 28,164.25] | 0.671 | 0.671 | ||
| Anti-S/RBD abs (median [IQR]) | 10,258.00 [7318.00, 10,535.00] | 13,251.00 [10,215.50, 28,164.25] | 0.157 | 0.236 | ||
| Type of SARS-CoV-2 Variant | Alpha and Delta | Delta and Omicron | Alpha and Omicron | Omicron and Omicron | Delta | Omicron | p-Values | BH Correction |
|---|---|---|---|---|---|---|---|---|
| n | 2 | 10 | 3 | 4 | 44 | 122 | ||
| Anti-N abs (median [IQR]) | 53.75 [46.14, 61.37] | 20.44 [9.49, 38.21] | 0.106 | 0.208 | ||||
| Anti-N abs (median [IQR]) | 99.22 [48.03, 166.85] | 20.44 [9.49, 38.21] | 0.002 | 0.016 | ||||
| Anti-N abs (median [IQR]) | 153.50 [138.65, 168.35] | 20.44 [9.49, 38.21] | 0.021 | 0.061 | ||||
| Anti-N abs (median [IQR]) | 15.98 [11.91, 16.63] | 20.44 [9.49, 38.21] | 0.296 | 0.382 | ||||
| Anti-N abs (median [IQR]) | 53.75 [46.14, 61.37] | 18.80 [7.78, 42.76] | 0.153 | 0.245 | ||||
| Anti-N abs (median [IQR]) | 99.22 [48.03, 166.85] | 18.80 [7.78, 42.76] | 0.002 | 0.016 | ||||
| Anti-N abs (median [IQR]) | 153.50 [138.65, 168.35] | 18.80 [7.78, 42.76] | 0.023 | 0.063 | ||||
| Anti-N abs (median [IQR]) | 15.98 [11.91, 16.63] | 18.80 [7.78, 42.76] | 0.278 | 0.426 | ||||
| Anti-S/RBD abs (median [IQR]) | 4682.00 [4493.50, 4870.50] | 16,064.50 [9669.50, 30,972.25] | 0.041 | 0.090 | ||||
| Anti-S/RBD abs (median [IQR]) | 19,780.00 [11,070.75, 27,964.50] | 16,064.50 [9669.50, 30,972.25] | 0.824 | 0.906 | ||||
| Anti-S/RBD abs (median [IQR]) | 10,258.00 [7318.00, 10,535.00] | 16,064.50 [9669.50, 30,972.25] | 0.117 | 0.214 | ||||
| Anti-S/RBD abs (median [IQR]) | 13,251.00 [10,215.50, 28,164.25] | 16,064.50 [9669.50, 30,972.25] | 0.970 | 0.970 | ||||
| Anti-S/RBD abs (median [IQR]) | 4682.00 [4493.50, 4870.50] | 22,494.00 [14,728.75, 39,342.25] | 0.021 | 0.063 | ||||
| Anti-S/RBD abs (median [IQR]) | 19,780.00 [11,070.75, 27,964.50] | 22,494.00 [14,728.75, 39,342.25] | 0.310 | 0.426 | ||||
| Anti-S/RBD abs (median [IQR]) | 10,258.00 [7318.00, 10,535.00] | 22,494.00 [14,728.75, 39,342.25] | 0.021 | 0.063 | ||||
| Anti-S/RBD abs (median [IQR]) | 13,251.00 [10,215.50, 28,164.25] | 22,494.00 [14,728.75, 39,342.25] | 0.381 | 0.466 | ||||
| Symptom | Delta (n = 44) (%) | Omicron (n = 126) (%) | p-Value | p-Value BH Correction |
|---|---|---|---|---|
| Fever | 18 (40.9) | 55 (43.7) | 0.860 | 0.983 |
| Night sweats | 5 (11.4) | 14 (11.1) | 1.000 | 1.000 |
| Myalgia | 4 (9.1) | 7 (5.6) | 0.478 | 0.695 |
| Headache | 32 (72.7) | 84 (66.7) | 0.573 | 0.764 |
| Cough | 22 (50.0) | 84 (66.7) | 0.070 | 0.187 |
| Throat pain | 17 (38.6) | 86 (68.3) | 0.001 | 0.005 |
| Dyspnea | 10 (22.7) | 48 (38.1) | 0.068 | 0.187 |
| Common cold | 24 (54.5) | 86 (68.3) | 0.142 | 0.325 |
| Diarrhea | 2 (4.5) | 4 (3.2) | 0.650 | 0.800 |
| Nausea | 1 (2.3) | 5 (4.0) | 1.000 | 1.00 |
| Loss of Appetite | 3 (6.8) | 5 (4.0) | 0.428 | 0.695 |
| Loss of concentration | 5 (11.4) | 33 (26.2) | 0.057 | 0.187 |
| Depression | 1 (2.3) | 1 (0.8) | 0.452 | 0.695 |
| Hair loss | 1 (2.3) | 0 (0.0) | 0.259 | 0.518 |
| Anosmia | 24 (54.5) | 21 (16.7) | <0.001 | 0.005 |
| Ageusia | 25 (56.8) | 21 (16.7) | <0.001 | 0.005 |
| Symptom | Individuals Exhibiting the Symptom Anti-S/RBD abs (Median [IQR]) | n | Individuals Not Exhibiting the Symptom Anti-S/RBD abs (Median [IQR]) | n | p-Value | p-Value BH Correction |
|---|---|---|---|---|---|---|
| Fever | 19,438 [11,140; 33,794.] | 106 | 21,861 [12,947; 38,839] | 78 | 0.233 | 0.414 |
| Night sweats | 21,483 [11,494; 35,856] | 156 | 20,975 [12,450; 37,735] | 20 | 0.747 | 0.750 |
| Myalgia | 20,719 [11,138; 35,453] | 173 | 31,884 [20,686; 39,585] | 12 | 0.411 | 0.658 |
| Headache | 20,404 [10,812; 38,307] | 61 | 21,655 [13,276; 35,067] | 124 | 0.725 | 0.750 |
| Cough | 17,099 [10,730; 30,334] | 70 | 22,452 [14,475; 39,675] | 115 | 0.015 | 0.116 |
| Throat pain | 17,468 [10,227; 30,334] | 70 | 22,668 [14,241; 41,173] | 115 | 0.005 | 0.080 |
| Dyspnea | 18,002 [10,812; 35,856] | 125 | 24,217 [15,966; 37,735] | 60 | 0.028 | 0.116 |
| Common cold | 20,719 [10,564; 30,180] | 63 | 22,003 [14,116; 39,179] | 112 | 0.128 | 0.386 |
| Diarrhea | 20,850 [11,141; 35,333] | 179 | 42,252 [35,215; 52,360] | 6 | 0.029 | 0.116 |
| Nausea | 20,975 [11,140; 36,368] | 178 | 33,829 [24,803; 37,419] | 7 | 0.211 | 0.414 |
| Loss of Appetite | 21,483 [11,878; 35,856] | 177 | 12,852 [8688; 43,2201] | 8 | 0.562 | 0.750 |
| Loss of Concentration | 20,225 [11,138; 35,453] | 145 | 24,259 [14,641; 41,187] | 40 | 0.169 | 0.386 |
| Depression | 21,247 [11,319; 36,197] | 183 | 33,899 [23,463; 44,334] | 2 | 0.652 | 0.750 |
| Hair loss | 21,174 [11,407, 36,027] | 184 | 54,769 [54,769; 54,769] | 1 | 0.160 | 0.386 |
| Anosmia | 21,483 [11,494; 38,487] | 137 | 20,452 [11,464; 33,845] | 48 | 0.574 | 0.750 |
| Ageusia | 21,365 [11,124; 38,602] | 136 | 21,100 [11,878; 33,675] | 49 | 0.750 | 0.750 |
| Symptom | Individuals Exhibiting the Symptom Anti-N abs (Median [IQR]) | n | Individuals Not Exhibiting the Symptom Anti-N abs (Median [IQR]) | n | p-Value | p-Value BH Correction |
|---|---|---|---|---|---|---|
| Fever | 19.09 [6.07, 43.27] | 106 | 23.39 [10.58, 54.00] | 78 | 0.278 | 0.789 |
| Night sweats | 20.44 [9.07, 49.96] | 156 | 23.19 [9.85, 35.13] | 20 | 0.528 | 0.789 |
| Myalgia | 20.14 [8.70, 45.14] | 173 | 35.13 [15.83, 41.05] | 12 | 0.263 | 0.789 |
| Headache | 17.59 [4.62, 45.14] | 61 | 22.84 [10.30, 45.07] | 124 | 0.398 | 0.789 |
| Cough | 16.94 [5.88, 48.46] | 70 | 23.72 [10.64, 44.00] | 115 | 0.245 | 0.789 |
| Throat pain | 20.44 [7.74, 43.93] | 70 | 21.78 [9.37, 46.88] | 115 | 0.689 | 0.789 |
| Dyspnea | 22.16 [7.80, 58.85] | 125 | 19.31 [10.62, 37.55] | 60 | 0.690 | 0.789 |
| Common cold | 18.90 [8.16, 32.29] | 63 | 24.79 [9.64, 59.37] | 112 | 0.100 | 0.789 |
| Diarrhea | 20.44 [9.19, 44.23] | 179 | 36.57 [14.73, 45.32] | 6 | 0.632 | 0.789 |
| Nausea | 20.59 [9.28, 44.24] | 178 | 26.79 [7.53, 82.97] | 7 | 0.859 | 0.883 |
| Loss of Appetite | 21.12 [8.38, 48.36] | 177 | 26.68 [15.87, 39.33] | 8 | 0.592 | 0.789 |
| Loss of concentration | 21.91 [9.16, 44.24] | 145 | 16.89 [8.69, 43.14] | 40 | 0.646 | 0.789 |
| Depression | 20.44 [8.89, 44.23] | 183 | 97.55 [67.78, 127.33] | 2 | 0.115 | 0.789 |
| Hair loss | 20.59 [8.98, 46.00] | 184 | 38.01 [38.01, 38.01] | 1 | 0.516 | 0.789 |
| Anosmia | 19.98 [8.37, 50.16] | 137 | 23.08 [11.45, 38.97] | 48 | 0.883 | 0.883 |
| Ageusia | 18.76 [7.80, 49.96] | 136 | 25.65 [13.04, 39.93] | 49 | 0.424 | 0.789 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gerhards, C.; Steingass, M.; Heininger, A.; Lange, B.; Hetjens, M.; Gerigk, M.; Neumaier, M.; Evliyaoglu, O.; Kittel, M. The Impact of Clinical Factors and SARS-CoV-2 Variants on Antibody Production in Vaccinated German Healthcare Professionals Infected Either with the Delta or the Omicron Variant. Vaccines 2024, 12, 163. https://doi.org/10.3390/vaccines12020163
Gerhards C, Steingass M, Heininger A, Lange B, Hetjens M, Gerigk M, Neumaier M, Evliyaoglu O, Kittel M. The Impact of Clinical Factors and SARS-CoV-2 Variants on Antibody Production in Vaccinated German Healthcare Professionals Infected Either with the Delta or the Omicron Variant. Vaccines. 2024; 12(2):163. https://doi.org/10.3390/vaccines12020163
Chicago/Turabian StyleGerhards, Catharina, Marlene Steingass, Alexandra Heininger, Bettina Lange, Michael Hetjens, Marlis Gerigk, Michael Neumaier, Osman Evliyaoglu, and Maximilian Kittel. 2024. "The Impact of Clinical Factors and SARS-CoV-2 Variants on Antibody Production in Vaccinated German Healthcare Professionals Infected Either with the Delta or the Omicron Variant" Vaccines 12, no. 2: 163. https://doi.org/10.3390/vaccines12020163
APA StyleGerhards, C., Steingass, M., Heininger, A., Lange, B., Hetjens, M., Gerigk, M., Neumaier, M., Evliyaoglu, O., & Kittel, M. (2024). The Impact of Clinical Factors and SARS-CoV-2 Variants on Antibody Production in Vaccinated German Healthcare Professionals Infected Either with the Delta or the Omicron Variant. Vaccines, 12(2), 163. https://doi.org/10.3390/vaccines12020163







