Minimal Determinants for Lifelong Antiviral Antibody Responses in Mice from a Single Exposure to Virus-like Immunogens at Low Doses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of SVLS
2.2. Mice Immunizations
2.3. Enzyme-Linked Immunosorbent Assay (ELISA)
2.4. Preparation of HIV-1 Virions Pseudotyped with SARS-CoV-2 Envelope
2.5. Virion Neutralization Assay
2.6. BioLayer Interferometry Measurements
2.7. Measurement of Ag-Specific IgG Concentration in Animal Sera Using Magnetic Beads
2.8. Statistical Analysis
3. Results
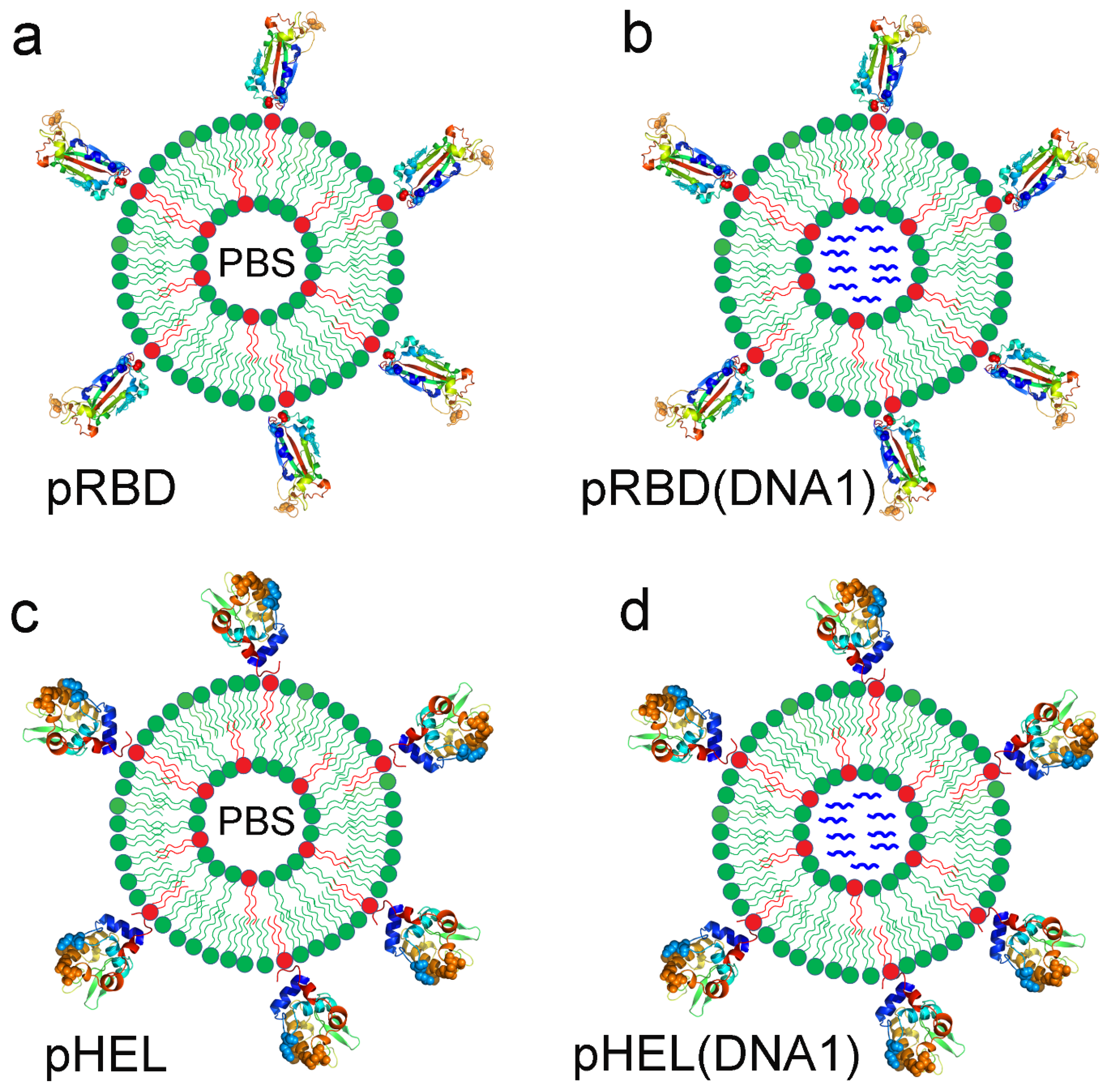
3.1. Four Sets of SVLS Chosen for Current Immunization Studies
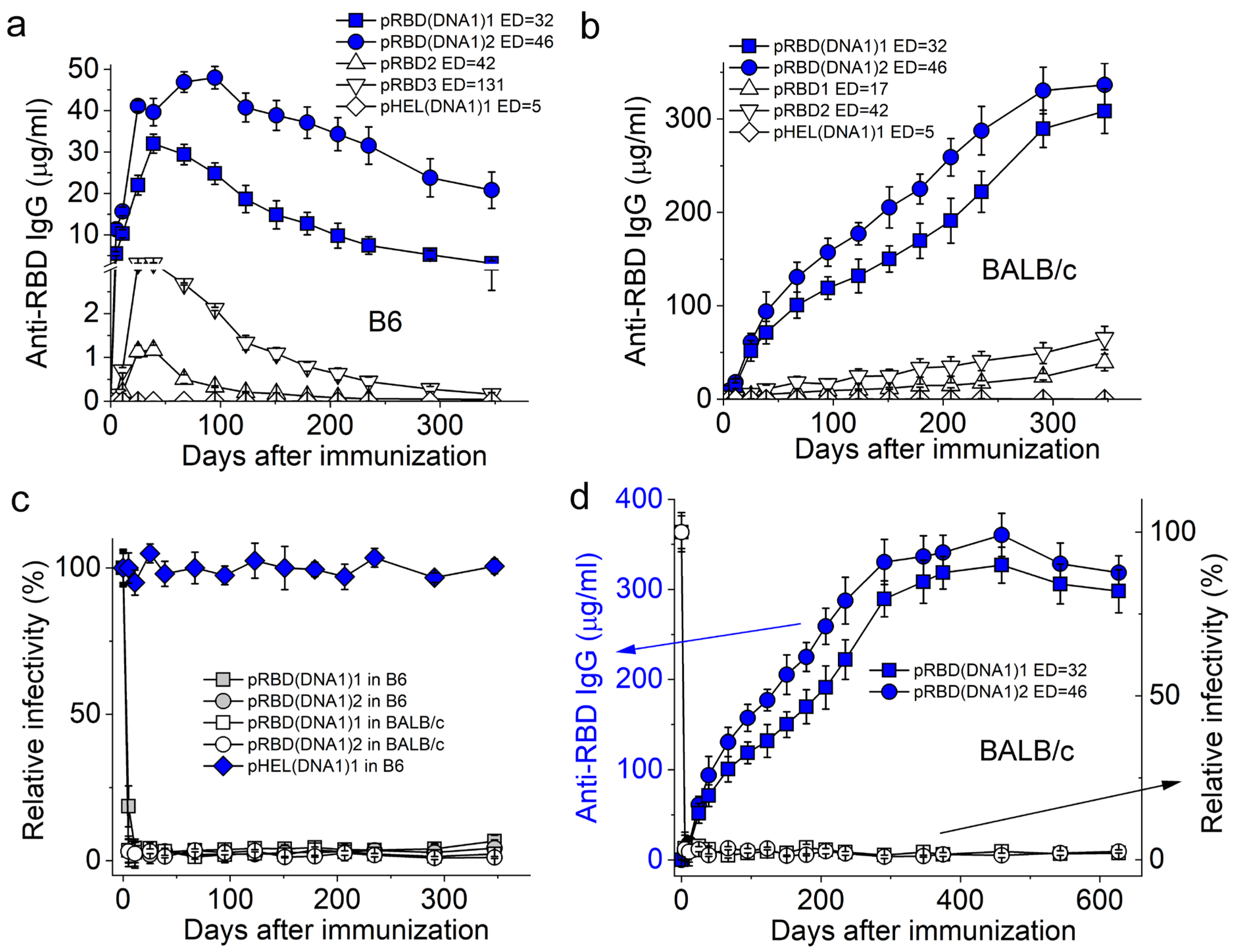
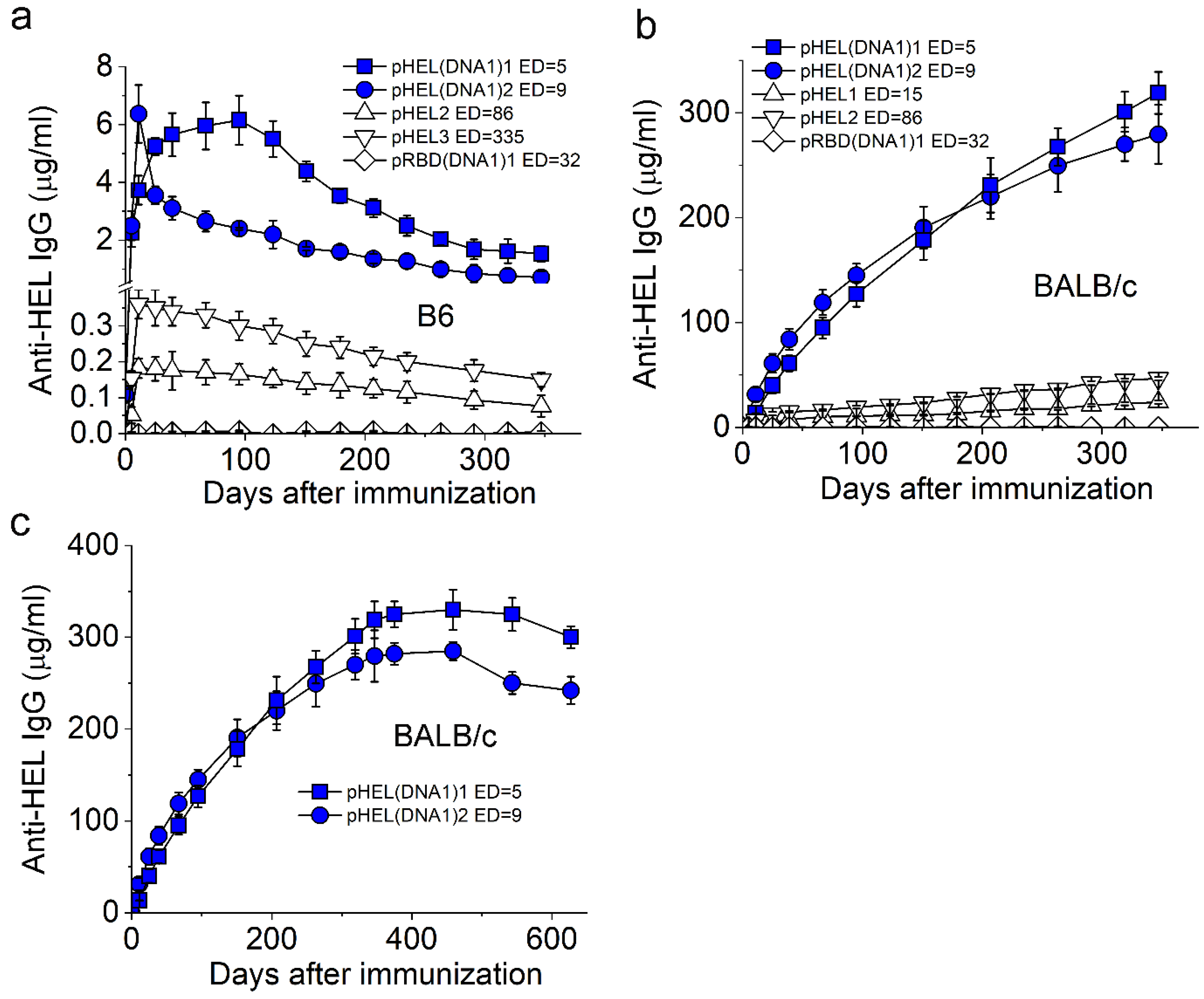
3.2. SVLS Induce Durable IgG Responses
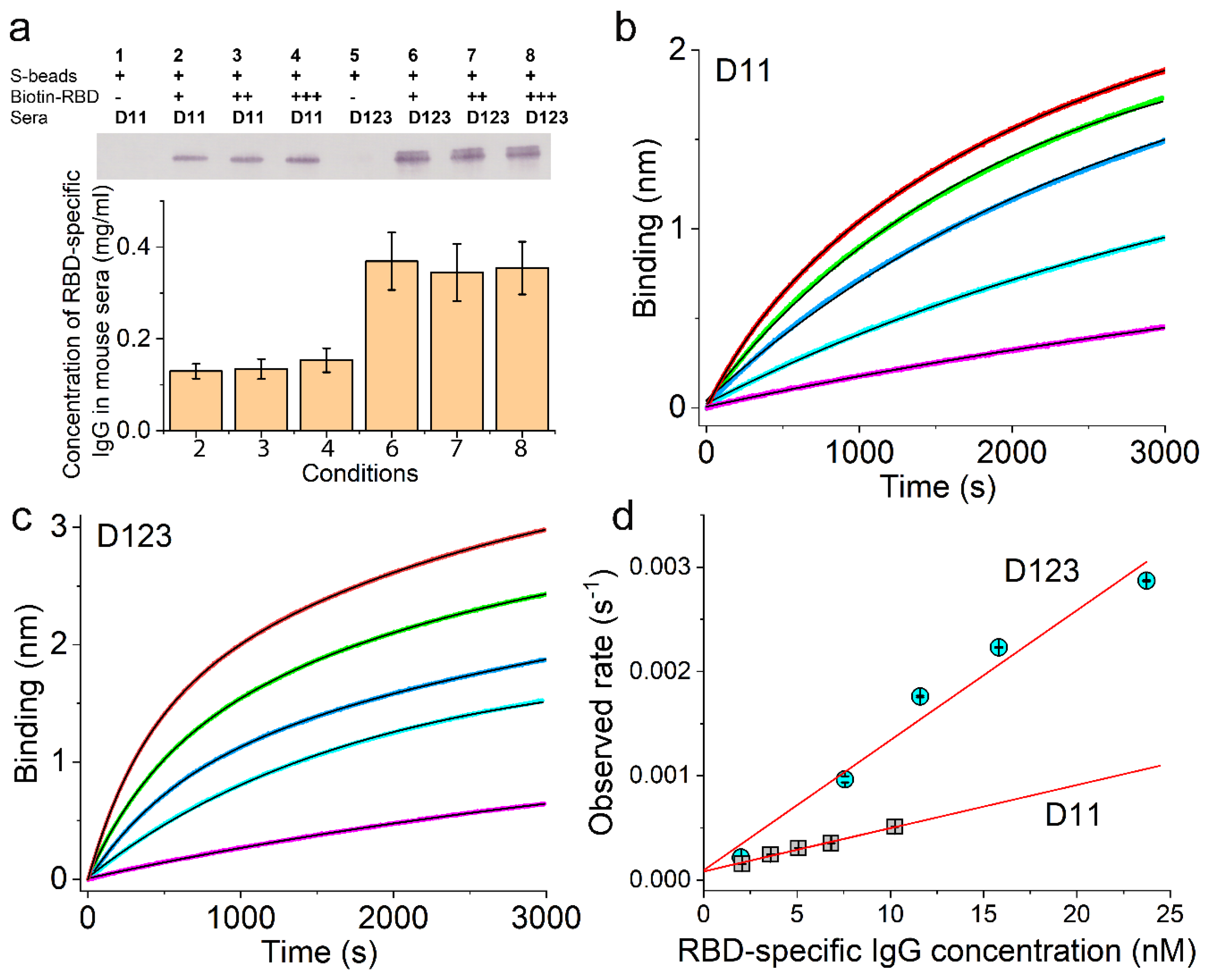
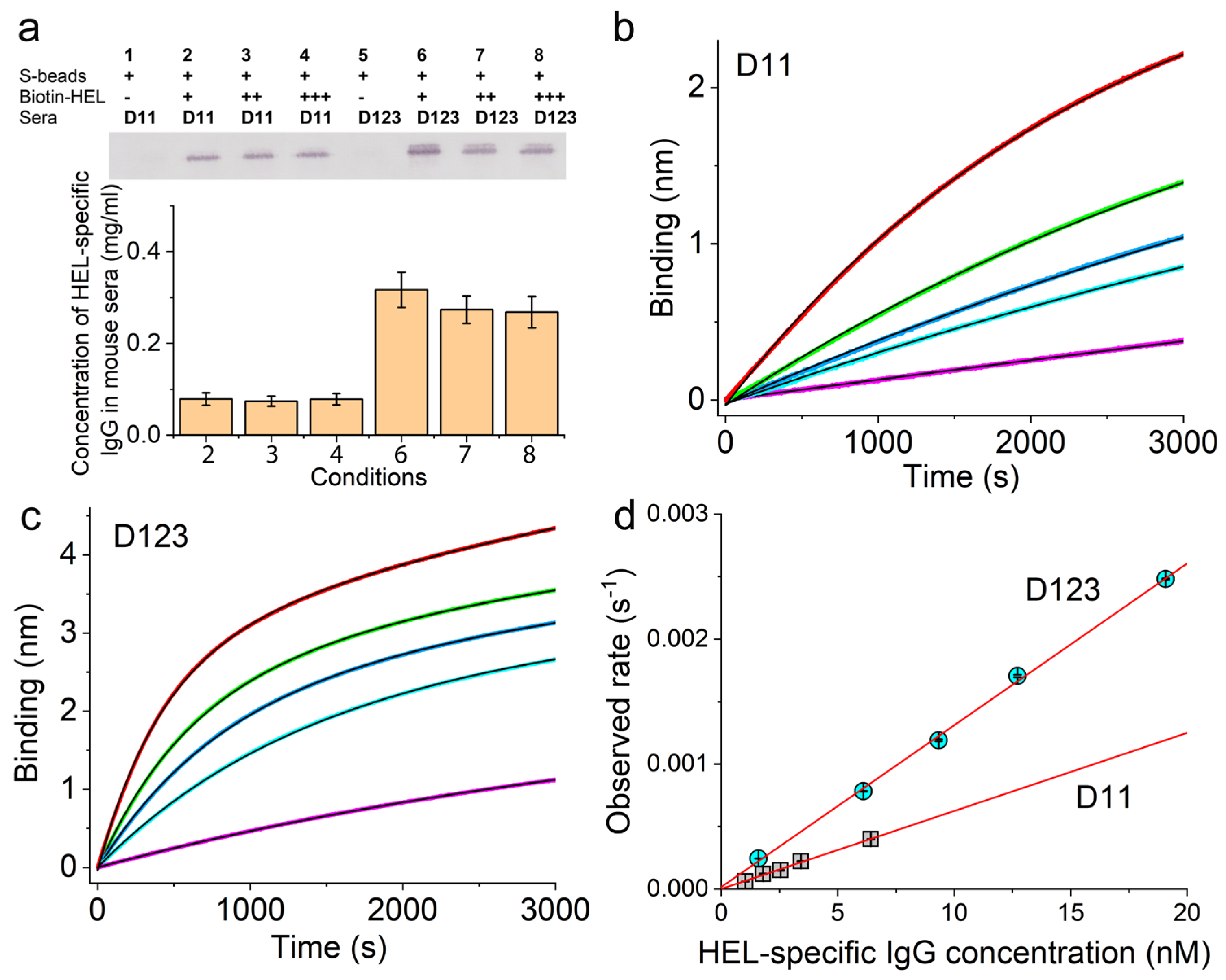
3.3. Validation of the Sensor Coating Density in BioLayer Interferometry
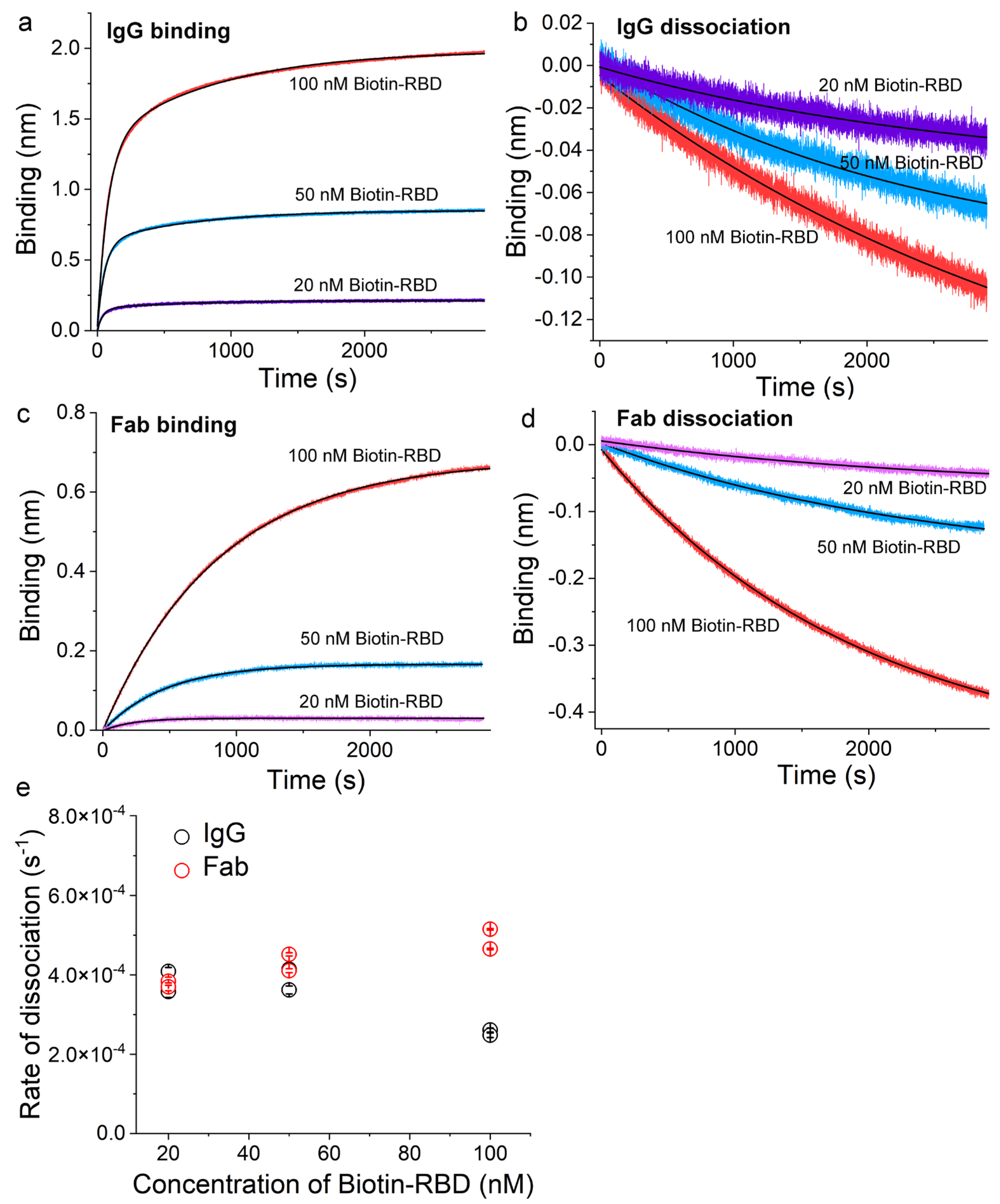
3.4. Ag-Specific Binding Kinetics Measured Using BioLayer Interferometry
3.5. SVLS Induce IgG Affinity Maturation in BALB/c Mice
4. Discussion
- (1)
- The minimal molecular determinants for a durable Ab response
- (2)
- Virus-like immunogens elicit Ag-specific IgG with very slow off-rates.
- (3)
- SVLS as a new platform for dissection of antiviral response and potential vaccines.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Palin, A.C.; Alter, G.; Crotty, S.; Ellebedy, A.H.; Lane, M.C.; Lee, F.E.; Locci, M.; Malaspina, A.; Mallia, C.; McElrath, M.J.; et al. The persistence of memory: Defining, engineering, and measuring vaccine durability. Nat. Immunol. 2022, 23, 1665–1668. [Google Scholar] [CrossRef]
- Amanna, I.J.; Carlson, N.E.; Slifka, M.K. Duration of humoral immunity to common viral and vaccine antigens. N. Engl. J. Med. 2007, 357, 1903–1915. [Google Scholar] [CrossRef] [PubMed]
- Widge, A.T.; Rouphael, N.G.; Jackson, L.A.; Anderson, E.J.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination. N. Engl. J. Med. 2021, 384, 80–82. [Google Scholar] [CrossRef]
- Anderson, M.; Stec, M.; Gosha, A.; Mohammad, T.; Boler, M.; Tojo Suarez, R.; Behun, D.; Landay, A.; Cloherty, G.; Moy, J. Longitudinal Severe Acute Respiratory Syndrome Coronavirus 2 Vaccine Antibody Responses and Identification of Vaccine Breakthrough Infections Among Healthcare Workers Using Nucleocapsid Immunoglobulin G. J. Infect. Dis. 2022, 226, 1934–1942. [Google Scholar] [CrossRef]
- Korosec, C.S.; Farhang-Sardroodi, S.; Dick, D.W.; Gholami, S.; Ghaemi, M.S.; Moyles, I.R.; Craig, M.; Ooi, H.K.; Heffernan, J.M. Long-term durability of immune responses to the BNT162b2 and mRNA-1273 vaccines based on dosage, age and sex. Sci. Rep. 2022, 12, 21232. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.L.; Mantus, G.; Manning, K.E.; Ellis, M.; Patel, M.; Ciric, C.R.; Lu, A.; Turner, J.S.; O’Halloran, J.A.; Presti, R.M.; et al. Loss of Pfizer (BNT162b2) Vaccine-Induced Antibody Responses against the SARS-CoV-2 Omicron Variant in Adolescents and Adults. J. Virol. 2022, 96, e0058222. [Google Scholar] [CrossRef] [PubMed]
- Slifka, M.K.; Ahmed, R. Long-term humoral immunity against viruses: Revisiting the issue of plasma cell longevity. Trends Microbiol. 1996, 4, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Slifka, M.K.; Amanna, I.J. Role of Multivalency and Antigenic Threshold in Generating Protective Antibody Responses. Front. Immunol. 2019, 10, 956. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, D. Instructing durable humoral immunity for COVID-19 and other vaccinable diseases. Immunity 2022, 55, 945–964. [Google Scholar] [CrossRef]
- Chackerian, B.; Lowy, D.R.; Schiller, J.T. Induction of autoantibodies to mouse CCR5 with recombinant papillomavirus particles. Proc. Natl. Acad. Sci. USA 1999, 96, 2373–2378. [Google Scholar] [CrossRef] [PubMed]
- Dintzis, H.M.; Dintzis, R.Z.; Vogelstein, B. Molecular determinants of immunogenicity: The immunon model of immune response. Proc. Natl. Acad. Sci. USA 1976, 73, 3671–3675. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Abbott, R.K.; Freeman, B.L.; Haupt, S.; Groschel, B.; Silva, M.; Menis, S.; Irvine, D.J.; Schief, W.R.; Crotty, S. Multifaceted Effects of Antigen Valency on B Cell Response Composition and Differentiation In Vivo. Immunity 2020, 53, 548–563.e8. [Google Scholar] [CrossRef] [PubMed]
- Ols, S.; Lenart, K.; Arcoverde Cerveira, R.; Miranda, M.C.; Brunette, N.; Kochmann, J.; Corcoran, M.; Skotheim, R.; Philomin, A.; Cagigi, A.; et al. Multivalent antigen display on nanoparticle immunogens increases B cell clonotype diversity and neutralization breadth to pneumoviruses. Immunity 2023, 56, 2425–2441.e14. [Google Scholar] [CrossRef] [PubMed]
- Brooks, J.F.; Riggs, J.; Mueller, J.L.; Mathenge, R.; Wholey, W.Y.; Meyer, A.R.; Yoda, S.T.; Vykunta, V.S.; Nielsen, H.V.; Cheng, W.; et al. Molecular basis for potent B cell responses to antigen displayed on particles of viral size. Nat. Immunol. 2023, 24, 1762–1777. [Google Scholar] [CrossRef]
- Sefik, E.; Qu, R.; Junqueira, C.; Kaffe, E.; Mirza, H.; Zhao, J.; Brewer, J.R.; Han, A.; Steach, H.R.; Israelow, B.; et al. Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature 2022, 606, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Merad, M.; Blish, C.A.; Sallusto, F.; Iwasaki, A. The immunology and immunopathology of COVID-19. Science 2022, 375, 1122–1127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bastard, P.; Effort, C.H.G.; Cobat, A.; Casanova, J.L. Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature 2022, 603, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Wholey, W.Y.; Mueller, J.L.; Tan, C.; Brooks, J.F.; Zikherman, J.; Cheng, W. Synthetic Liposomal Mimics of Biological Viruses for the Study of Immune Responses to Infection and Vaccination. Bioconjug. Chem. 2020, 31, 685–697. [Google Scholar] [CrossRef]
- Wholey, W.Y.; Yoda, S.T.; Cheng, W. Site-Specific and Stable Conjugation of the SARS-CoV-2 Receptor-Binding Domain to Liposomes in the Absence of Any Other Adjuvants Elicits Potent Neutralizing Antibodies in BALB/c Mice. Bioconjug. Chem. 2021, 32, 2497–2506. [Google Scholar] [CrossRef]
- Chen, Z.; Moon, J.J.; Cheng, W. Quantitation and Stability of Protein Conjugation on Liposomes for Controlled Density of Surface Epitopes. Bioconjug. Chem. 2018, 29, 1251–1260. [Google Scholar] [CrossRef]
- Wholey, W.Y.; Meyer, A.R.; Yoda, S.T.; Mueller, J.L.; Mathenge, R.; Chackerian, B.; Zikherman, J.; Cheng, W. An integrated signaling threshold initiates IgG response towards virus-like immunogens. bioRxiv 2024. [Google Scholar] [CrossRef]
- Chen, Z.; Wholey, W.Y.; Hassani Najafabadi, A.; Moon, J.J.; Grigorova, I.; Chackerian, B.; Cheng, W. Self-Antigens Displayed on Liposomal Nanoparticles above a Threshold of Epitope Density Elicit Class-Switched Autoreactive Antibodies Independent of T Cell Help. J. Immunol. 2020, 204, 335–347. [Google Scholar] [CrossRef]
- Knipe, D.M.; Howley, P.M. Fields Virology; Wolters Kluwer/Lippincott Williams & Wilkins Health: Philadelphia, PA, USA, 2013. [Google Scholar]
- Klein, J.S.; Bjorkman, P.J. Few and far between: How HIV may be evading antibody avidity. PLoS Pathog. 2010, 6, e1000908. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W. The Density Code for the Development of a Vaccine? J. Pharm. Sci. 2016, 105, 3223–3232. [Google Scholar] [CrossRef] [PubMed]
- Mombaerts, P.; Clarke, A.R.; Rudnicki, M.A.; Iacomini, J.; Itohara, S.; Lafaille, J.J.; Wang, L.; Ichikawa, Y.; Jaenisch, R.; Hooper, M.L.; et al. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature 1992, 360, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183, 739–751.e8. [Google Scholar] [CrossRef] [PubMed]
- Connor, R.I.; Chen, B.K.; Choe, S.; Landau, N.R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 1995, 206, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Sercarz, E.E.; Lehmann, P.V.; Ametani, A.; Benichou, G.; Miller, A.; Moudgil, K. Dominance and crypticity of T cell antigenic determinants. Annu. Rev. Immunol. 1993, 11, 729–766. [Google Scholar] [CrossRef]
- Watson, D.S.; Endsley, A.N.; Huang, L. Design considerations for liposomal vaccines: Influence of formulation parameters on antibody and cell-mediated immune responses to liposome associated antigens. Vaccine 2012, 30, 2256–2272. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Kondo, H.; Shiroishi, M.; Matsushima, M.; Tsumoto, K.; Kumagai, I. Crystal structure of anti-Hen egg white lysozyme antibody (HyHEL-10) Fv-antigen complex. Local structural changes in the protein antigen and water-mediated interactions of Fv-antigen and light chain-heavy chain interfaces. J. Biol. Chem. 1999, 274, 27623–27631. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.E., Jr.; Bertram, C.D. Lymphatic System Flows. Annu. Rev. Fluid Mech. 2018, 50, 459–482. [Google Scholar] [CrossRef] [PubMed]
- Junt, T.; Moseman, E.A.; Iannacone, M.; Massberg, S.; Lang, P.A.; Boes, M.; Fink, K.; Henrickson, S.E.; Shayakhmetov, D.M.; Di Paolo, N.C.; et al. Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature 2007, 450, 110–114. [Google Scholar] [CrossRef]
- Fehr, T.; Skrastina, D.; Pumpens, P.; Zinkernagel, R.M. T cell-independent type I antibody response against B cell epitopes expressed repetitively on recombinant virus particles. Proc. Natl. Acad. Sci. USA 1998, 95, 9477–9481. [Google Scholar] [CrossRef]
- Ochsenbein, A.F.; Pinschewer, D.D.; Odermatt, B.; Ciurea, A.; Hengartner, H.; Zinkernagel, R.M. Correlation of T cell independence of antibody responses with antigen dose reaching secondary lymphoid organs: Implications for splenectomized patients and vaccine design. J. Immunol. 2000, 164, 6296–6302. [Google Scholar] [CrossRef]
- Iwasaki, A.; Medzhitov, R. Regulation of adaptive immunity by the innate immune system. Science 2010, 327, 291–295. [Google Scholar] [CrossRef]
- Iwasaki, A.; Medzhitov, R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015, 16, 343–353. [Google Scholar] [CrossRef]
- Schmidt, F.; Weisblum, Y.; Muecksch, F.; Hoffmann, H.H.; Michailidis, E.; Lorenzi, J.C.C.; Mendoza, P.; Rutkowska, M.; Bednarski, E.; Gaebler, C.; et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020, 217, e20201181. [Google Scholar] [CrossRef]
- Goodrick, C.L. Life-span and the inheritance of longevity of inbred mice. J. Gerontol. 1975, 30, 257–263. [Google Scholar] [CrossRef]
- Bortnick, A.; Chernova, I.; Quinn, W.J., 3rd; Mugnier, M.; Cancro, M.P.; Allman, D. Long-lived bone marrow plasma cells are induced early in response to T cell-independent or T cell-dependent antigens. J. Immunol. 2012, 188, 5389–5396. [Google Scholar] [CrossRef] [PubMed]
- Taillardet, M.; Haffar, G.; Mondiere, P.; Asensio, M.J.; Gheit, H.; Burdin, N.; Defrance, T.; Genestier, L. The thymus-independent immunity conferred by a pneumococcal polysaccharide is mediated by long-lived plasma cells. Blood 2009, 114, 4432–4440. [Google Scholar] [CrossRef] [PubMed]
- Bates, J.T.; Keefer, C.J.; Utley, T.J.; Correia, B.E.; Schief, W.R.; Crowe, J.E., Jr. Reversion of somatic mutations of the respiratory syncytial virus-specific human monoclonal antibody Fab19 reveal a direct relationship between association rate and neutralizing potency. J. Immunol. 2013, 190, 3732–3739. [Google Scholar] [CrossRef]
- Foote, J.; Eisen, H.N. Kinetic and affinity limits on antibodies produced during immune responses. Proc. Natl. Acad. Sci. USA 1995, 92, 1254–1256. [Google Scholar] [CrossRef]
- Slifka, M.K.; Matloubian, M.; Ahmed, R. Bone marrow is a major site of long-term antibody production after acute viral infection. J. Virol. 1995, 69, 1895–1902. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.H.; Baumgarth, N. The Multifaceted B Cell Response to Influenza Virus. J. Immunol. 2019, 202, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.J.; Ding, Z.; Dowling, M.R.; Hill, D.L.; Webster, R.H.; McKenzie, C.; Pitt, C.; O’Donnell, K.; Mulder, J.; Brodie, E.; et al. Intrinsically determined turnover underlies broad heterogeneity in plasma-cell lifespan. Immunity 2023, 56, 1596–1612.e4. [Google Scholar] [CrossRef]
- Pishesha, N.; Harmand, T.J.; Ploegh, H.L. A guide to antigen processing and presentation. Nat. Rev. Immunol. 2022, 22, 751–764. [Google Scholar] [CrossRef]
- Kalinke, U.; Bucher, E.M.; Ernst, B.; Oxenius, A.; Roost, H.P.; Geley, S.; Kofler, R.; Zinkernagel, R.M.; Hengartner, H. The role of somatic mutation in the generation of the protective humoral immune response against vesicular stomatitis virus. Immunity 1996, 5, 639–652. [Google Scholar] [CrossRef]
- Lam, J.H.; Smith, F.L.; Baumgarth, N. B Cell Activation and Response Regulation During Viral Infections. Viral. Immunol. 2020, 33, 294–306. [Google Scholar] [CrossRef]
- Roost, H.P.; Bachmann, M.F.; Haag, A.; Kalinke, U.; Pliska, V.; Hengartner, H.; Zinkernagel, R.M. Early high-affinity neutralizing anti-viral IgG responses without further overall improvements of affinity. Proc. Natl. Acad. Sci. USA 1995, 92, 1257–1261. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.H.; Baumgarth, N. Toll-like receptor mediated inflammation directs B cells towards protective antiviral extrafollicular responses. Nat. Commun. 2023, 14, 3979. [Google Scholar] [CrossRef] [PubMed]
- Galimidi, R.P.; Klein, J.S.; Politzer, M.S.; Bai, S.; Seaman, M.S.; Nussenzweig, M.C.; West, A.P., Jr.; Bjorkman, P.J. Intra-spike crosslinking overcomes antibody evasion by HIV-1. Cell 2015, 160, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Meining, W.; Fischer, M.; Bacher, A.; Ladenstein, R. X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 A resolution: Determinants of thermostability revealed from structural comparisons. J. Mol. Biol. 2001, 306, 1099–1114. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, I.; Iwahori, K.; Kumagai, S. Ferritin in the field of nanodevices. Biochim. Biophys. Acta 2010, 1800, 846–857. [Google Scholar] [CrossRef]
- King, N.P.; Sheffler, W.; Sawaya, M.R.; Vollmar, B.S.; Sumida, J.P.; Andre, I.; Gonen, T.; Yeates, T.O.; Baker, D. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science 2012, 336, 1171–1174. [Google Scholar] [CrossRef] [PubMed]
- Bridges, C.B.; Harper, S.A.; Fukuda, K.; Uyeki, T.M.; Cox, N.J.; Singleton, J.A.; Advisory Committee on Immunization, P. Prevention and control of influenza. Recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm. Rep. 2003, 52, 1–34. [Google Scholar] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wholey, W.-Y.; Meyer, A.R.; Yoda, S.-T.; Chackerian, B.; Zikherman, J.; Cheng, W. Minimal Determinants for Lifelong Antiviral Antibody Responses in Mice from a Single Exposure to Virus-like Immunogens at Low Doses. Vaccines 2024, 12, 405. https://doi.org/10.3390/vaccines12040405
Wholey W-Y, Meyer AR, Yoda S-T, Chackerian B, Zikherman J, Cheng W. Minimal Determinants for Lifelong Antiviral Antibody Responses in Mice from a Single Exposure to Virus-like Immunogens at Low Doses. Vaccines. 2024; 12(4):405. https://doi.org/10.3390/vaccines12040405
Chicago/Turabian StyleWholey, Wei-Yun, Alexander R. Meyer, Sekou-Tidiane Yoda, Bryce Chackerian, Julie Zikherman, and Wei Cheng. 2024. "Minimal Determinants for Lifelong Antiviral Antibody Responses in Mice from a Single Exposure to Virus-like Immunogens at Low Doses" Vaccines 12, no. 4: 405. https://doi.org/10.3390/vaccines12040405






