Elucidating the Onset of Cross-Protective Immunity after Intranasal Vaccination with the Attenuated African Swine Fever Vaccine Candidate BA71ΔCD2
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Viruses
2.3. Animal Experiment
2.4. Quantitative PCR for the Detection of ASFV
2.5. Enzyme-Linked Immunosorbent Assay (ELISA)
2.6. Multiplex Luminex Assay
2.7. RNA-Seq Library Preparation and Sequencing
2.8. Bioinformatic Analysis
2.9. Statistical Analyses
3. Results
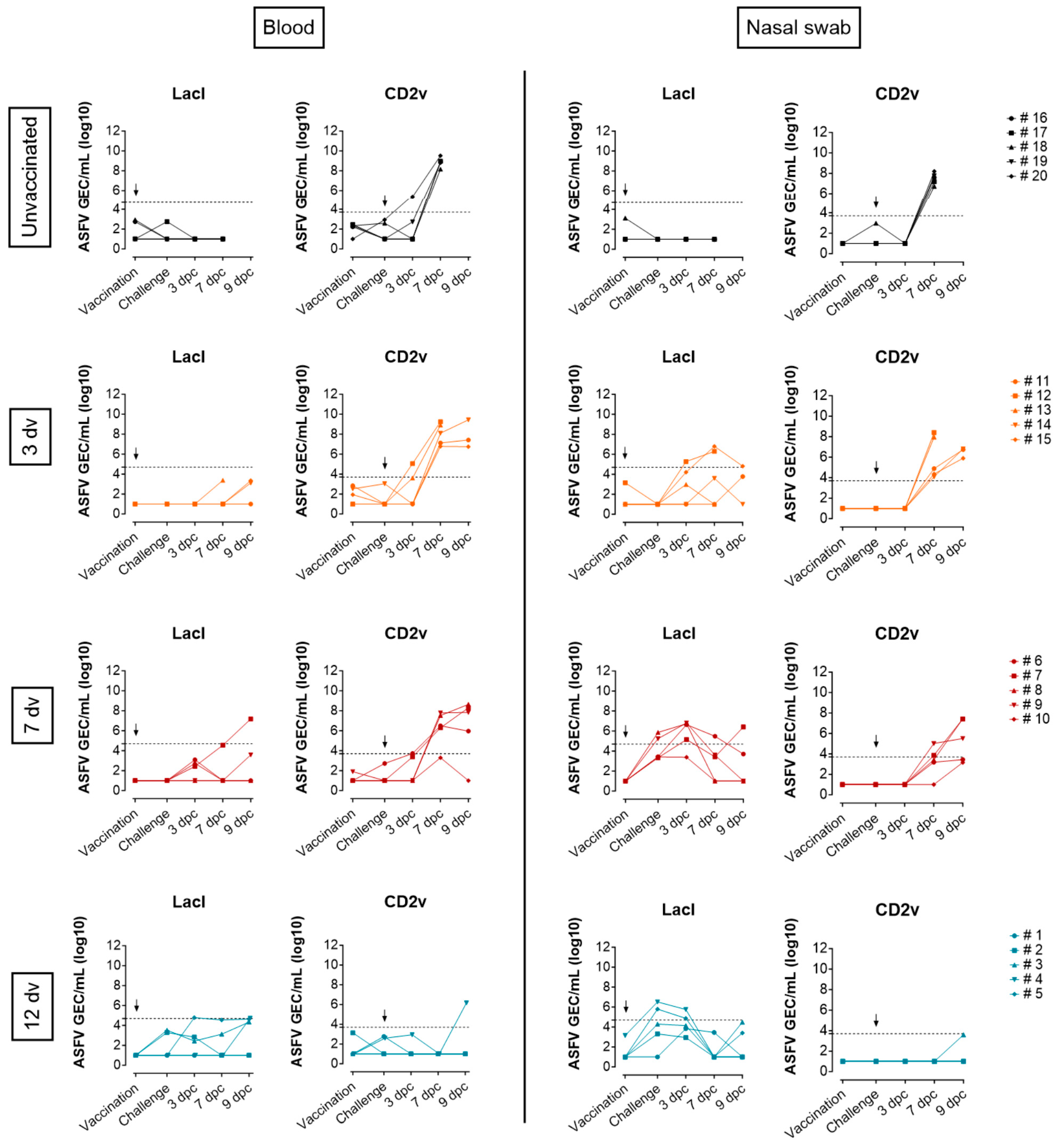
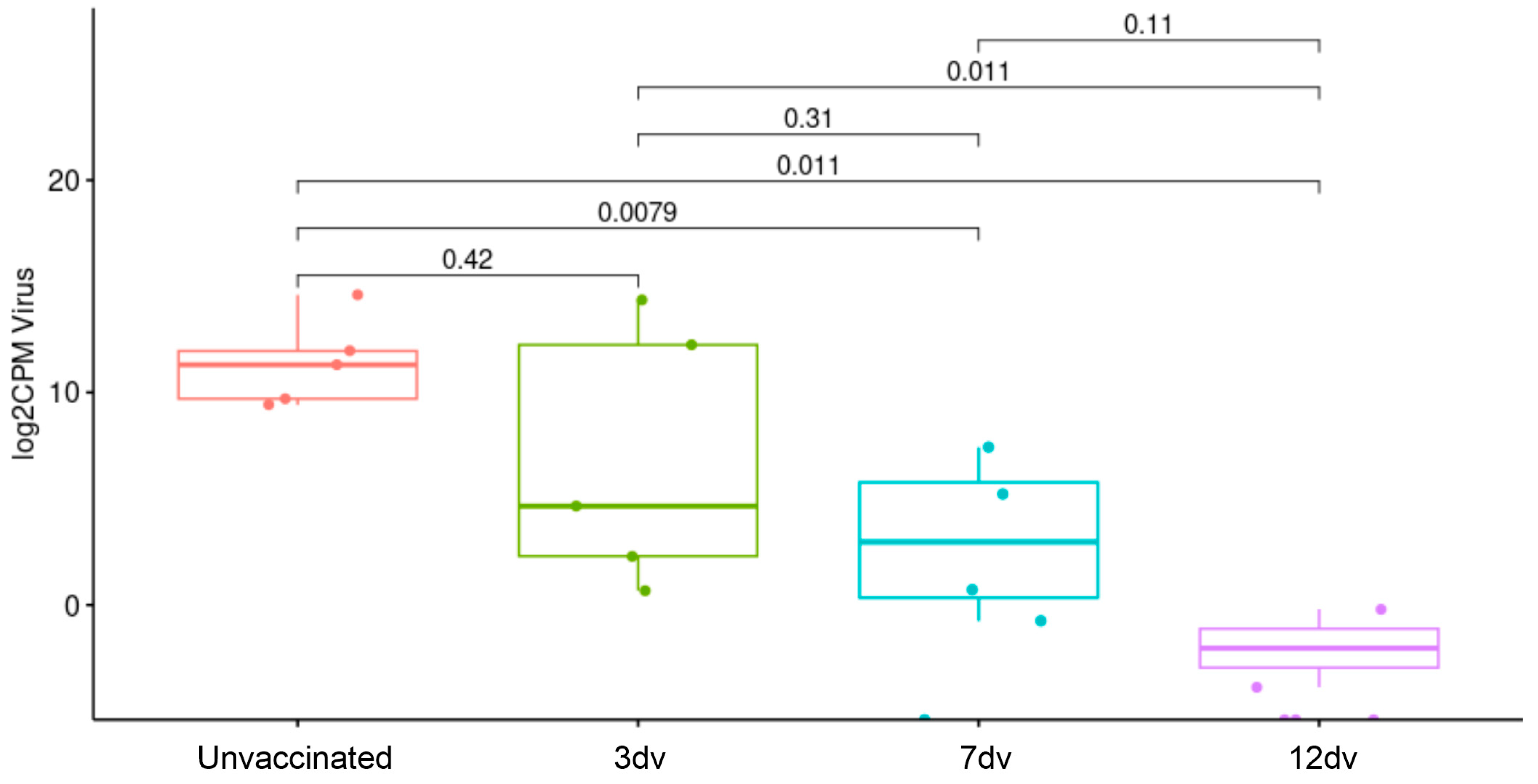
3.1. Onset of Cross-Protection Induced in Pigs Intranasally Vaccinated with BA71ΔCD2
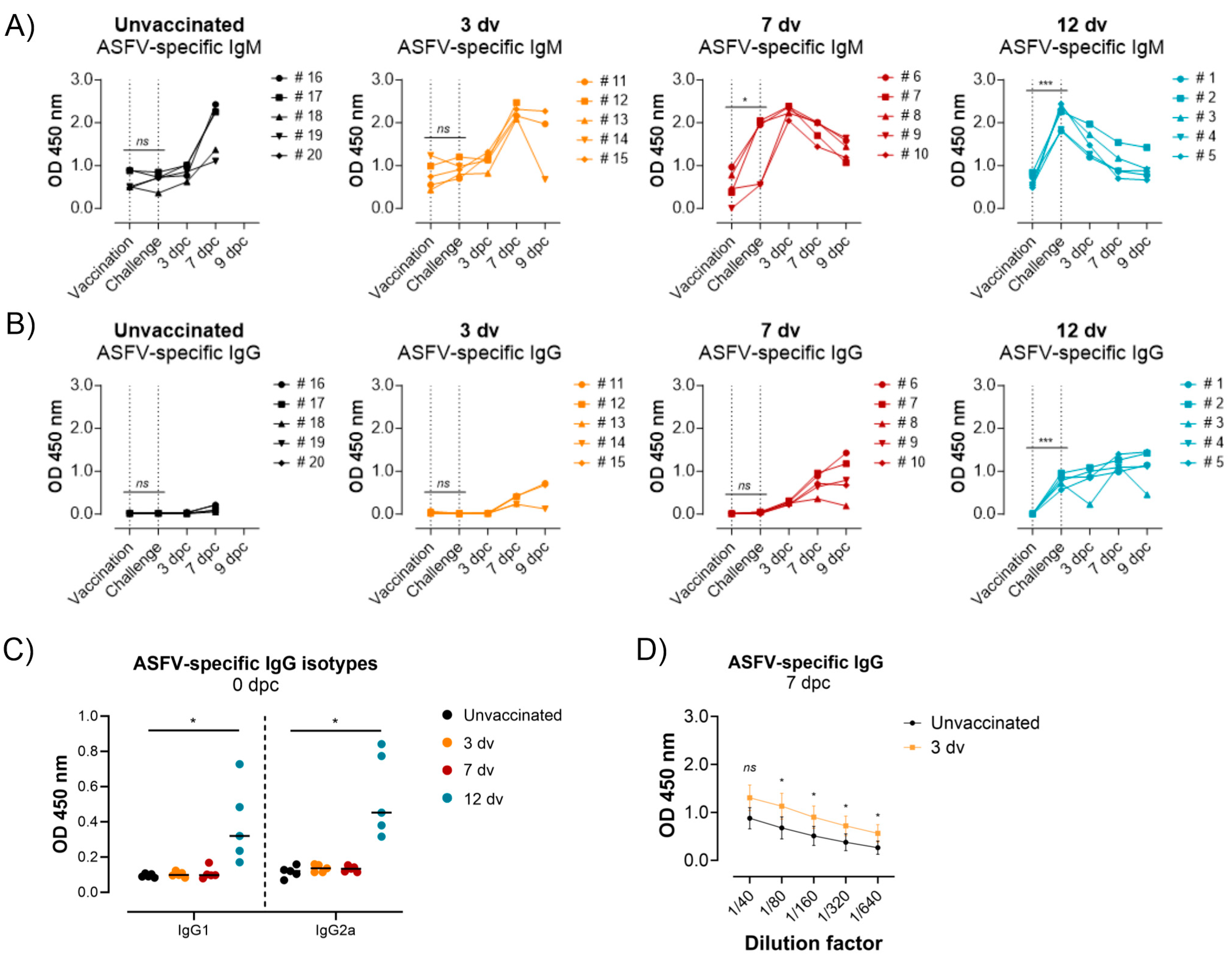
3.2. Presence of ASFV-Specific IgG Antibodies before Challenge Is Associated with Control of Infection
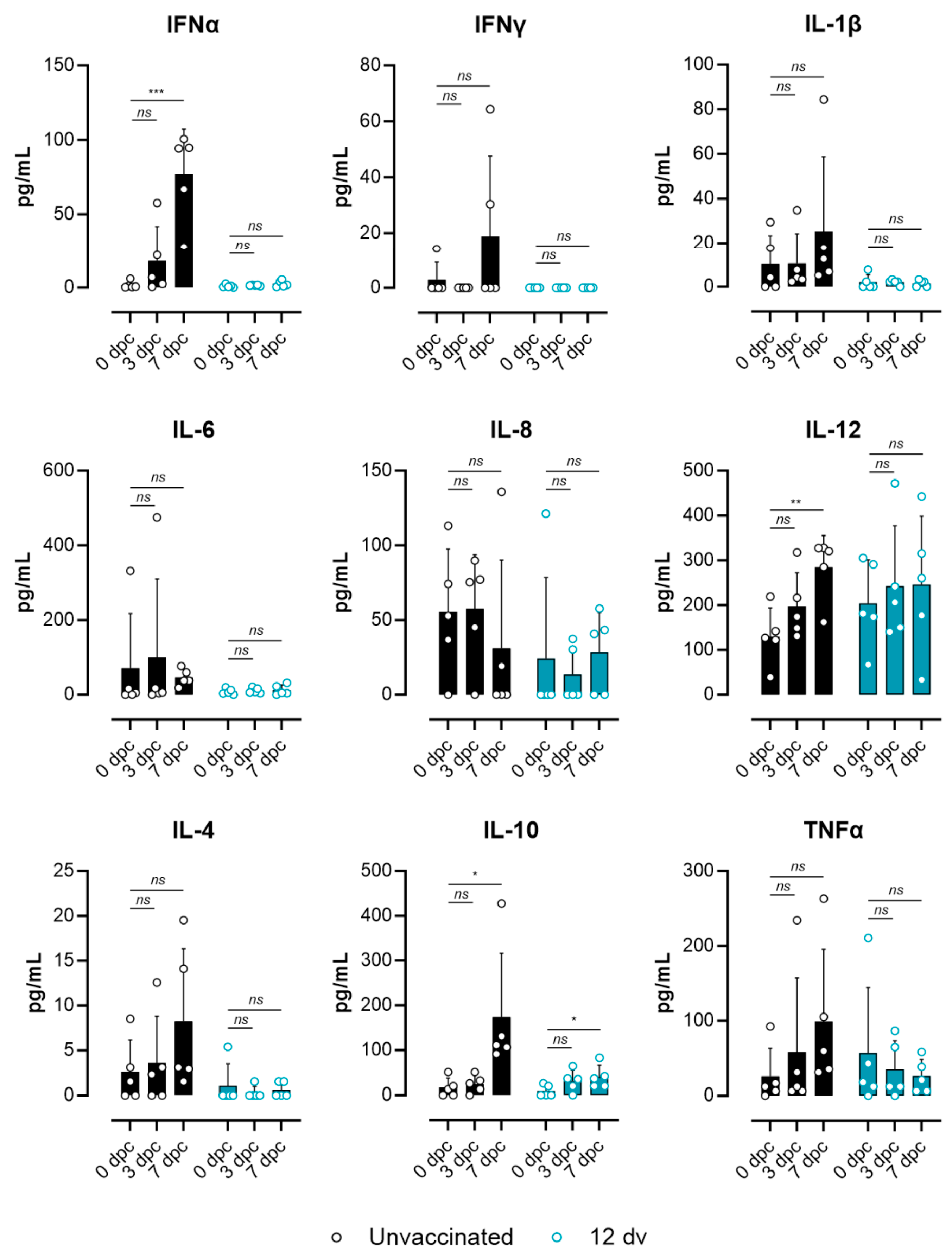
3.3. Lack of ASF-Associated Apoptosis and Immunopathology Markers in Pigs Vaccinated 12 Days Prior to the Challenge
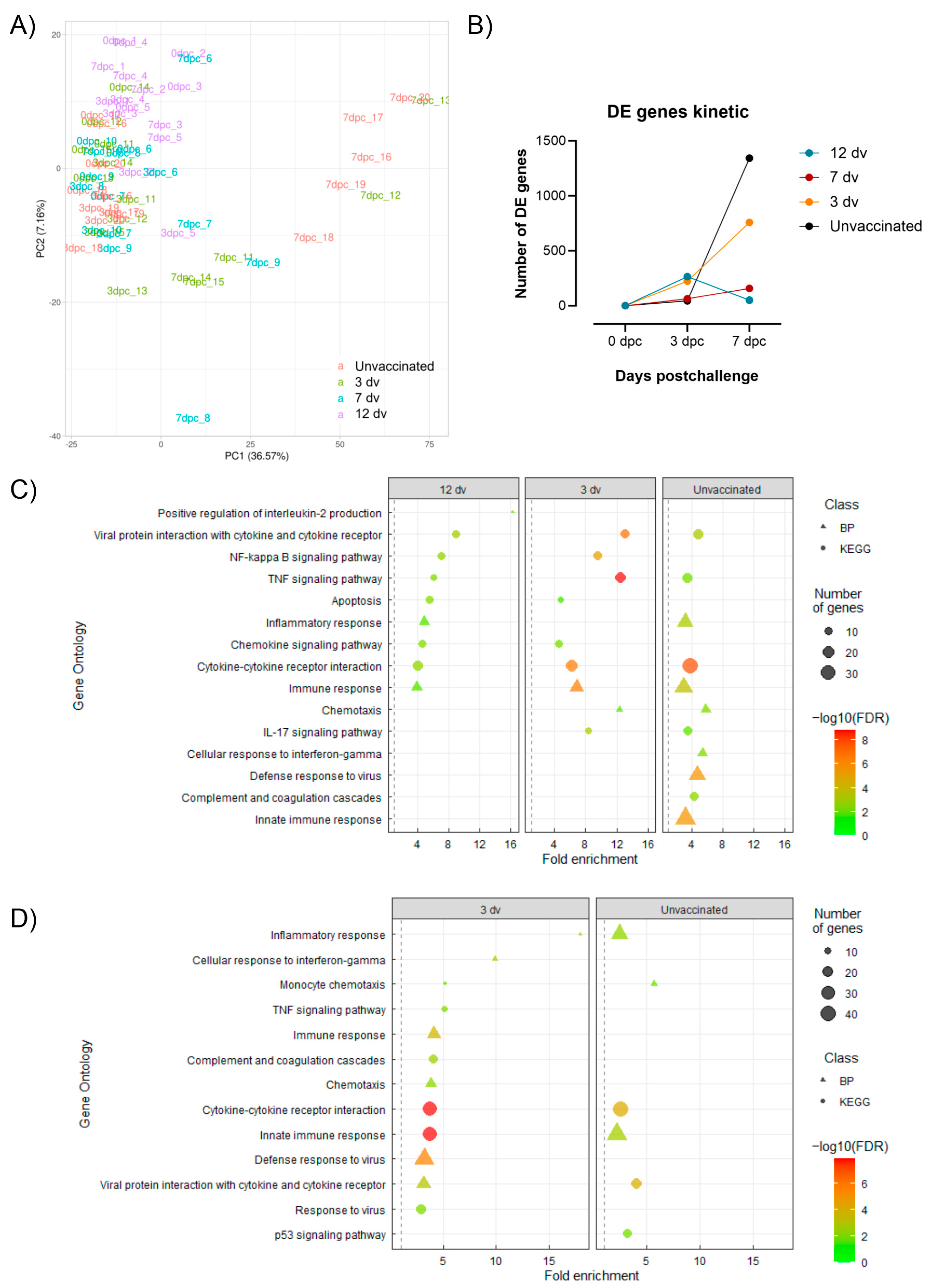
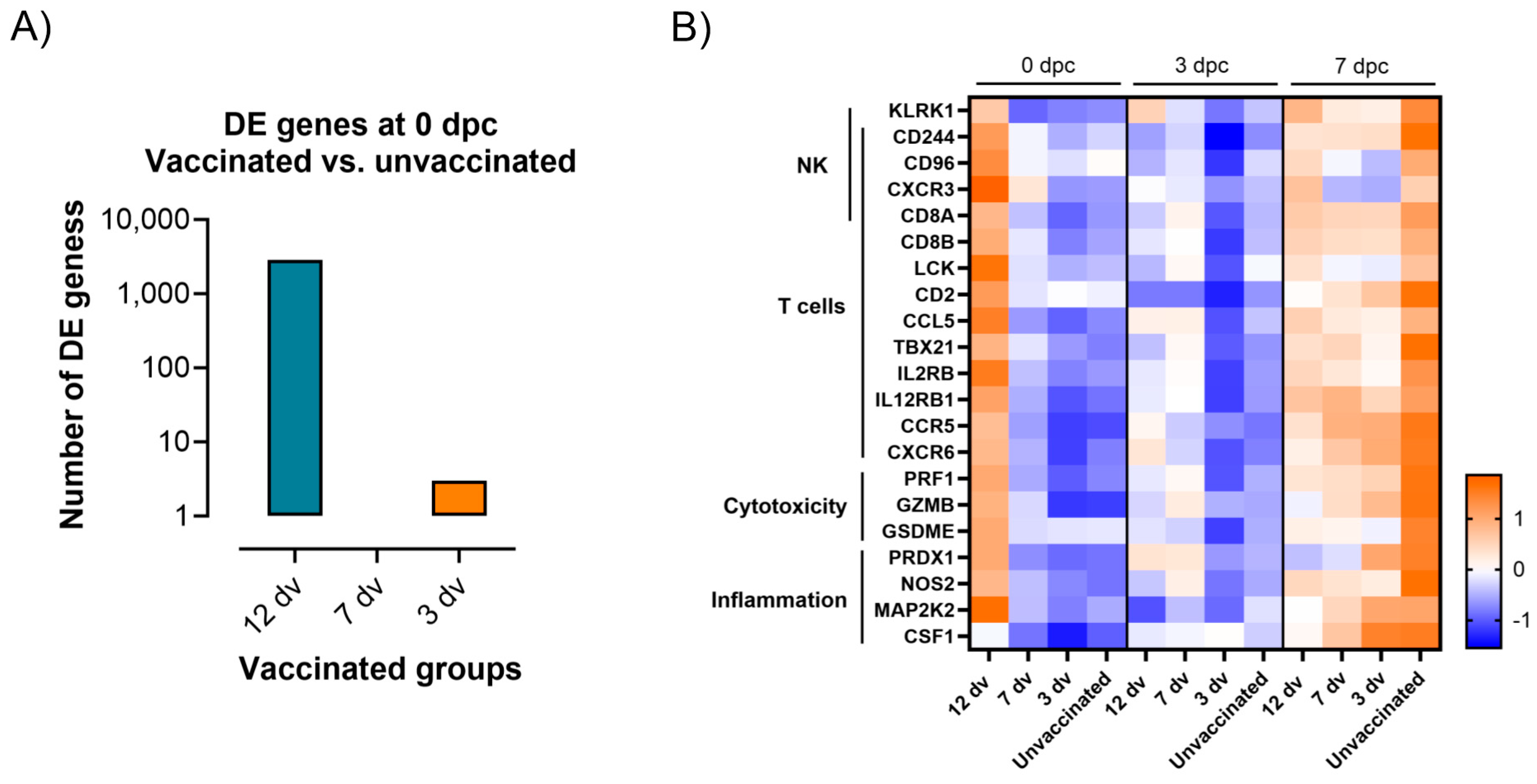
3.4. Control of Infection Is Associated with a Blood Cytotoxic Transcriptomic Signature before Challenge
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Penrith, M.L.; Vosloo, W. Review of African Swine Fever: Transmission, Spread and Control. J. S. Afr. Vet. Assoc. 2009, 80, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Blome, S.; Gabriel, C.; Beer, M. Pathogenesis of African Swine Fever in Domestic Pigs and European Wild Boar. Virus Res. 2013, 173, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Karalyan, Z.; Zakaryan, H.; Arzumanyan, H.; Sargsyan, K.; Voskanyan, H.; Hakobyan, L.; Abroyan, L.; Avetisyan, A.; Karalova, E. Pathology of Porcine Peripheral White Blood Cells during Infection with African Swine Fever Virus. BMC Vet. Res. 2012, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Franzoni, G.; Pedrera, M.; Sánchez-Cordón, P.J. African Swine Fever Virus Infection and Cytokine Response In Vivo: An Update. Viruses 2023, 15, 233. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, J.; Zhang, Y.; Yang, J.; Wang, L.; Qi, Y.; Han, X.; Zhou, X.; Miao, F.; Chen, T.; et al. Cytokine Storm in Domestic Pigs Induced by Infection of Virulent African Swine Fever Virus. Front. Vet. Sci. 2020, 7, 601641. [Google Scholar] [CrossRef] [PubMed]
- Salguero, F.J.; Sánchez-Cordón, P.J.; Núñez, A.; Fernández de Marco, M.; Gómez-Villamandos, J.C. Proinflammatory Cytokines Induce Lymphocyte Apoptosis in Acute African Swine Fever Infection. J. Comp. Pathol. 2005, 132, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Juszkiewicz, M.; Walczak, M.; Woźniakowski, G.; Podgórska, K. African Swine Fever: Transmission, Spread, and Control through Biosecurity and Disinfection, Including Polish Trends. Viruses 2023, 15, 2275. [Google Scholar] [CrossRef]
- Bosch-Camós, L.; López, E.; Rodriguez, F. African Swine Fever Vaccines: A Promising Work Still in Progress. Porcine Health Manag. 2020, 6, 17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, S.; Zhang, H.; Qin, Z.; Shan, H.; Cai, X. Vaccines for African Swine Fever: An Update. Front. Microbiol. 2023, 14, 1139494. [Google Scholar] [CrossRef]
- Tran, X.H.; Le, T.T.P.; Nguyen, Q.H.; Do, T.T.; Nguyen, V.D.; Gay, C.G.; Borca, M.V.; Gladue, D.P. African Swine Fever Virus Vaccine Candidate ASFV-G-ΔI177L Efficiently Protects European and Native Pig Breeds against Circulating Vietnamese Field Strain. Transbound. Emerg. Dis. 2021, 69, e497–e504. [Google Scholar]
- Wang, Z.; Zhang, J.; Li, F.; Zhang, Z.; Chen, W.; Zhang, X.; Sun, E.; Zhu, Y.; Liu, R.; He, X.; et al. The Attenuated African Swine Fever Vaccine HLJ/18-7GD Provides Protection against Emerging Prevalent Genotype II Variants in China. Emerg. Microbes Infect. 2024, 13, 2300464. [Google Scholar] [CrossRef]
- Zhao, D.; Sun, E.; Huang, L.; Ding, L.; Zhu, Y.; Zhang, J.; Shen, D.; Zhang, X.; Zhang, Z.; Ren, T.; et al. Highly Lethal Genotype I and II Recombinant African Swine Fever Viruses Detected in Pigs. Nat. Commun. 2023, 14, 3096. [Google Scholar] [CrossRef] [PubMed]
- Monteagudo, P.L.; Lacasta, A.; López, E.; Bosch, L.; Collado, J.; Pina-Pedrero, S.; Correa-Fiz, F.; Accensi, F.; Navas, M.J.; Vidal, E.; et al. BA71ΔCD2: A New Recombinant Live Attenuated African Swine Fever Virus with Cross-Protective Capabilities. J. Virol. 2017, 91, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Bosch-Camós, L.; Alonso, U.; Esteve-Codina, A.; Chang, C.-Y.; Martín-Mur, B.; Accensi, F.; Muñoz, M.; Navas, M.J.; Dabad, M.; Vidal, E.; et al. Cross-Protection against African Swine Fever Virus upon Intranasal Vaccination Is Associated with an Adaptive-Innate Immune Crosstalk. PLoS Pathog. 2022, 18, e1010931. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.B.; Krug, P.W.; Ramirez-Medina, E.; Valladares, A.; Rai, A.; Espinoza, N.; Gladue, D.P.; Borca, M.V. The Presence of Virus Neutralizing Antibodies Is Highly Associated with Protection against Virulent Challenge in Domestic Pigs Immunized with ASFV Live Attenuated Vaccine Candidates. Pathogens 2022, 11, 1311. [Google Scholar] [CrossRef] [PubMed]
- Argilaguet, J.M.; Pérez-Martín, E.; Nofrarías, M.; Gallardo, C.; Accensi, F.; Lacasta, A.; Mora, M.; Ballester, M.; Galindo-Cardiel, I.; López-Soria, S.; et al. DNA Vaccination Partially Protects against African Swine Fever Virus Lethal Challenge in the Absence of Antibodies. PLoS ONE 2012, 7, e40942. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A Simple Method of Estimating Fifty per Cent endpoints12. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Galindo-Cardiel, I.; Ballester, M.; Solanes, D.; Nofrarías, M.; López-Soria, S.; Argilaguet, J.M.; Lacasta, A.; Accensi, F.; Rodríguez, F.; Segalés, J. Standardization of Pathological Investigations in the Framework of Experimental ASFV Infections. Virus Res. 2013, 173, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Soler, A.; Nieto, R.; Carrascosa, A.L.; De Mia, G.M.; Bishop, R.P.; Martins, C.; Fasina, F.O.; Couacy-Hymman, E.; Heath, L.; et al. Comparative Evaluation of Novel African Swine Fever Virus (ASF) Antibody Detection Techniques Derived from Specific ASF Viral Genotypes with the OIE Internationally Prescribed Serological Tests. Vet. Microbiol. 2013, 162, 32–43. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision Weights Unlock Linear Model Analysis Tools for RNA-Seq Read Counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Carlson, J.; O’Donnell, V.; Alfano, M.; Velazquez Salinas, L.; Holinka, L.G.; Krug, P.W.; Gladue, D.P.; Higgs, S.; Borca, M.V. Association of the Host Immune Response with Protection Using a Live Attenuated African Swine Fever Virus Model. Viruses 2016, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, V.; Risatti, G.R.; Holinka, L.G.; Krug, P.W. Simultaneous Deletion of the 9GL and UK Genes from the African Swine Fever Virus Georgia 2007 Isolate Offers Increased Safety and Protection against Homologous Challenge. J. Virol. 2016, 91, e01760-16. [Google Scholar] [CrossRef] [PubMed]
- Radulovic, E.; Mehinagic, K.; Wüthrich, T.; Hilty, M.; Posthaus, H.; Summerfield, A.; Ruggli, N.; Benarafa, C. The Baseline Immunological and Hygienic Status of Pigs Impact Disease Severity of African Swine Fever. PLoS Pathog. 2022, 18, e1010522. [Google Scholar] [CrossRef] [PubMed]
- Weiss, M.; Blazek, K.; Byrne, A.J.; Perocheau, D.P.; Udalova, I.A. IRF5 Is a Specific Marker of Inflammatory Macrophages in Vivo. Mediat. Inflamm. 2013, 2013, 245804. [Google Scholar] [CrossRef] [PubMed]
- Paun, A.; Bankoti, R.; Joshi, T.; Pitha, P.M.; Stäger, S. Critical Role of IRF-5 in the Development of T Helper 1 Responses to Leishmania Donovani Infection. PLoS Pathog. 2011, 7, e1001246. [Google Scholar] [CrossRef]
- Sun, E.; Huang, L.; Zhang, X.; Zhang, J.; Shen, D.; Zhang, Z.; Wang, Z.; Huo, H.; Wang, W.; Huangfu, H.; et al. Genotype I African Swine Fever Viruses Emerged in Domestic Pigs in China and Caused Chronic Infection. Emerg. Microbes Infect. 2021, 10, 2183–2193. [Google Scholar] [CrossRef]
- Graham, S.P.; Everett, H.E.; Haines, F.J.; Johns, H.L.; Sosan, O.A.; Salguero, F.J.; Clifford, D.J.; Steinbach, F.; Drew, T.W.; Crooke, H.R. Challenge of Pigs with Classical Swine Fever Viruses after C-Strain Vaccination Reveals Remarkably Rapid Protection and Insights into Early Immunity. PLoS ONE 2012, 7, e29310. [Google Scholar] [CrossRef]
- Bohórquez, J.A.; Wang, M.; Díaz, I.; Alberch, M.; Pérez-Simó, M.; Rosell, R.; Gladue, D.P.; Borca, M.V.; Ganges, L. The FlagT4G Vaccine Confers a Strong and Regulated Immunity and Early Virological Protection against Classical Swine Fever. Viruses 2022, 14, 1954. [Google Scholar] [CrossRef]
- Horsington, J.; Perez, C.B.; Maradei, E.; Novo, S.G.; Gonzales, J.L.; Singanallur, N.B.; Bonastre, P.; Vosloo, W. Protective Effects of High-Potency FMDV O1 Manisa Monovalent Vaccine in Cattle Challenged with FMDV O/SKR/2010 at 7 or 4 Days Post Vaccination. Vaccine 2017, 35, 5179–5185. [Google Scholar] [CrossRef]
- Cox, S.J.; Parida, S.; Voyce, C.; Reid, S.M.; Hamblin, P.A.; Hutchings, G.; Paton, D.J.; Barnett, P.V. Further Evaluation of Higher Potency Vaccines for Early Protection of Cattle against FMDV Direct Contact Challenge. Vaccine 2007, 25, 7687–7695. [Google Scholar] [CrossRef] [PubMed]
- Eblé, P.L.; Bouma, A.; de Bruin, M.G.M.; van Hemert-Kluitenberg, F.; van Oirschot, J.T.; Dekker, A. Vaccination of Pigs Two Weeks before Infection Significantly Reduces Transmission of Foot-and-Mouth Disease Virus. Vaccine 2004, 22, 1372–1378. [Google Scholar] [CrossRef] [PubMed]
- Onisk, D.V.; Borca, M.V.; Kutish, G.; Kramer, E.; Irusta, P.; Rock, D.L. Passively Transferred African Swine Fever Virus Antibodies Protect Swine against Lethal Infection. Virology 1994, 198, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Schlafer, D.H.; Mebus, C.A.; McVicar, J.W. African Swine Fever in Neonatal Pigs: Passively Acquired Protection from Colostrum or Serum of Recovered Pigs. Am. J. Vet. Res. 1984, 45, 1367–1372. [Google Scholar]
- Leitão, A.; Cartaxeiro, C.; Coelho, R.; Cruz, B.; Parkhouse, R.M.E.; Portugal, F.C.; Vigário, J.D.; Martins, C.L.V. The Non-Haemadsorbing African Swine Fever Virus Isolate ASFV/NH/P68 Provides a Model for Defining the Protective Anti-Virus Immune Response. J. Gen. Virol. 2001, 82, 513–523. [Google Scholar] [CrossRef]
- Oura, C.A.L.; Denyer, M.S.; Takamatsu, H.; Parkhouse, R.M.E. In Vivo Depletion of CD8+ T Lymphocytes Abrogates Protective Immunity to African Swine Fever Virus. J. Gen. Virol. 2005, 86, 2445–2450. [Google Scholar] [CrossRef]
- Takamatsu, H.-H.; Denyer, M.S.; Lacasta, A.; Stirling, C.M.A.; Argilaguet, J.M.; Netherton, C.L.; Oura, C.A.L.; Martins, C.; Rodríguez, F. Cellular Immunity in ASFV Responses. Virus Res. 2013, 173, 110–121. [Google Scholar] [CrossRef]
- Webster, R.G.; Askonas, B.A. Cross-Protection and Cross-Reactive Cytotoxic T Cells Induced by Influenza Virus Vaccines in Mice. Eur. J. Immunol. 1980, 10, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Müllbacher, A.; Lobigs, M.; Alsharifi, M.; Regner, M. Cytotoxic T-Cell Immunity as a Target for Influenza Vaccines. Lancet Infect. Dis. 2006, 6, 255–256. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, A.; Franzoni, G.; Netherton, C.L.; Hartmann, L.; Blome, S.; Blohm, U. Adaptive Cellular Immunity against African Swine Fever Virus Infections. Pathogens 2022, 11, 274. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marín-Moraleda, D.; Muñoz-Basagoiti, J.; Tort-Miró, A.; Navas, M.J.; Muñoz, M.; Vidal, E.; Cobos, À.; Martín-Mur, B.; Meas, S.; Motuzova, V.; et al. Elucidating the Onset of Cross-Protective Immunity after Intranasal Vaccination with the Attenuated African Swine Fever Vaccine Candidate BA71ΔCD2. Vaccines 2024, 12, 517. https://doi.org/10.3390/vaccines12050517
Marín-Moraleda D, Muñoz-Basagoiti J, Tort-Miró A, Navas MJ, Muñoz M, Vidal E, Cobos À, Martín-Mur B, Meas S, Motuzova V, et al. Elucidating the Onset of Cross-Protective Immunity after Intranasal Vaccination with the Attenuated African Swine Fever Vaccine Candidate BA71ΔCD2. Vaccines. 2024; 12(5):517. https://doi.org/10.3390/vaccines12050517
Chicago/Turabian StyleMarín-Moraleda, David, Jordana Muñoz-Basagoiti, Aida Tort-Miró, María Jesús Navas, Marta Muñoz, Enric Vidal, Àlex Cobos, Beatriz Martín-Mur, Sochanwattey Meas, Veronika Motuzova, and et al. 2024. "Elucidating the Onset of Cross-Protective Immunity after Intranasal Vaccination with the Attenuated African Swine Fever Vaccine Candidate BA71ΔCD2" Vaccines 12, no. 5: 517. https://doi.org/10.3390/vaccines12050517
APA StyleMarín-Moraleda, D., Muñoz-Basagoiti, J., Tort-Miró, A., Navas, M. J., Muñoz, M., Vidal, E., Cobos, À., Martín-Mur, B., Meas, S., Motuzova, V., Chang, C.-Y., Gut, M., Accensi, F., Pina-Pedrero, S., Núñez, J. I., Esteve-Codina, A., Gavrilov, B., Rodriguez, F., Liu, L., & Argilaguet, J. (2024). Elucidating the Onset of Cross-Protective Immunity after Intranasal Vaccination with the Attenuated African Swine Fever Vaccine Candidate BA71ΔCD2. Vaccines, 12(5), 517. https://doi.org/10.3390/vaccines12050517








