Flow Cytometry-Based Measurement of Antibodies Specific for Cell Surface-Expressed Folded SARS-CoV-2 Receptor-Binding Domains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Control Subjects
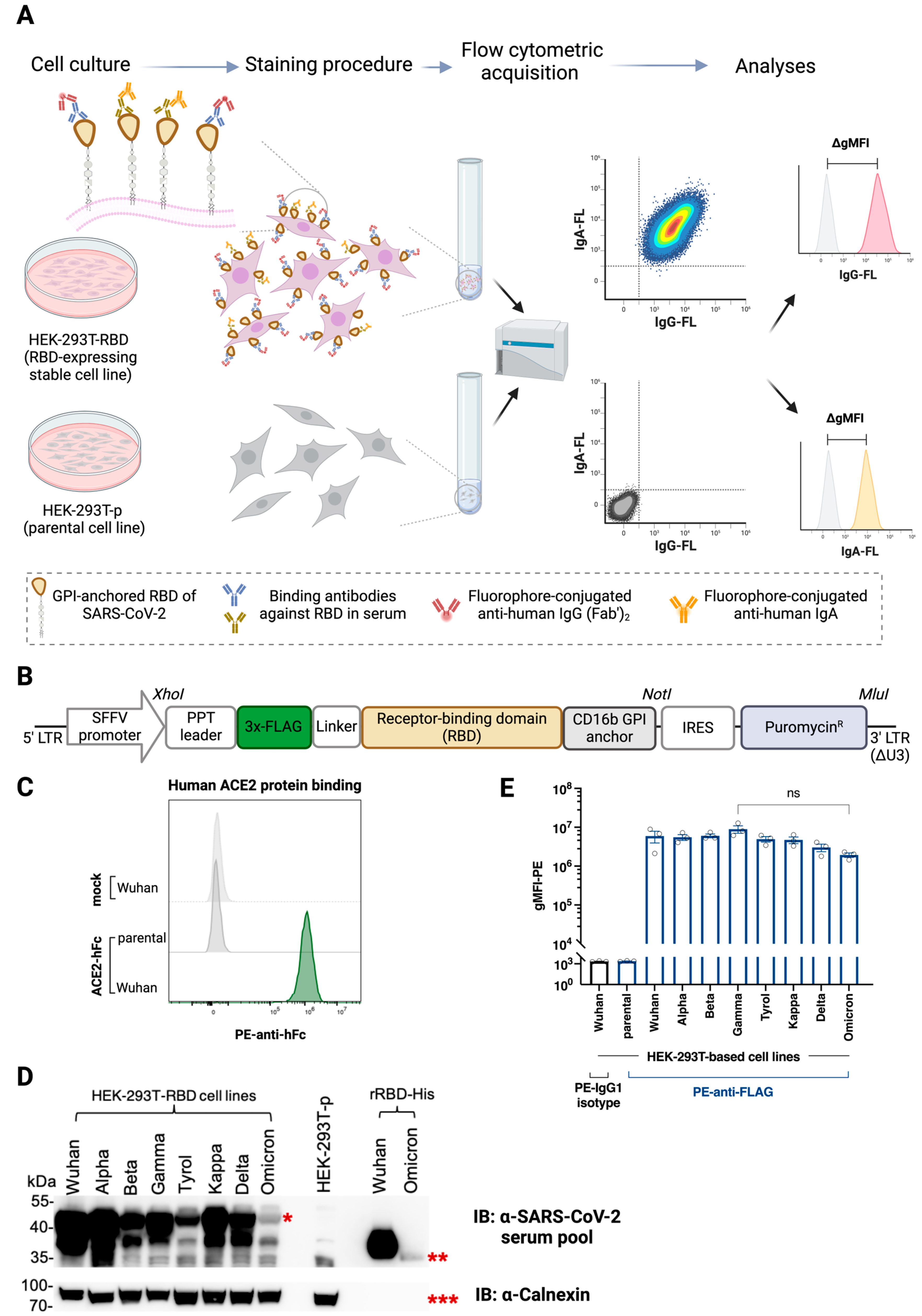
2.2. Generation of SARS-CoV-2 RBD (and Spike) Expression Constructs
2.3. Development of Stable Cell Lines
2.4. Flow Cytometry-Based Cellular Assay (FCCA)
2.5. Enzyme-Linked Immunosorbent Assay (ELISA)
2.6. FCCA Adsorption Experiments
2.7. Western Blot Analyses
2.8. Data and Statistical Analyses
3. Results
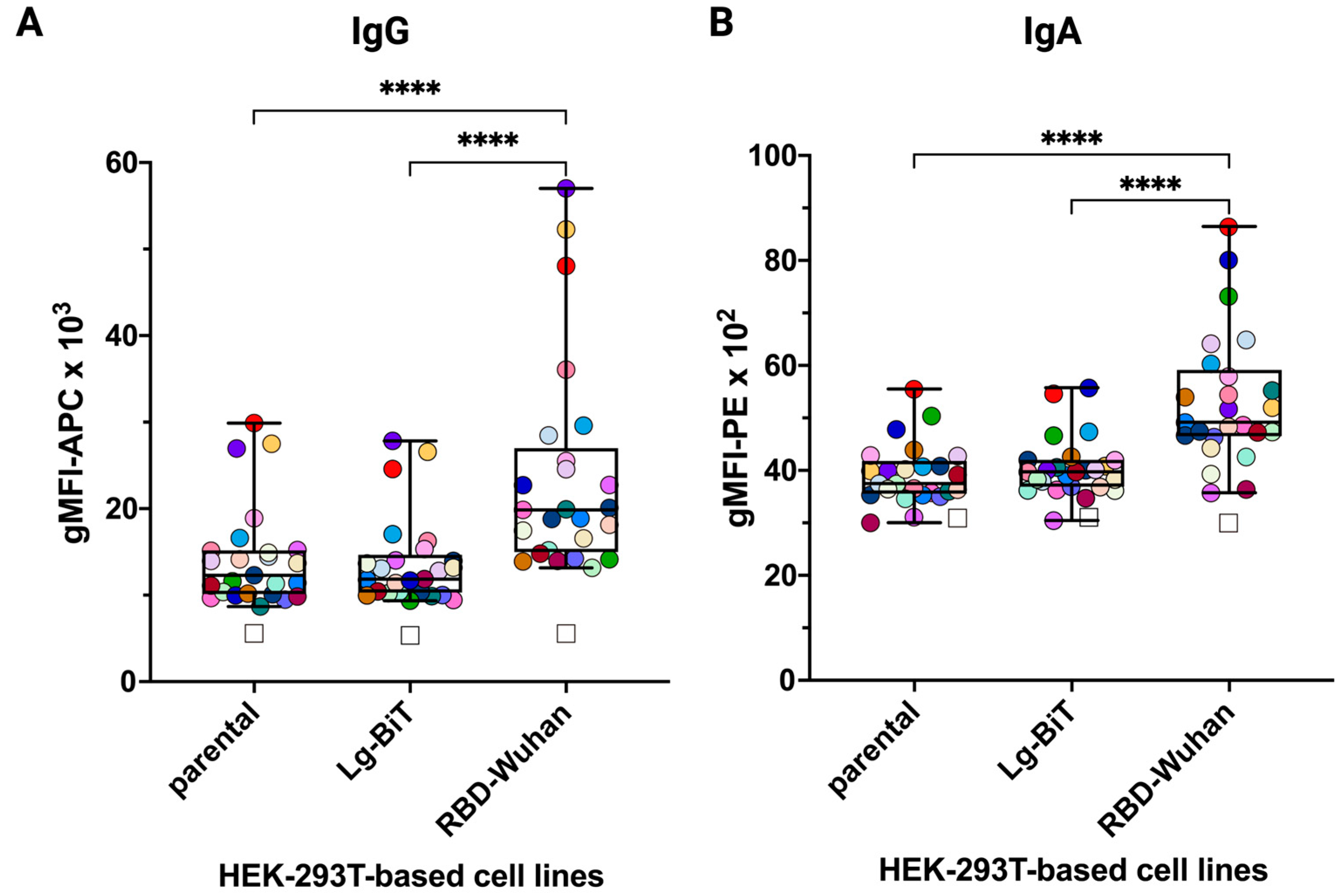
3.1. HEK-293T Transfectants Expressing GPI-Anchored RBD of SARS-CoV-2 Variants Bind Human ACE2 and Anti-SARS-CoV-2 Antibodies
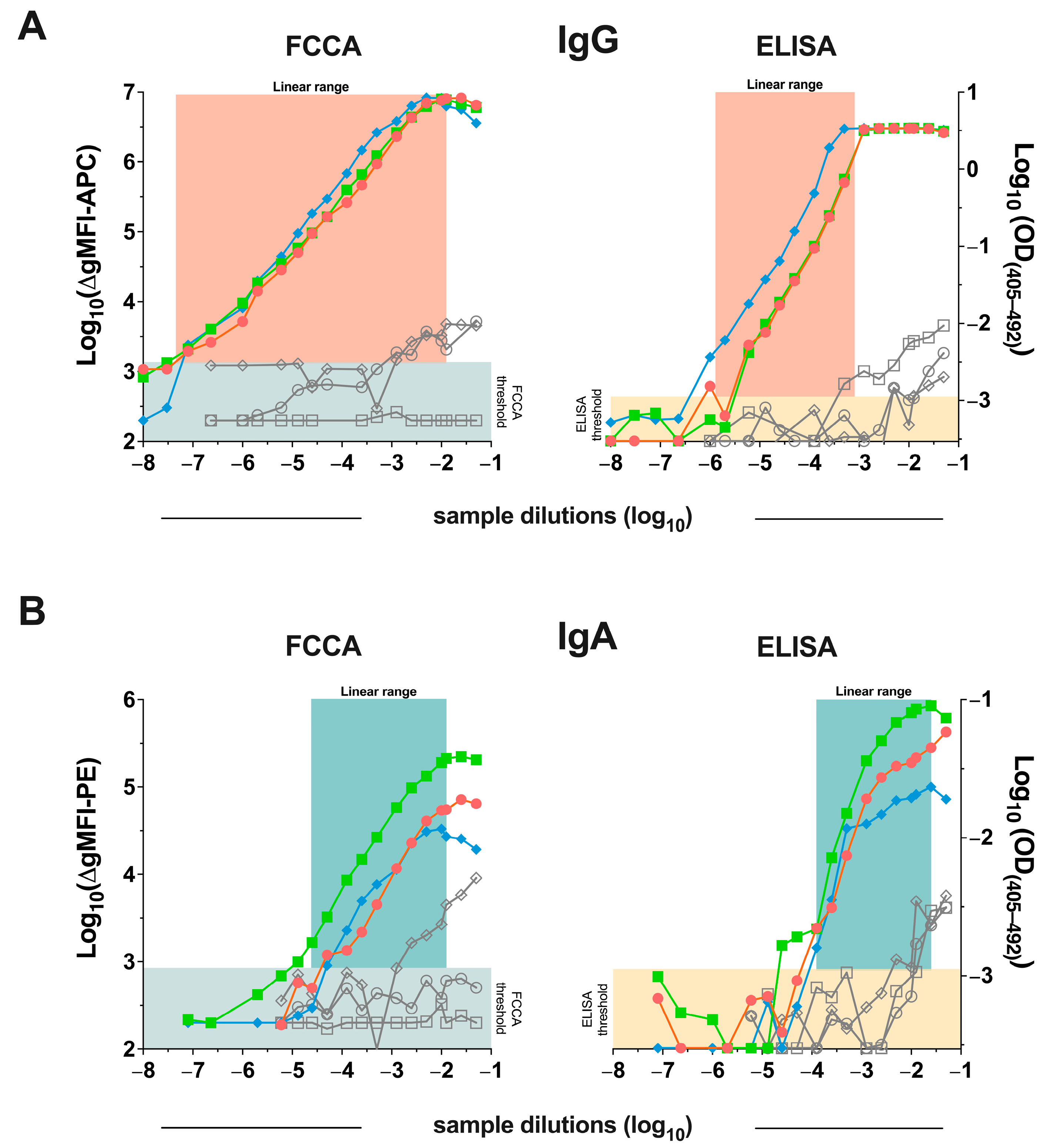
3.2. Detection of SARS-CoV-2 Anti-RBD Antibodies Using the RBD Transfectants Revealed That the FCCA Has a Wide Linear Performance Range
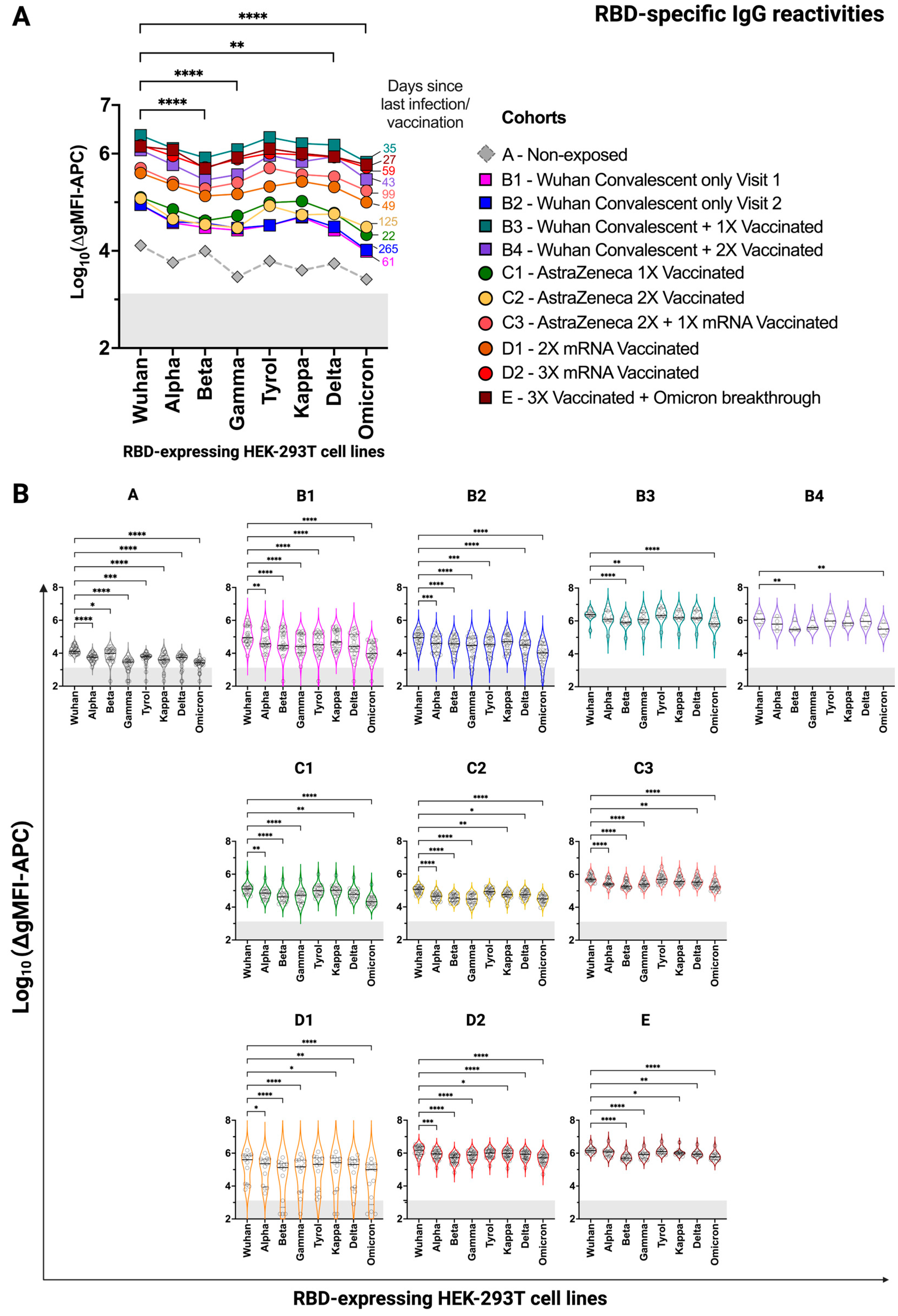
3.3. HEK-293T Cells Expressing Surface-Anchored SARS-CoV-2 RBD Versions Are Suitable for Comparing Specific Antibody Levels in Convalescent and/or Vaccinated Individuals
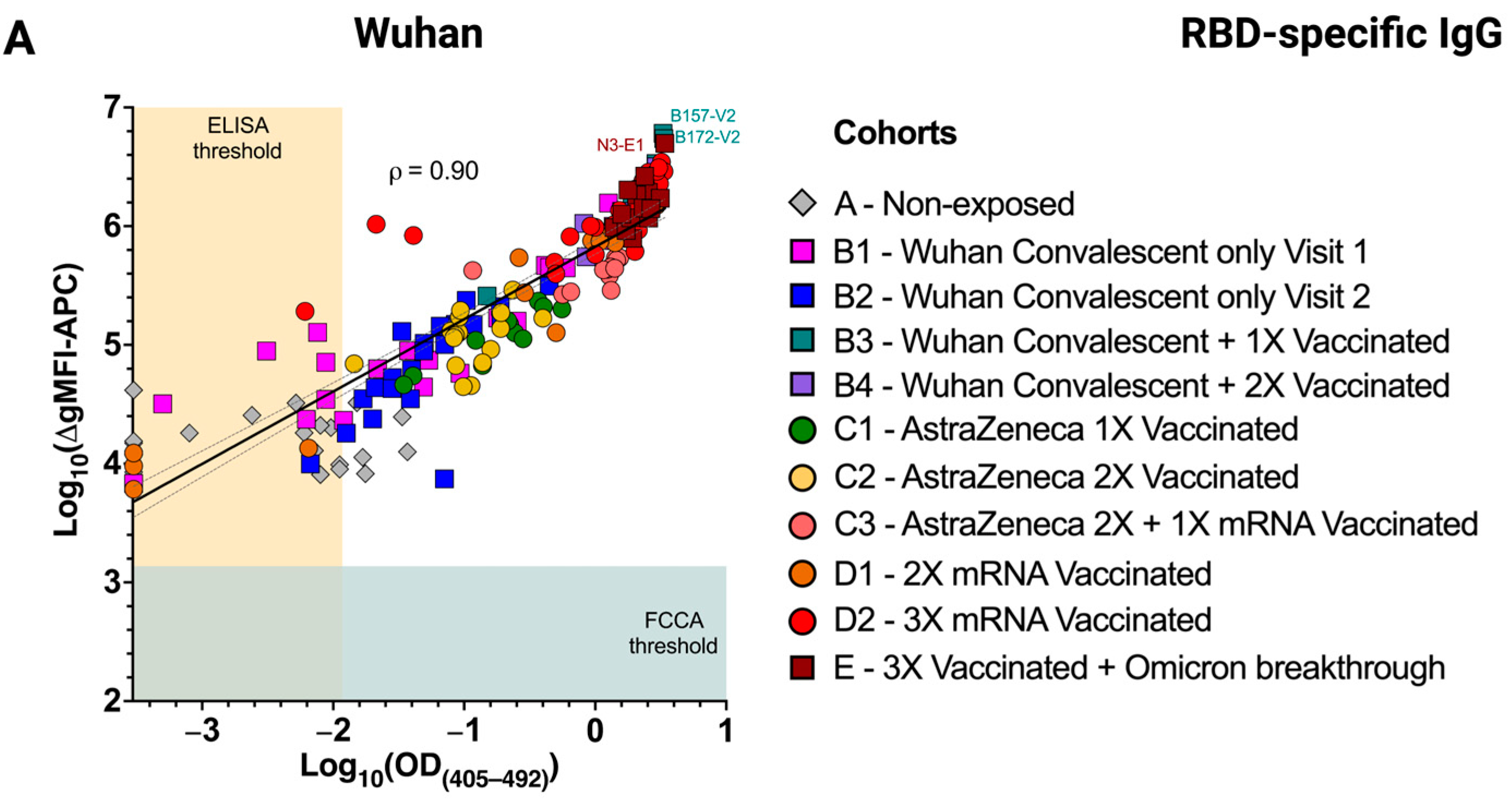
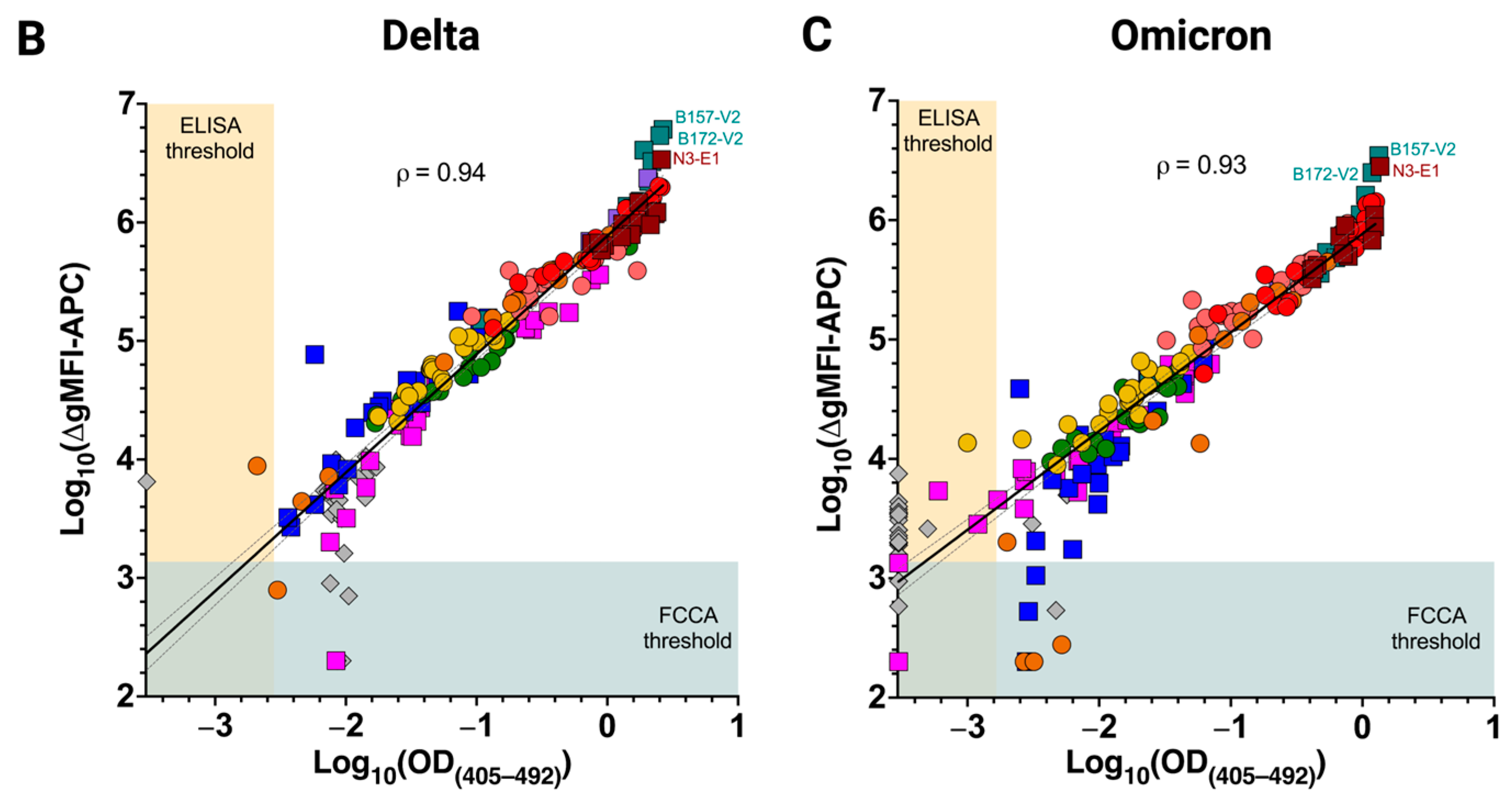
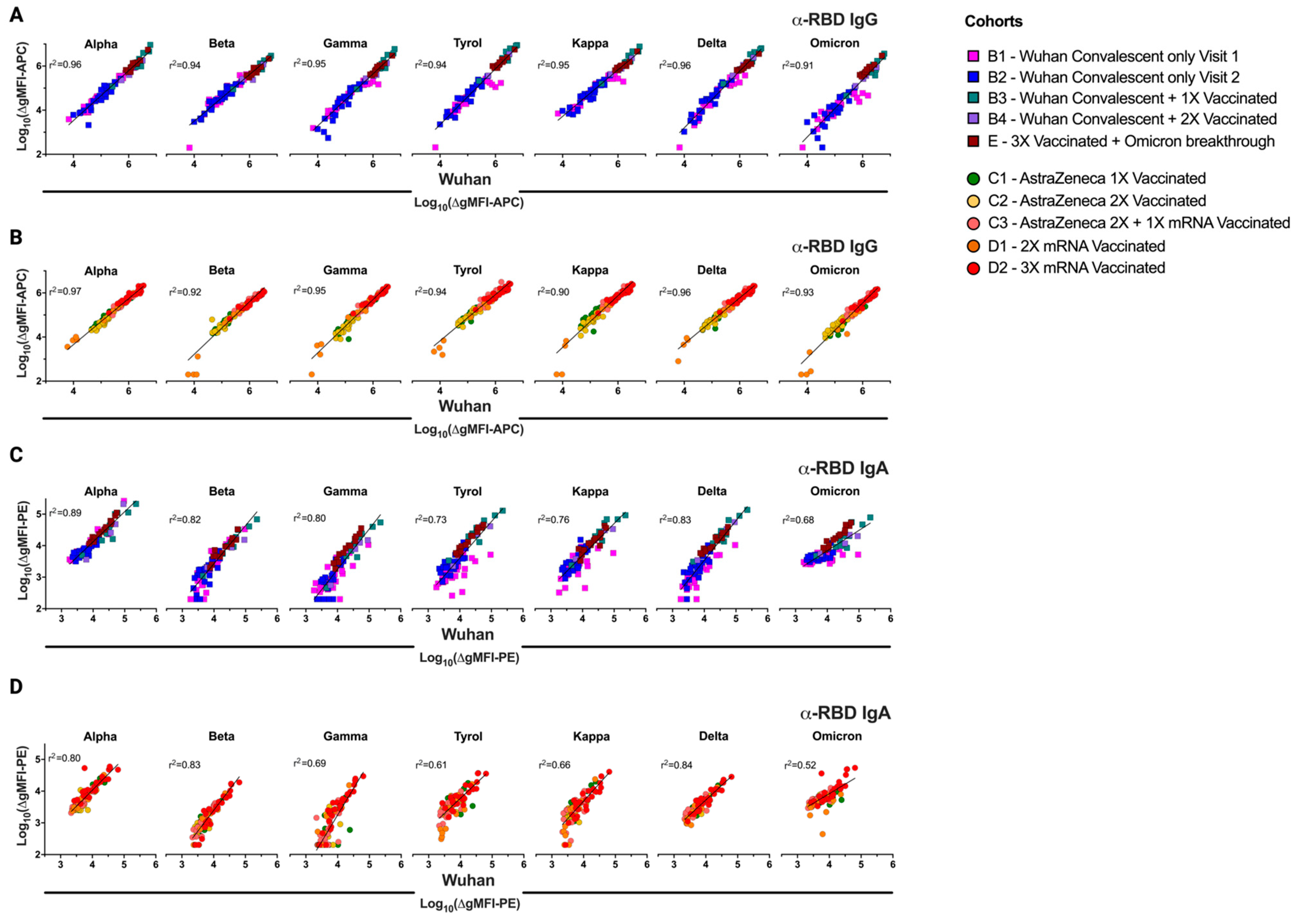
3.4. Excellent Correlation of the FCCA with the ELISA Results
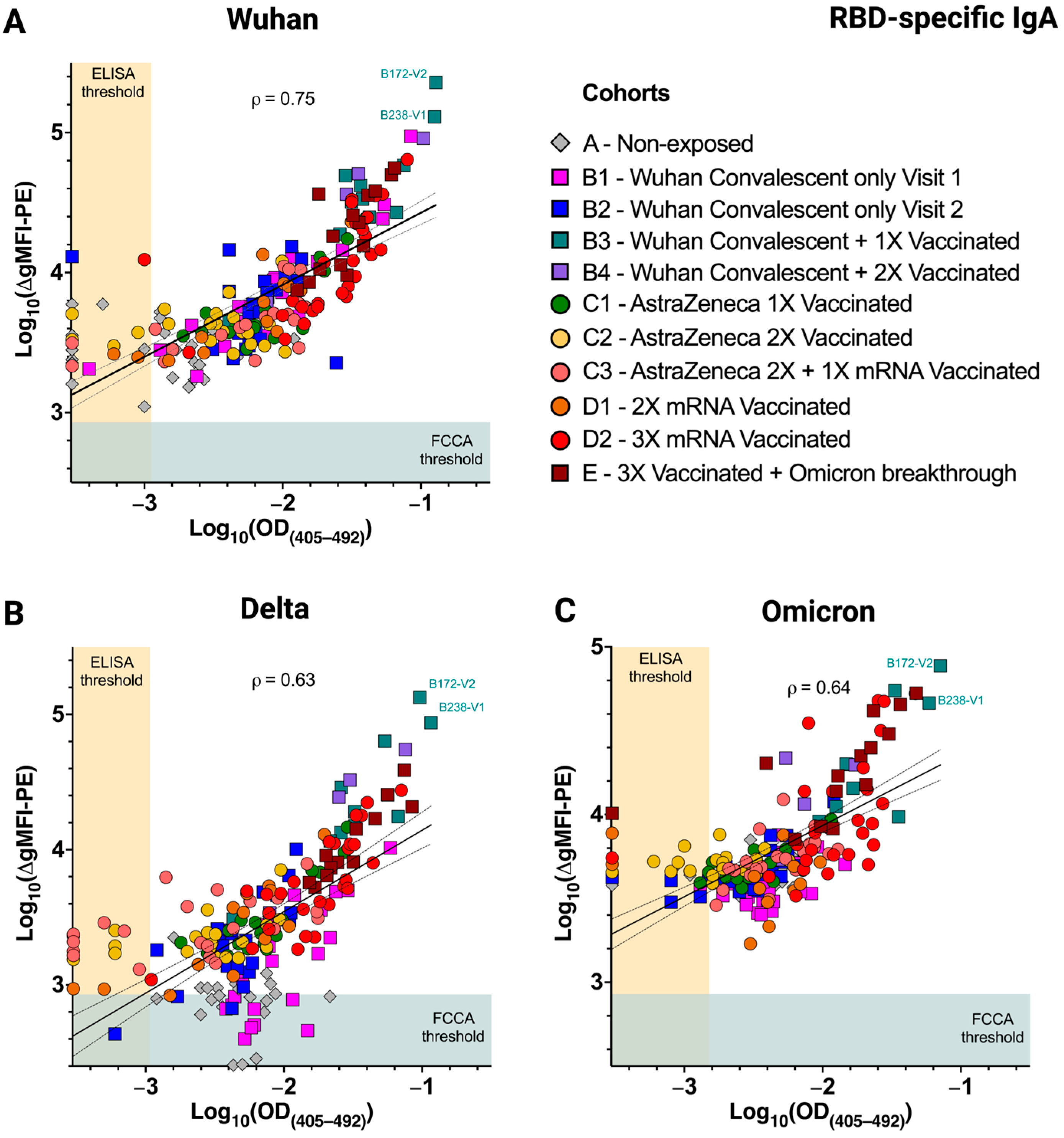
3.5. Differences in Infection- and Vaccine-Induced Correlations between RBD-Wuhan-Hu-1 IgG and IgA Reactivity
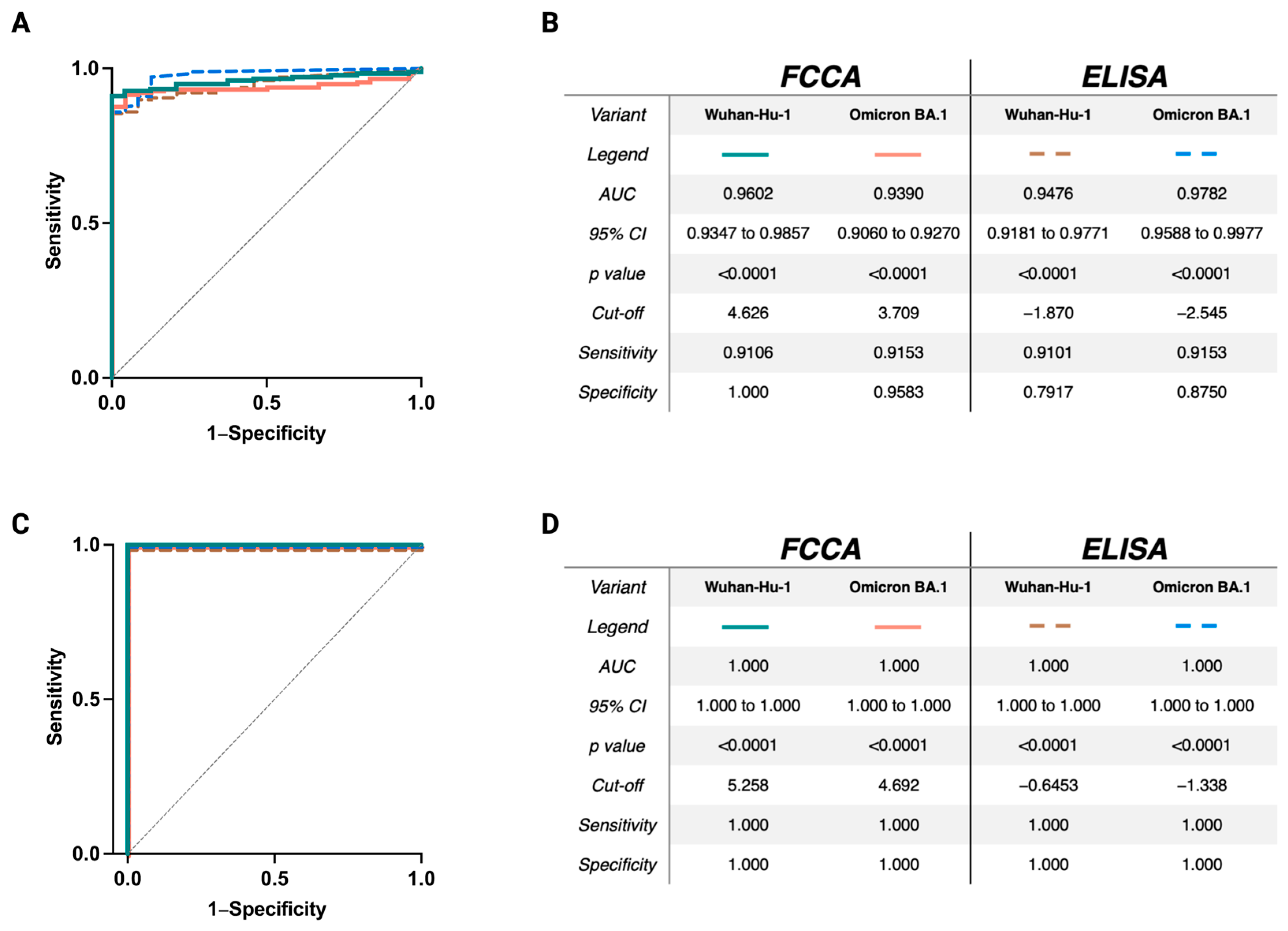
3.6. Excellent Sensitivity and Specificity of the FCCA Underscores Its Usefulness as a SARS-CoV-2-Specific Diagnostic Test
3.7. The FCCA Platform May Identify SARS-CoV-2-Specific Antibodies in Individuals without Evidence of a SARS-CoV-2 Infection
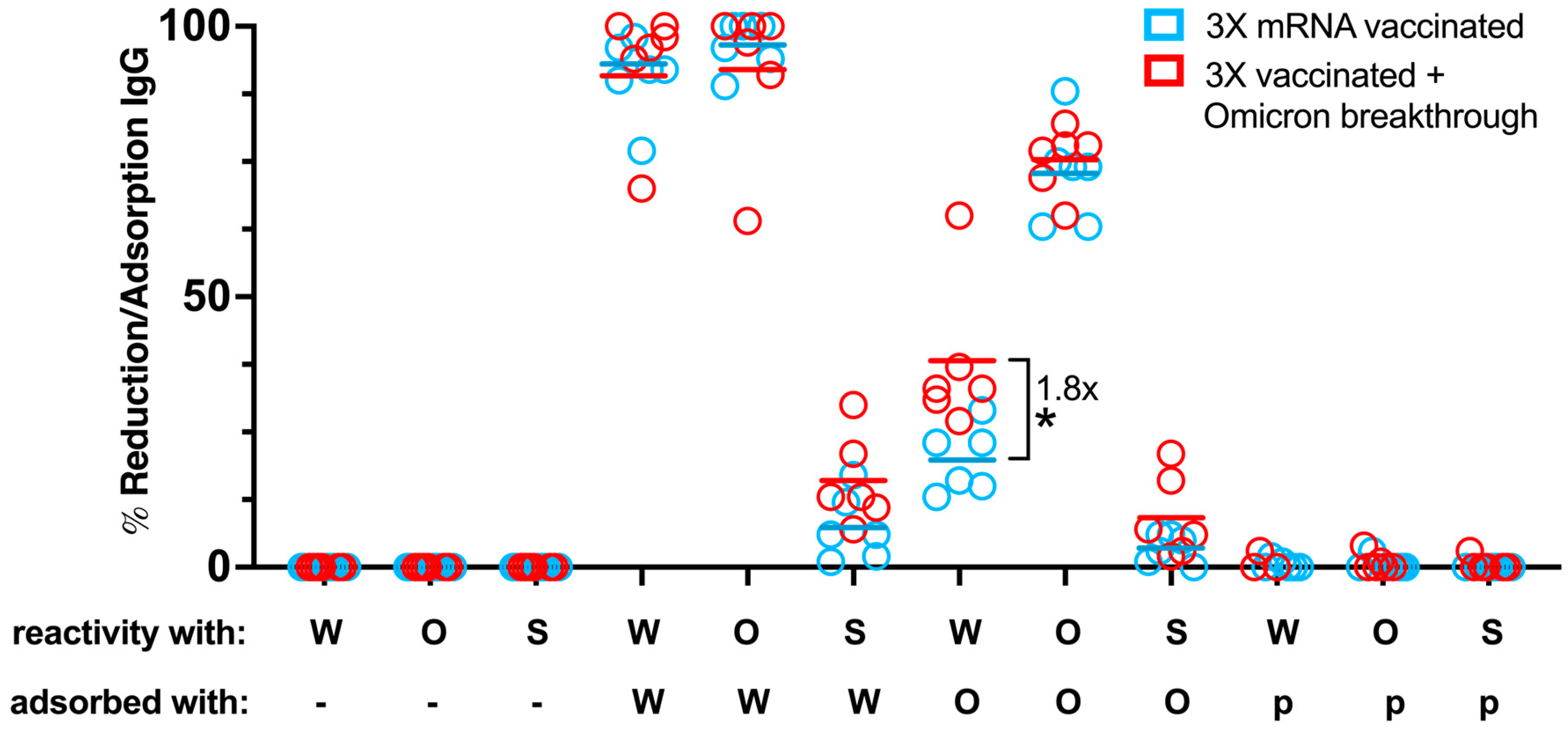
3.8. Adsorption Studies with RBD Transfectants Indicate Differences in RBD-Specific Antibody Composition in Vaccinated Individuals with and without Breakthrough Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACE2 | angiotensin-converting enzyme 2 |
| ADCC | antibody-dependent cellular cytotoxicity |
| FCCA | flow cytometry-based cellular assay |
| gMFI | geometric mean fluorescence intensity |
| GPI | glycosylphosphatidylinositol |
| HAT | hemagglutination-based test |
| IRES | internal ribosome entry site |
| OD | optical density |
| PANGO | phylogenetic assignment of named global outbreak lineages |
| PPT | pre-pro-trypsin |
| Puro | puromycin resistance gene |
| RBD | receptor-binding domain |
| ROC | receiver-operating characteristic |
| S | spike (SARS-CoV-2 protein) |
| SCCs | single-cell clones |
| SD | standard deviation |
| SI | stimulation index |
| VOC | variant of concern |
| VOI | variant of interest |
| WHO | World Health Organization |
References
- Otto, S.P.; Day, T.; Arino, J.; Colijn, C.; Dushoff, J.; Li, M.; Mechai, S.; Van Domselaar, G.; Wu, J.; Earn, D.J.D.; et al. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr. Biol. 2021, 31, R918–R929. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Roy, S.S.; Rastogi, R.; Arora, K.; Undale, A.; Gupta, R.; Arora, N.M.; Kundu, P.K. VLP-ELISA for the Detection of IgG Antibodies against Spike, Envelope, and Membrane Antigens of SARS-CoV-2 in Indian Population. Vaccines 2023, 11, 743. [Google Scholar] [CrossRef] [PubMed]
- Gattinger, P.; Ohradanova-Repic, A.; Valenta, R. Importance, Applications and Features of Assays Measuring SARS-CoV-2 Neutralizing Antibodies. Int. J. Mol. Sci. 2023, 24, 5352. [Google Scholar] [CrossRef] [PubMed]
- Rocha, V.P.C.; Quadros, H.C.; Fernandes, A.M.S.; Gonçalves, L.P.; Badaró, R.J.d.S.; Soares, M.B.P.; Machado, B.A.S. An Overview of the Conventional and Novel Methods Employed for SARS-CoV-2 Neutralizing Antibody Measurement. Viruses 2023, 15, 1504. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, K.; Cheng, S.; Binder, R.A.; Mantis, N.J.; Crawford, J.M.; Okoye, N.; Braun, J.G.; Joung, S.; Wang, M.; Lozanski, G.; et al. Clinical Utility of SARS-CoV-2 Serological Testing and Defining a Correlate of Protection. Vaccines 2023, 11, 1644. [Google Scholar] [CrossRef] [PubMed]
- Laurie, M.T.; Liu, J.; Sunshine, S.; Peng, J.; Black, D.; Mitchell, A.M.; Mann, S.A.; Pilarowski, G.; Zorn, K.C.; Rubio, L.; et al. SARS-CoV-2 Variant Exposures Elicit Antibody Responses With Differential Cross-Neutralization of Established and Emerging Strains Including Delta and Omicron. J. Infect. Dis. 2022, 225, 1909–1914. [Google Scholar] [CrossRef] [PubMed]
- Röltgen, K.; Nielsen, S.C.; Silva, O.; Younes, S.F.; Zaslavsky, M.; Costales, C.; Yang, F.; Wirz, O.F.; Solis, D.; Hoh, R.A.; et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell 2022, 185, 1025–1040.e1014. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.; Garner-Spitzer, E.; Schötta, A.-M.; Orola, M.; Wessely, A.; Zwazl, I.; Ohradanova-Repic, A.; Weseslindtner, L.; Tajti, G.; Gebetsberger, L.; et al. SARS-CoV-2-mRNA Booster Vaccination Reverses Non-Responsiveness and Early Antibody Waning in Immunocompromised Patients—A Phase Four Study Comparing Immune Responses in Patients With Solid Cancers, Multiple Myeloma and Inflammatory Bowel Disease. Front. Immunol. 2022, 13, 889138. [Google Scholar] [CrossRef] [PubMed]
- Barrière, J.; Carles, M.; Audigier-Valette, C.; Re, D.; Adjtoutah, Z.; Seitz-Polski, B.; Gounant, V.; Descamps, D.; Zalcman, G. Third dose of anti-SARS-CoV-2 vaccine for patients with cancer: Should humoral responses be monitored? A position article. Eur. J. Cancer 2021, 162, 182–193. [Google Scholar] [CrossRef]
- Garner-Spitzer, E.; Wagner, A.; Gudipati, V.; Schoetta, A.-M.; Orola-Taus, M.; Kundi, M.; Kunert, R.; Mayrhofer, P.; Huppa, J.B.; Stockinger, H.; et al. Lower magnitude and faster waning of antibody responses to SARS-CoV-2 vaccination in anti-TNF-α-treated IBD patients are linked to lack of activation and expansion of cTfh1 cells and impaired B memory cell formation. EBioMedicine 2023, 96, 104788. [Google Scholar] [CrossRef]
- Kratzer, B.; Trapin, D.; Gattinger, P.; Oberhofer, T.; Sehgal, A.N.A.; Waidhofer-Söllner, P.; Rottal, A.; Körmöczi, U.; Grabmeier-Pfistershammer, K.; Kopetzky, G.H.; et al. Lack of Induction of RBD-Specific Neutralizing Antibodies despite Repeated Heterologous SARS-CoV-2 Vaccination Leading to Seroconversion and Establishment of T Cell-Specific Memory in a Patient in Remission of Multiple Myeloma. Vaccines 2022, 10, 374. [Google Scholar] [CrossRef] [PubMed]
- Gattinger, P.; Borochova, K.; Dorofeeva, Y.; Henning, R.; Kiss, R.; Kratzer, B.; Mühl, B.; Perkmann, T.; Trapin, D.; Trella, M.; et al. Antibodies in serum of convalescent patients following mild COVID-19 do not always prevent virus-receptor binding. Allergy 2021, 76, 878–883. [Google Scholar] [CrossRef]
- Wajnberg, A.; Amanat, F.; Firpo, A.; Altman, D.R.; Bailey, M.J.; Mansour, M.; McMahon, M.; Meade, P.; Mendu, D.R.; Muellers, K.; et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 2020, 370, 1227–1230. [Google Scholar] [CrossRef]
- McLean, G.R.; Zhang, Y.; Ndoyi, R.; Martin, A.; Winer, J. Rapid Quantification of SARS-CoV-2 Neutralising Antibodies Using Time-Resolved Fluorescence Immunoassay. Vaccines 2022, 10, 2149. [Google Scholar] [CrossRef]
- Graninger, M.; Jani, C.M.; Reuberger, E.; Prüger, K.; Gaspar, P.; Springer, D.N.; Borsodi, C.; Weidner, L.; Rabady, S.; Puchhammer-Stöckl, E.; et al. Comprehensive Comparison of Seven SARS-CoV-2-Specific Surrogate Virus Neutralization and Anti-Spike IgG Antibody Assays Using a Live-Virus Neutralization Assay as a Reference. Microbiol. Spectr. 2023, 11, e0231422. [Google Scholar] [CrossRef]
- Gattinger, P.; Niespodziana, K.; Stiasny, K.; Sahanic, S.; Tulaeva, I.; Borochova, K.; Dorofeeva, Y.; Schlederer, T.; Sonnweber, T.; Hofer, G.; et al. Neutralization of SARS-CoV-2 requires antibodies against conformational receptor-binding domain epitopes. Allergy 2022, 77, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Kratzer, B.; Trapin, D.; Ettel, P.; Körmöczi, U.; Rottal, A.; Tuppy, F.; Feichter, M.; Gattinger, P.; Borochova, K.; Dorofeeva, Y.; et al. Immunological imprint of COVID-19 on human peripheral blood leukocyte populations. Allergy 2021, 76, 751–765. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Stevenson, B.J.; Hagenbüchle, O.; Wellauer, P.K. Sequence organisation and transcriptional regulation of the mouse elastase II and trypsln genes. Nucleic Acids Res. 1986, 14, 8307–8330. [Google Scholar] [CrossRef]
- Hopp, T.P.; Prickett, K.S.; Price, V.L.; Libby, R.T.; March, C.J.; Cerretti, D.P.; Urdal, D.L.; Conlon, P.J. A Short Polypeptide Marker Sequence Useful for Recombinant Protein Identification and Purification. Nat. Bio/Technology 1988, 6, 1204–1210. [Google Scholar] [CrossRef]
- Chen, J. Serial analysis of binding elements for human transcription factors. Nat. Protoc. 2006, 1, 1481–1493. [Google Scholar] [CrossRef] [PubMed]
- Derdak, S.V.; Kueng, H.J.; Leb, V.M.; Neunkirchner, A.; Schmetterer, K.G.; Bielek, E.; Majdic, O.; Knapp, W.; Seed, B.; Pickl, W.F. Direct stimulation of T lymphocytes by immunosomes: Virus-like particles decorated with T cell receptor/CD3 ligands plus costimulatory molecules. Proc. Natl. Acad. Sci. USA 2006, 103, 13144–13149. [Google Scholar] [CrossRef]
- Naldini, L.; Blömer, U.; Gallay, P.; Ory, D.; Mulligan, R.; Gage, F.H.; Verma, I.M.; Trono, D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 1996, 272, 263–267. [Google Scholar] [CrossRef]
- Hobbs, S.; Jitrapakdeeb, S.; Wallace, J.C. Development of a bicistronic vector driven by the human polypeptide chain elongation factor 1α promoter for creation of stable mammalian cell lines that express very high levels of recombinant proteins. Biochem. Biophys. Res. Commun. 1998, 252, 368–372. [Google Scholar] [CrossRef] [PubMed]
- Witt, H.; Luck, W.; Becker, M. A signal peptide cleavage site mutation in the cationic trypsinogen gene is strongly associated with chronic pancreatitis. Gastroenterology 1999, 117, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.; Schallhorn, A.; Wurm, F.M. Transfecting Mammalian Cells: Optimization of Critical Parameters Affecting Calcium-Phosphate Precipitate Formation. Nucleic Acids Res. 1996, 24, 596–601. [Google Scholar] [CrossRef]
- Ory, D.S.; Neugeboren, B.A.; Mulligan, R.C. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA 1996, 93, 11400–11406. [Google Scholar] [CrossRef]
- Hunter, M.R.; Grimsey, N.L.; Glass, M. Sulfation of the FLAG epitope is affected by co-expression of G protein-coupled receptors in a mammalian cell model. Sci. Rep. 2016, 6, 27316. [Google Scholar] [CrossRef]
- Tan, J.L.; Ueda, N.; Mercer, A.A.; Fleming, S.B. Investigation of orf virus structure and morphogenesis using recombinants expressing FLAG-tagged envelope structural proteins: Evidence for wrapped virus particles and egress from infected cells. J. Gen. Virol. 2009, 90, 614–625. [Google Scholar] [CrossRef]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef]
- Gattinger, P.; Kratzer, B.; Tulaeva, I.; Niespodziana, K.; Ohradanova-Repic, A.; Gebetsberger, L.; Borochova, K.; Garner-Spitzer, E.; Trapin, D.; Hofer, G.; et al. Vaccine based on folded receptor binding domain-PreS fusion protein with potential to induce sterilizing immunity to SARS-CoV-2 variants. Allergy 2022, 77, 2431–2445. [Google Scholar] [CrossRef]
- Gattinger, P.; Tulaeva, I.; Borochova, K.; Kratzer, B.; Trapin, D.; Kropfmüller, A.; Pickl, W.F.; Valenta, R. Omicron: A SARS-CoV-2 variant of real concern. Allergy 2022, 77, 1616–1620. [Google Scholar] [CrossRef] [PubMed]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119. [Google Scholar] [CrossRef]
- Robbiani, D.F.; Gaebler, C.; Muecksch, F.; Lorenzi, J.C.C.; Wang, Z.; Cho, A.; Agudelo, M.; Barnes, C.O.; Gazumyan, A.; Finkin, S.; et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020, 584, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Suntronwong, N.; Assawakosri, S.; Kanokudom, S.; Yorsaeng, R.; Auphimai, C.; Thongmee, T.; Vichaiwattana, P.; Duangchinda, T.; Chantima, W.; Pakchotanon, P.; et al. Strong Correlations between the Binding Antibodies against Wild-Type and Neutralizing Antibodies against Omicron BA.1 and BA.2 Variants of SARS-CoV-2 in Individuals Following Booster (Third-Dose) Vaccination. Diagnostics 2022, 12, 1781. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.; Guzek, A.; Ruff, J.; Jasinska, J.; Scheikl, U.; Zwazl, I.; Kundi, M.; Stockinger, H.; Farcet, M.R.; Kreil, T.R.; et al. Neutralising SARS-CoV-2 RBD-specific antibodies persist for at least six months independently of symptoms in adults. Commun. Med. 2021, 1, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kontopoulou, K.; Nakas, C.T.; Papazisis, G. Significant Increase in Antibody Titers after the 3rd Booster Dose of the Pfizer–BioNTech mRNA COVID-19 Vaccine in Healthcare Workers in Greece. Vaccines 2022, 10, 876. [Google Scholar] [CrossRef]
- Tran, T.T.; Vaage, E.B.; Mehta, A.; Chopra, A.; Tietze, L.; Kolderup, A.; Anthi, A.; König, M.; Nygaard, G.; Lind, A.; et al. Titers of antibodies against ancestral SARS-CoV-2 correlate with levels of neutralizing antibodies to multiple variants. NPJ Vaccines 2022, 7, 174. [Google Scholar] [CrossRef]
- Immordino, P.; Pisciotta, V.; Amodio, E.; Bonura, C.; Bonura, F.; Cacioppo, F.; Calamusa, G.; Capra, G.; Casuccio, A.; De Grazia, S.; et al. An Analysis of the Neutralizing Antibodies against the Main SARS-CoV-2 Variants in Healthcare Workers (HCWs) Vaccinated against or Infected by SARS-CoV-2. Vaccines 2023, 11, 1702. [Google Scholar] [CrossRef]
- Muranishi, K.; Kinoshita, M.; Inoue, K.; Ohara, J.; Mihara, T.; Sudo, K.; Ishii, K.J.; Sawa, T.; Ishikura, H. Antibody Response Following the Intranasal Administration of SARS-CoV-2 Spike Protein-CpG Oligonucleotide Vaccine. Vaccines 2023, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Havervall, S.; Marking, U.; Svensson, J.; Greilert-Norin, N.; Bacchus, P.; Nilsson, P.; Hober, S.; Gordon, M.; Blom, K.; Klingström, J.; et al. Anti-Spike Mucosal IgA Protection against SARS-CoV-2 Omicron Infection. New Engl. J. Med. 2022, 387, 1333–1336. [Google Scholar] [CrossRef]
- Lam, J.-Y.; Wong, W.-M.; Yuen, C.-K.; Ng, Y.-Y.; San, C.-H.; Yuen, K.-Y.; Kok, K.-H. An RNA-Scaffold Protein Subunit Vaccine for Nasal Immunization. Vaccines 2023, 11, 1550. [Google Scholar] [CrossRef]
- Shrwani, K.; Sharma, R.; Krishnan, M.; Jones, T.; Mayora-Neto, M.; Cantoni, D.; Temperton, N.J.; Dobson, S.L.; Subramaniam, K.; McNamara, P.S.; et al. Detection of Serum Cross-Reactive Antibodies and Memory Response to SARS-CoV-2 in Prepandemic and Post–COVID-19 Convalescent Samples. J. Infect. Dis. 2021, 224, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.N.; Choo, E.M.; Nakamura, Y.; Suzuki, R.; Shiina, T.; Shin-I, T.; Fukuta, M.; Nguyen, C.T.; Nguyen, T.T.T.; Nguyen, L.K.H.; et al. Pre-existing cross-reactive neutralizing activity against SARS-CoV-2 and seasonal coronaviruses prior to the COVID-19 pandemic (2014–2019) with limited immunity against recent emerging SARS-CoV-2 variants, Vietnam. Int. J. Infect. Dis. 2024, 139, 109–117. [Google Scholar] [CrossRef]
- Gumanova, N.G.; Gorshkov, A.U.; Bogdanova, N.L.; Korolev, A.I.; Drapkina, O.M. Detection of Anti-SARS-CoV-2-S1 RBD-Specific Antibodies Prior to and during the Pandemic in 2011–2021 and COVID-19 Observational Study in 2019–2021. Vaccines 2022, 10, 581. [Google Scholar] [CrossRef]
- Sim, K.-Y.; Ko, G.-H.; Bae, S.-E.; Choi, K.Y.; Lee, J.S.; Kim, B.C.; Lee, K.H.; Song, M.-R.; Park, S.-G. Two Opposing Roles of SARS-CoV-2 RBD-Reactive Antibodies in Pre-Pandemic Plasma Samples From Elderly People in ACE2-Mediated Pseudovirus Infection. Front. Immunol. 2021, 12, 813240. [Google Scholar] [CrossRef] [PubMed]
- González, F.E.; Gleisner, A.; Falcón-Beas, F.; Osorio, F.; López, M.N.; Salazar-Onfray, F. Tumor cell lysates as immunogenic sources for cancer vaccine design. Hum. Vaccines Immunother. 2014, 10, 3261–3269. [Google Scholar] [CrossRef]
- Meng, Z.; Zhang, Y.; Zhou, X.; Ji, J.; Liu, Z. Nanovaccines with cell-derived components for cancer immunotherapy. Adv. Drug Deliv. Rev. 2022, 182, 114107. [Google Scholar] [CrossRef]
- Sarkar, N.H.; Moore, D.H. Immunization of mice against murine mammary tumor virus infection and mammary tumor development. Cancer Res. 1978, 38, 1468–1472. [Google Scholar]
- Cacciottolo, M.; Li, Y.; Nice, J.B.; LeClaire, M.J.; Twaddle, R.; Mora, C.L.; Adachi, S.Y.; Young, M.; Angeles, J.; Elliott, K.; et al. Nanograms of SARS-CoV-2 spike protein delivered by exosomes induce potent neutralization of both delta and omicron variants. PLoS ONE 2023, 18, e0290046. [Google Scholar] [CrossRef]
- Serretiello, E.; Ballini, A.; Smimmo, A.; Acunzo, M.; Raimo, M.; Cantore, S.; Di Domenico, M. Extracellular Vesicles as a Translational Approach for the Treatment of COVID-19 Disease: An Updated Overview. Viruses 2023, 15, 1976. [Google Scholar] [CrossRef] [PubMed]
- Dogan, M.; Kozhaya, L.; Placek, L.; Gunter, C.; Yigit, M.; Hardy, R.; Plassmeyer, M.; Coatney, P.; Lillard, K.; Bukhari, Z.; et al. SARS-CoV-2 specific antibody and neutralization assays reveal the wide range of the humoral immune response to virus. Commun. Biol. 2021, 4, 129. [Google Scholar] [CrossRef] [PubMed]
- Egia-Mendikute, L.; Bosch, A.; Prieto-Fernández, E.; Lee, S.Y.; Jiménez-Lasheras, B.; del Río, A.G.; Antoñana-Vildosola, A.; Bruzzone, C.; Bizkarguenaga, M.; Embade, N.; et al. Sensitive detection of SARS-CoV-2 seroconversion by flow cytometry reveals the presence of nucleoprotein-reactive antibodies in unexposed individuals. Commun. Biol. 2021, 4, 486. [Google Scholar] [CrossRef]
- Anand, S.P.; Prévost, J.; Richard, J.; Perreault, J.; Tremblay, T.; Drouin, M.; Fournier, M.J.; Lewin, A.; Bazin, R.; Finzi, A. High-throughput detection of antibodies targeting the SARS-CoV-2 Spike in longitudinal convalescent plasma samples. Transfusion 2021, 61, 1377–1382. [Google Scholar] [CrossRef]
- Grzelak, L.; Temmam, S.; Planchais, C.; Demeret, C.; Tondeur, L.; Huon, C.; Guivel-Benhassine, F.; Staropoli, I.; Chazal, M.; Dufloo, J.; et al. A comparison of four serological assays for detecting anti–SARS-CoV-2 antibodies in human serum samples from different populations. Sci. Transl. Med. 2020, 12, eabc3103. [Google Scholar] [CrossRef] [PubMed]
- Horndler, L.; Delgado, P.; Abia, D.; Balabanov, I.; Martínez-Fleta, P.; Cornish, G.; A Llamas, M.; Serrano-Villar, S.; Sánchez-Madrid, F.; Fresno, M.; et al. Flow cytometry multiplexed method for the detection of neutralizing human antibodies to the native SARS-CoV-2 spike protein. EMBO Mol. Med. 2021, 13, e13549. [Google Scholar] [CrossRef]
- Piñero, P.; De La Calle, F.M.M.; Horndler, L.; Alarcón, B.; Barrientos, M.U.; Sarmiento, H.; Tarín, F. Flow cytometry detection of sustained humoral immune response (IgG + IgA) against native spike glycoprotein in asymptomatic/mild SARS-CoV-2 infection. Sci. Rep. 2021, 11, 1–5. [Google Scholar] [CrossRef]
- Lapuente, D.; Maier, C.; Irrgang, P.; Hübner, J.; Peter, A.S.; Hoffmann, M.; Ensser, A.; Ziegler, K.; Winkler, T.H.; Birkholz, T.; et al. Rapid response flow cytometric assay for the detection of antibody responses to SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 751–759. [Google Scholar] [CrossRef]
- Simard, C.; Richard, J.; Bazin, R.; Finzi, A.; Trépanier, P. Standardization of a flow cytometry SARS-CoV-2 serologic test. Cytotechnology 2022, 74, 99–103. [Google Scholar] [CrossRef]
- Bodary, S.C.; McLean, J.W. The integrin beta 1 subunit associates with the vitronectin receptor alpha v subunit to form a novel vitronectin receptor in a human embryonic kidney cell line. J. Biol. Chem. 1990, 265, 5938–5941. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Kawamura, K.; Li, Q.; Okamoto, S.; Tada, Y.; Tatsumi, K.; Shimada, H.; Hiroshima, K.; Yamaguchi, N.; Tagawa, M. Mesenchymal stem cells are efficiently transduced with adenoviruses bearing type 35-derived fibers and the transduced cells with the IL-28A gene produces cytotoxicity to lung carcinoma cells co-cultured. BMC Cancer 2014, 14, 713. [Google Scholar] [CrossRef] [PubMed]


| Age | Time to Last Event # (days) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | Group | Type of SARS-CoV-2 Exposure | Individuals (n) | Female (%) | Range | Mean | SD * | Range | Median |
| 1 | A | Non-exposed | 25 | 16 (64) | 19–68 | 51 | 13 | n.a. | |
| 2 | B1 | Wuhan-Hu-1 Convalescent only (Visit 1) | 25 | 10 (40) | 22–76 | 49 | 15 | 37–246 | 61 |
| 3 | B2 | Wuhan-Hu-1 Convalescent only (Visit 2) | 25 | 9 (36) | 18–78 | 49 | 15 | 248–306 | 265 |
| 4 | B3 | Wuhan-Hu-1 Convalescent + 1X Vaccinated | 11 | 5 (45) | 22–77 | 49 | 19 | 20–89 | 35 |
| 5 | B4 | Wuhan-Hu-1 Convalescent + 2X Vaccinated | 4 | 2 (50) | 39–68 | 49 | 13 | 21–103 | 43 |
| 6 | C1 | AstraZeneca 1X Vaccinated | 17 | 13 (76) | 22–61 | 40 | 14 | 21–29 | 22 |
| 7 | C2 | AstraZeneca 2X Vaccinated | 21 | 14 (67) | 22–59 | 40 | 13 | 96–188 | 125 |
| 8 | C3 | AstraZeneca 2X + 1X mRNA Vaccinated | 22 | 16 (73) | 24–69 | 37 | 14 | 23–134 | 99 |
| 9 | D1 | 2X mRNA Vaccinated | 13 | 5 (38) | 21–85 | 49 | 18 | 21–213 | 49 |
| 10 | D2 | 3X mRNA Vaccinated | 27 | 15 (56) | 21–68 | 39 | 14 | 20–122 | 59 |
| 11 | E | 3X Vaccinated + Omicron breakthrough | 14 | 7 (50) | 23–62 | 38 | 13 | 22–74 | 27 |
| Protein | Amino Acid Sequence * |
|---|---|
| Pre-pro-trypsin (PPT) leader sequence † | MNPLLILTFVAAALA |
| FLAG-tag | DYKDHDGDYKDHDIDYKDDDDK |
| Linker sequence | GGGGS |
| RBD (Wuhan-Hu-1) | PNITNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKL NDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDS KVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTN GVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNK KFLPFQQFGRDIADTTDAVRDPQTLE |
| Minimal GPI-anchor acceptor sequence of human CD16b # | SLAVSTISSFSPPGYQVSFCLVMVLLFAVDTGLYFSVKTNI |
| Age | Time to Last Event # (days) | |||||||
|---|---|---|---|---|---|---|---|---|
| Group | Type of SARS-CoV-2 Exposure | Individuals (n) | Female (%) | Range | Mean | SD ** | Range | Median |
| D2 * | 3X mRNA Vaccinated | 6 | 3 (50) | 23–45 | 34 | 7 | 20–50 | 29 |
| E * | 3X Vaccinated + Omicron breakthrough | 6 | 3 (50) | 23–43 | 32 | 7 | 23–47 | 26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sehgal, A.N.A.; Safran, J.; Kratzer, B.; Gattinger, P.; Stieger, R.B.; Musiejovsky, L.; Trapin, D.; Ettel, P.; Körmöczi, U.; Rottal, A.; et al. Flow Cytometry-Based Measurement of Antibodies Specific for Cell Surface-Expressed Folded SARS-CoV-2 Receptor-Binding Domains. Vaccines 2024, 12, 377. https://doi.org/10.3390/vaccines12040377
Sehgal ANA, Safran J, Kratzer B, Gattinger P, Stieger RB, Musiejovsky L, Trapin D, Ettel P, Körmöczi U, Rottal A, et al. Flow Cytometry-Based Measurement of Antibodies Specific for Cell Surface-Expressed Folded SARS-CoV-2 Receptor-Binding Domains. Vaccines. 2024; 12(4):377. https://doi.org/10.3390/vaccines12040377
Chicago/Turabian StyleSehgal, Al Nasar Ahmed, Jera Safran, Bernhard Kratzer, Pia Gattinger, Robert B. Stieger, Laszlo Musiejovsky, Doris Trapin, Paul Ettel, Ulrike Körmöczi, Arno Rottal, and et al. 2024. "Flow Cytometry-Based Measurement of Antibodies Specific for Cell Surface-Expressed Folded SARS-CoV-2 Receptor-Binding Domains" Vaccines 12, no. 4: 377. https://doi.org/10.3390/vaccines12040377







