A Unique mRNA Vaccine Elicits Protective Efficacy against the SARS-CoV-2 Omicron Variant and SARS-CoV
Abstract
1. Introduction
2. Materials and Methods
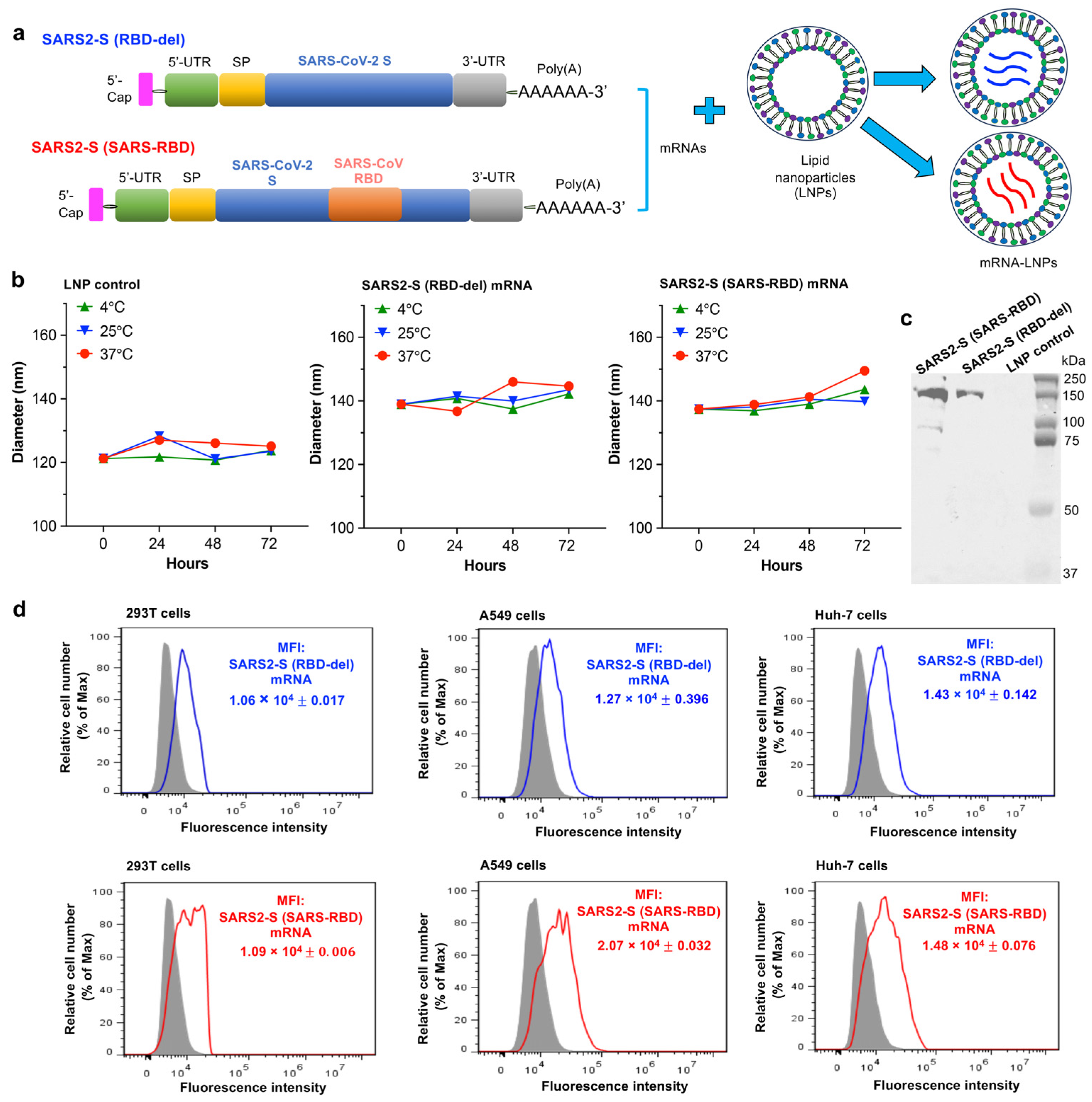
2.1. Design and Construct of mRNA Vaccines
2.2. Synthesis of mRNA Vaccines
2.3. mRNA Formulation and Characterization
2.4. mRNA Expression by Western Blot
2.5. mRNA Expression by Flow Cytometry
2.6. Multiplex Assaycat
2.7. Enzyme-Linked Immunoassay (ELISA)
2.8. Pseudovirus Preparation and Pseudovirus Neutralization Assay
2.9. Plaque Reduction Neutralization Assay
2.10. Plaque Assay for the Detection of Viral Titers
2.11. Real-Time Quantitative PCR (qRT-PCR)
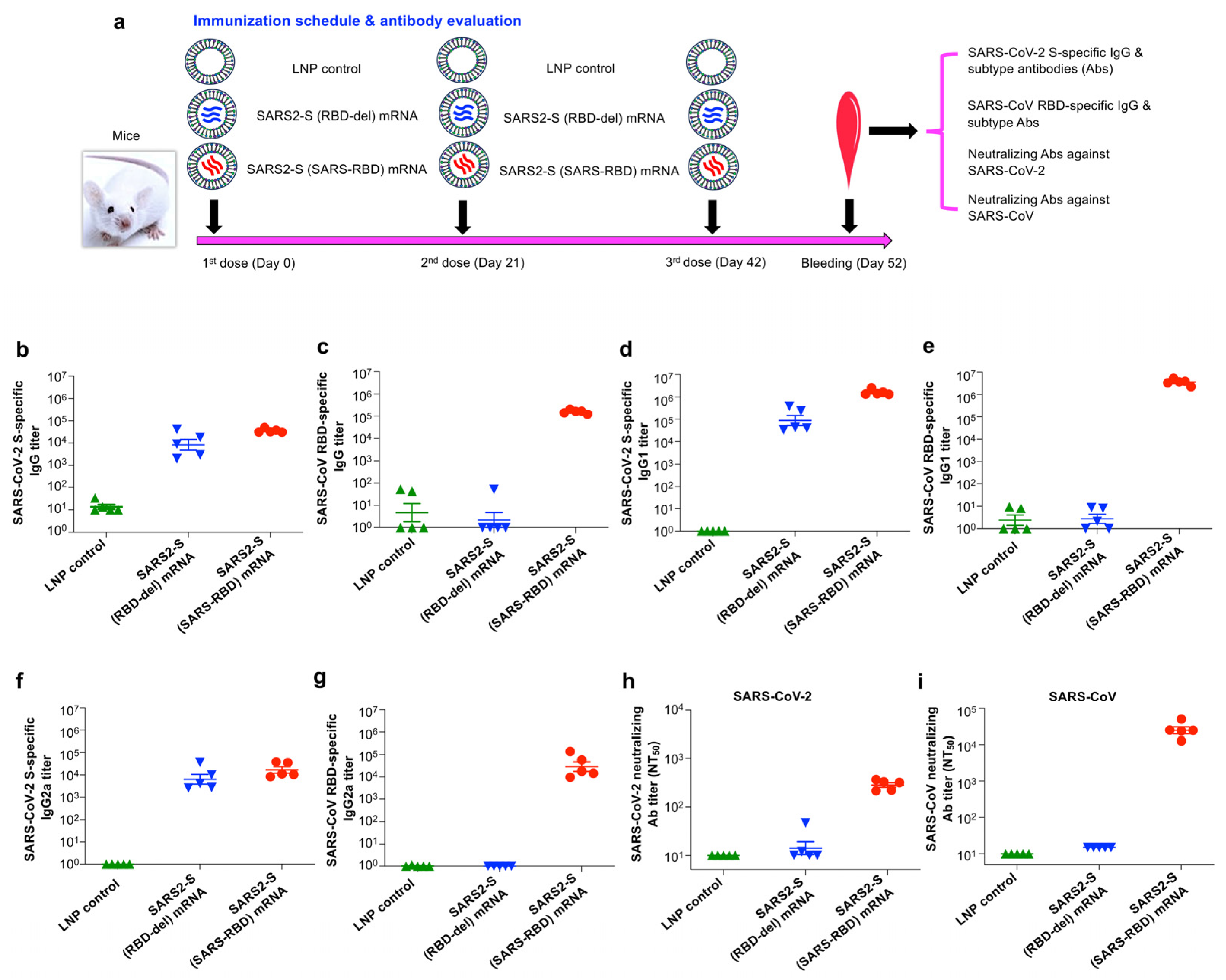
2.12. Mouse Immunization Procedures
2.13. Challenge of Immunized Mice with SARS-CoV-2 Omicron Variant
2.14. Challenge of Immunized Mice with SARS-CoV
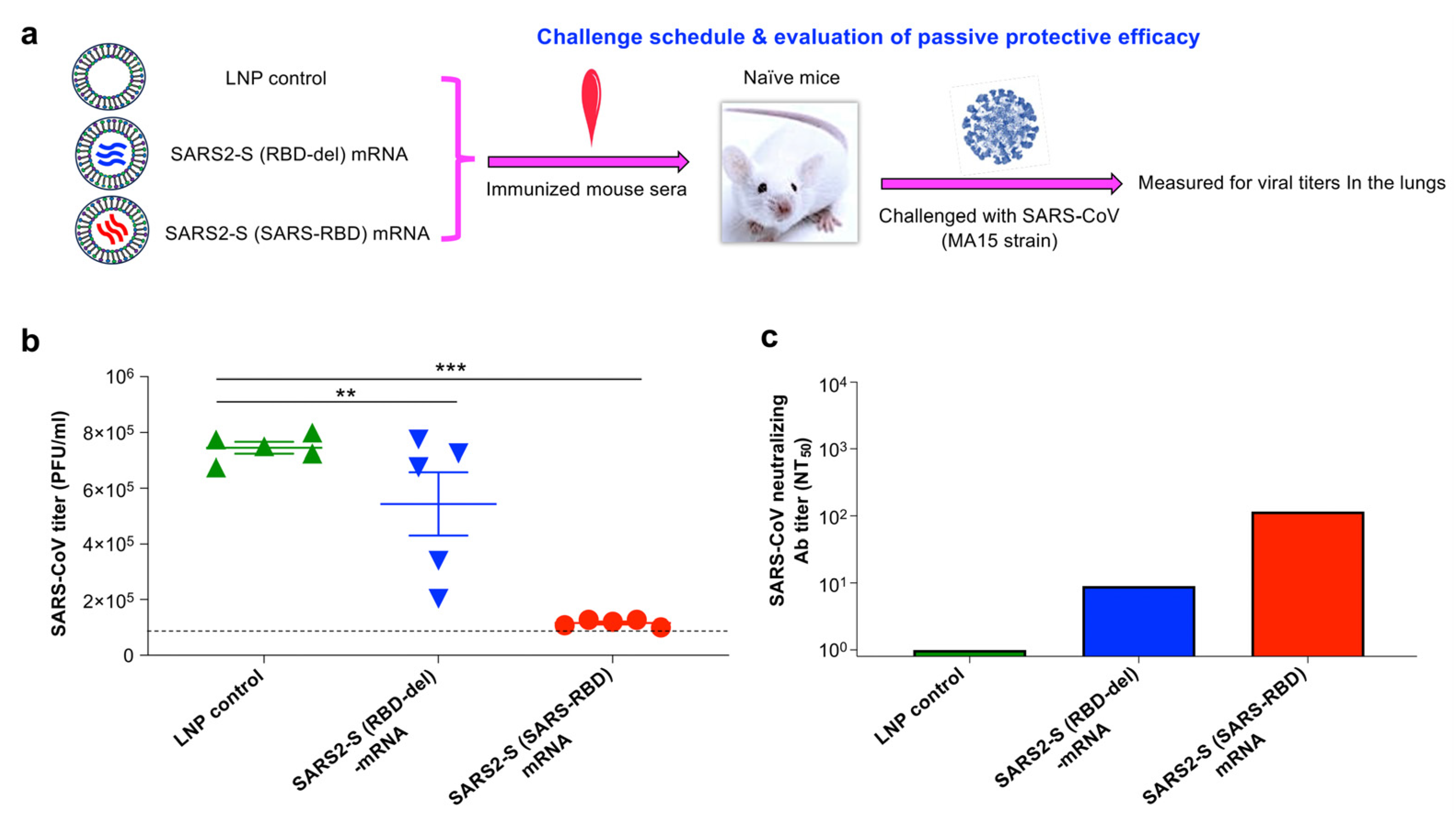
2.15. Challenge of Immune Serum-Transferred Mice with SARS-CoV
2.16. Statistical Analysis
3. Results
3.1. Construction and Characterization of mRNA Vaccines
3.2. SARS2-S (SARS-RBD) mRNA Vaccine Elicited Effective Antibodies with Neutralizing Activity against SARS-CoV-2 and SARS-CoV
3.3. SARS2-S (SARS-RBD) mRNA Vaccine Elicited Durable SARS-CoV-2 S and SARS-CoV RBD-Specific T-Cell Responses
3.4. SARS2-S (SARS-RBD) mRNA Vaccine-Protected Mice against Challenge with SARS-CoV-2 Omicron Variant and SARS-CoV
3.5. SARS2-S (SARS-RBD) mRNA-Immune Sera Protected Naïve Mice against SARS-CoV-2 Infection
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- World Health Organization. Coronavirus (COVID-19) Dashboard. 2023. Available online: https://covid19.who.int/ (accessed on 28 April 2024).
- Stadler, K.; Masignani, V.; Eickmann, M.; Becker, S.; Abrignani, S.; Klenk, H.D.; Rappuoli, R. SARS—beginning to understand a new virus. Nat. Rev. Microbiol. 2003, 1, 209–218. [Google Scholar] [CrossRef]
- Fronteira, I.; Sidat, M.; Magalhães, J.P.; de Barros, F.P.C.; Delgado, A.P.; Correia, T.; Daniel-Ribeiro, C.T.; Ferrinho, P. The SARS-CoV-2 pandemic: A syndemic perspective. One Health 2021, 12, 100228. [Google Scholar] [CrossRef]
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W.; et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1953–1966. [Google Scholar] [CrossRef]
- Shi, J.; Wang, G.; Zheng, J.; Verma, A.K.; Guan, X.; Malisheni, M.M.; Geng, Q.; Li, F.; Perlman, S.; Du, L. Effective vaccination strategy using SARS-CoV-2 spike cocktail against Omicron and other variants of concern. NPJ Vaccines 2022, 7, 169. [Google Scholar] [CrossRef]
- World Health Organization. Tracking SARS-CoV-2 Variants. 2023. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 4 May 2024).
- Wrenn, J.O.; Pakala, S.B.; Vestal, G.; Shilts, M.H.; Brown, H.M.; Bowen, S.M.; Strickland, B.A.; Williams, T.; Mallal, S.A.; Jones, I.D.; et al. COVID-19 severity from Omicron and Delta SARS-CoV-2 variants. Influenza Other Respir. Viruses 2022, 16, 832–836. [Google Scholar] [CrossRef]
- Rahimi, F.; Talebi Bezmin Abadi, A. Omicron: A highly transmissible SARS-CoV-2 variant. Gene Rep. 2022, 27, 101549. [Google Scholar] [CrossRef]
- Sun, P.; Balinsky, C.A.; Jiang, L.; Jani, V.; Long, T.K.; Cheng, Y.; Serote, M.A.; Smith, A.B.; Fears, B.F.; Gatrell, S.K.; et al. Antibody responses to the SARS-CoV-2 ancestral strain and Omicron variants in Moderna mRNA-1273 vaccinated active-Duty US Navy Sailors and Marines. J. Infect. Dis. 2023, 228, 149–159. [Google Scholar] [CrossRef]
- Edara, V.V.; Manning, K.E.; Ellis, M.; Lai, L.; Moore, K.M.; Foster, S.L.; Floyd, K.; Davis-Gardner, M.E.; Mantus, G.; Nyhoff, L.E.; et al. mRNA-1273 and BNT162b2 mRNA vaccines have reduced neutralizing activity against the SARS-CoV-2 omicron variant. Cell Rep. Med. 2022, 3, 100529. [Google Scholar] [CrossRef]
- Shi, J.; Zheng, J.; Zhang, X.; Tai, W.; Odle, A.E.; Perlman, S.; Du, L. RBD-mRNA vaccine induces broadly neutralizing antibodies against Omicron and multiple other variants and protects mice from SARS-CoV-2 challenge. Transl. Res. 2022, 248, 11–21. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef]
- Wang, N.; Shang, J.; Jiang, S.; Du, L. Subunit vaccines against emerging pathogenic human coronaviruses. Front. Microbiol. 2020, 11, 298. [Google Scholar] [CrossRef]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef]
- He, Y.; Li, J.; Li, W.; Lustigman, S.; Farzan, M.; Jiang, S. Cross-neutralization of human and palm civet severe acute respiratory syndrome coronaviruses by antibodies targeting the receptor-binding domain of spike protein. J. Immunol. 2006, 176, 6085–6092. [Google Scholar] [CrossRef]
- Han, P.; Li, L.; Liu, S.; Wang, Q.; Zhang, D.; Xu, Z.; Li, X.; Peng, Q.; Su, C.; Huang, B.; et al. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell 2022, 185, 630–640.e610. [Google Scholar] [CrossRef]
- McCallum, M.; Czudnochowski, N.; Rosen, L.E.; Zepeda, S.K.; Bowen, J.E.; Walls, A.C.; Hauser, K.; Joshi, A.; Stewart, C.; Dillen, J.R.; et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science 2022, 375, 864–868. [Google Scholar] [CrossRef]
- Mannar, D.; Saville, J.W.; Zhu, X.; Srivastava, S.S.; Berezuk, A.M.; Tuttle, K.S.; Marquez, A.C.; Sekirov, I.; Subramaniam, S. SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science 2022, 375, 760–764. [Google Scholar] [CrossRef]
- Lasrado, N.; Collier, A.Y.; Miller, J.; Hachmann, N.P.; Liu, J.; Anand, T.; Bondzie, E.A.; Fisher, J.L.; Mazurek, C.R.; Patio, R.C.; et al. Waning immunity and IgG4 responses following bivalent mRNA boosting. Sci. Adv. 2024, 10, eadj9945. [Google Scholar] [CrossRef]
- Feikin, D.R.; Higdon, M.M.; Andrews, N.; Collie, S.; Deloria Knoll, M.; Kwong, J.C.; Link-Gelles, R.; Pilishvili, T.; Patel, M.K. Assessing COVID-19 vaccine effectiveness against Omicron subvariants: Report from a meeting of the World Health Organization. Vaccine 2023, 41, 2329–2338. [Google Scholar] [CrossRef]
- da Costa, C.H.S.; de Freitas, C.A.B.; Alves, C.N.; Lameira, J. Assessment of mutations on RBD in the Spike protein of SARS-CoV-2 Alpha, Delta and Omicron variants. Sci. Rep. 2022, 12, 8540. [Google Scholar] [CrossRef]
- Chaudhary, N.; Weissman, D.; Whitehead, K.A. mRNA vaccines for infectious diseases: Principles, delivery and clinical translation. Nat. Rev. Drug Discov. 2021, 20, 817–838. [Google Scholar] [CrossRef]
- Hogan, M.J.; Pardi, N. mRNA vaccines in the COVID-19 pandemic and beyond. Annu. Rev. Med. 2022, 73, 17–39. [Google Scholar] [CrossRef]
- Uraki, R.; Imai, M.; Ito, M.; Yamayoshi, S.; Kiso, M.; Jounai, N.; Miyaji, K.; Iwatsuki-Horimoto, K.; Takeshita, F.; Kawaoka, Y. An mRNA vaccine encoding the SARS-CoV-2 receptor-binding domain protects mice from various Omicron variants. NPJ Vaccines 2024, 9, 4. [Google Scholar] [CrossRef]
- Liang, Q.; Wang, Y.; Zhang, S.; Sun, J.; Sun, W.; Li, J.; Liu, Y.; Li, M.; Cheng, L.; Jiang, Y.; et al. RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants. iScience 2022, 25, 104043. [Google Scholar] [CrossRef]
- Stewart-Jones, G.B.E.; Elbashir, S.M.; Wu, K.; Lee, D.; Renzi, I.; Ying, B.; Koch, M.; Sein, C.E.; Choi, A.; Whitener, B.; et al. Domain-based mRNA vaccines encoding spike protein N-terminal and receptor binding domains confer protection against SARS-CoV-2. Sci. Transl. Med. 2023, 15, eadf4100. [Google Scholar] [CrossRef]
- Tai, W.; Zhang, X.; Drelich, A.; Shi, J.; Hsu, J.C.; Luchsinger, L.; Hillyer, C.D.; Tseng, C.K.; Jiang, S.; Du, L. A novel receptor-binding domain (RBD)-based mRNA vaccine against SARS-CoV-2. Cell Res. 2020, 30, 932–935. [Google Scholar] [CrossRef]
- Wang, G.; Shi, J.; Verma, A.K.; Guan, X.; Perlman, S.; Du, L. mRNA vaccines elicit potent neutralization against multiple SARS-CoV-2 omicron subvariants and other variants of concern. iScience 2022, 25, 105690. [Google Scholar] [CrossRef]
- Tai, W.; Zhang, X.; Yang, Y.; Zhu, J.; Du, L. Advances in mRNA and other vaccines against MERS-CoV. Transl. Res. 2021, 242, 20–37. [Google Scholar] [CrossRef]
- Tai, W.; Zhang, X.; He, Y.; Jiang, S.; Du, L. Identification of SARS-CoV RBD-targeting monoclonal antibodies with cross-reactive or neutralizing activity against SARS-CoV-2. Antiviral Res. 2020, 179, 104820. [Google Scholar] [CrossRef]
- Guan, X.; Verma, A.K.; Wang, G.; Shi, J.; Perlman, S.; Du, L. Glycosylated Delta-receptor-binding domain mucosal vaccine elicits broadly neutralizing antibodies with protection against SARS-CoV-2 challenge. iScience 2023, 26, 108033. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zheng, J.; Tai, W.; Verma, A.K.; Zhang, X.; Geng, Q.; Wang, G.; Guan, X.; Malisheni, M.M.; Odle, A.E.; et al. A glycosylated RBD protein induces enhanced neutralizing antibodies against Omicron and other variants with improved protection against SARS-CoV-2 infection. J Virol 2022, 96, e0011822. [Google Scholar] [CrossRef] [PubMed]
- Mohsin, M.; Mahmud, S. Omicron SARS-CoV-2 variant of concern: A review on its transmissibility, immune evasion, reinfection, and severity. Medicine 2022, 101, e29165. [Google Scholar] [CrossRef]
- Tada, T.; Zhou, H.; Dcosta, B.M.; Samanovic, M.I.; Chivukula, V.; Herati, R.S.; Hubbard, S.R.; Mulligan, M.J.; Landau, N.R. Increased resistance of SARS-CoV-2 Omicron variant to neutralization by vaccine-elicited and therapeutic antibodies. EBioMedicine 2022, 78, 103944. [Google Scholar] [CrossRef]
- Starr, T.N.; Czudnochowski, N.; Liu, Z.; Zatta, F.; Park, Y.J.; Addetia, A.; Pinto, D.; Beltramello, M.; Hernandez, P.; Greaney, A.J.; et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 2021, 597, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Thye, A.Y.; Law, J.W.; Pusparajah, P.; Letchumanan, V.; Chan, K.G.; Lee, L.H. Emerging SARS-CoV-2 variants of concern (VOCs): An impending global crisis. Biomedicines 2021, 9, 1303. [Google Scholar] [CrossRef]
- Ding, C.; He, J.; Zhang, X.; Jiang, C.; Sun, Y.; Zhang, Y.; Chen, Q.; He, H.; Li, W.; Xie, J.; et al. Crucial mutations of spike protein on SARS-CoV-2 evolved to variant strains escaping neutralization of convalescent plasmas and RBD-specific monoclonal antibodies. Front. Immunol. 2021, 12, 693775. [Google Scholar] [CrossRef]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef]
- Yan, H.; Jiao, H.; Liu, Q.; Zhang, Z.; Xiong, Q.; Wang, B.J.; Wang, X.; Guo, M.; Wang, L.F.; Lan, K.; et al. ACE2 receptor usage reveals variation in susceptibility to SARS-CoV and SARS-CoV-2 infection among bat species. Nat. Ecol. Evol. 2021, 5, 600–608. [Google Scholar] [CrossRef]
- Zhang, S.; Qiao, S.; Yu, J.; Zeng, J.; Shan, S.; Tian, L.; Lan, J.; Zhang, L.; Wang, X. Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution. Nat. Commun. 2021, 12, 1607. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.F.; Shi, Z.; Zhang, S.; Field, H.; Daszak, P.; Eaton, B.T. Review of bats and SARS. Emerg. Infect. Dis. 2006, 12, 1834–1840. [Google Scholar] [CrossRef] [PubMed]
- Chalkias, S.; Harper, C.; Vrbicky, K.; Walsh, S.R.; Essink, B.; Brosz, A.; McGhee, N.; Tomassini, J.E.; Chen, X.; Chang, Y.; et al. A bivalent Omicron-containing booster vaccine against COVID-19. N. Engl. J. Med. 2022, 387, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Hause, A.M.; Marquez, P.; Zhang, B.; Myers, T.R.; Gee, J.; Su, J.R.; Blanc, P.G.; Thomas, A.; Thompson, D.; Shimabukuro, T.T.; et al. Safety monitoring of bivalent COVID-19 mRNA vaccine booster doses among persons aged ≥12 years-United States, August 31-October 23, 2022. MMWR Morb. Mortal Wkly. Rep. 2022, 71, 1401–1406. [Google Scholar] [CrossRef] [PubMed]
- Branche, A.R.; Rouphael, N.G.; Diemert, D.J.; Falsey, A.R.; Losada, C.; Baden, L.R.; Frey, S.E.; Whitaker, J.A.; Little, S.J.; Anderson, E.J.; et al. Comparison of bivalent and monovalent SARS-CoV-2 variant vaccines: The phase 2 randomized open-label COVAIL trial. Nat. Med. 2023, 29, 2334–2346. [Google Scholar] [CrossRef] [PubMed]
- Chalkias, S.; Eder, F.; Essink, B.; Khetan, S.; Nestorova, B.; Feng, J.; Chen, X.; Chang, Y.; Zhou, H.; Montefiori, D.; et al. Safety, immunogenicity and antibody persistence of a bivalent Beta-containing booster vaccine against COVID-19: A phase 2/3 trial. Nat. Med. 2022, 28, 2388–2397. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, S.M.; O’Shea, K.J.; John, D.C.; Strych, U.; Bottazzi, M.E.; Martinez, M.F.; Ciciriello, A.; Chin, K.L.; Weatherwax, C.; Velmurugan, K.; et al. The potential epidemiologic, clinical, and economic value of a universal coronavirus vaccine: A modelling study. eClinicalMedicine 2024, 68, 102369. [Google Scholar] [CrossRef]
- Lamb, Y.N. BNT162b2 mRNA COVID-19 vaccine: First approval. Drugs 2021, 81, 495–501. [Google Scholar] [CrossRef]
- Milane, L.; Amiji, M. Clinical approval of nanotechnology-based SARS-CoV-2 mRNA vaccines: Impact on translational nanomedicine. Drug Deliv. Transl. Res. 2021, 11, 1309–1315. [Google Scholar] [CrossRef]
- Hajnik, R.L.; Plante, J.A.; Liang, Y.; Alameh, M.G.; Tang, J.; Bonam, S.R.; Zhong, C.; Adam, A.; Scharton, D.; Rafael, G.H.; et al. Dual spike and nucleocapsid mRNA vaccination confer protection against SARS-CoV-2 Omicron and Delta variants in preclinical models. Sci. Transl. Med. 2022, 14, eabq1945. [Google Scholar] [CrossRef]
- Kaplonek, P.; Cizmeci, D.; Fischinger, S.; Collier, A.R.; Suscovich, T.; Linde, C.; Broge, T.; Mann, C.; Amanat, F.; Dayal, D.; et al. mRNA-1273 and BNT162b2 COVID-19 vaccines elicit antibodies with differences in Fc-mediated effector functions. Sci. Transl. Med. 2022, 14, eabm2311. [Google Scholar] [CrossRef]
- Zhang, J.; Han, Z.B.; Liang, Y.; Zhang, X.F.; Jin, Y.Q.; Du, L.F.; Shao, S.; Wang, H.; Hou, J.W.; Xu, K.; et al. A mosaic-type trimeric RBD-based COVID-19 vaccine candidate induces potent neutralization against Omicron and other SARS-CoV-2 variants. Elife 2022, 11, e78633. [Google Scholar] [CrossRef]
- Altay Benetti, A.; Tan, E.Y.Z.; Chang, Z.W.; Bae, K.H.; Thwin, M.T.; Muthuramalingam, R.P.K.; Liao, K.C.; Wan, Y.; Ng, L.F.P.; Renia, L.; et al. Design and characterization of a new formulation for the delivery of COVID-19-mRNA vaccine to the nasal mucosa. Vaccines 2024, 12, 409. [Google Scholar] [CrossRef]
- Baldeon Vaca, G.; Meyer, M.; Cadete, A.; Hsiao, C.J.; Golding, A.; Jeon, A.; Jacquinet, E.; Azcue, E.; Guan, C.M.; Sanchez-Felix, X.; et al. Intranasal mRNA-LNP vaccination protects hamsters from SARS-CoV-2 infection. Sci. Adv. 2023, 9, eadh1655. [Google Scholar] [CrossRef]
- Alsoussi, W.B.; Malladi, S.K.; Zhou, J.Q.; Liu, Z.; Ying, B.; Kim, W.; Schmitz, A.J.; Lei, T.; Horvath, S.C.; Sturtz, A.J.; et al. SARS-CoV-2 Omicron boosting induces de novo B cell response in humans. Nature 2023, 617, 592–598. [Google Scholar] [CrossRef]
- Munro, A.P.S.; Janani, L.; Cornelius, V.; Aley, P.K.; Babbage, G.; Baxter, D.; Bula, M.; Cathie, K.; Chatterjee, K.; Dodd, K.; et al. Safety and immunogenicity of seven COVID-19 vaccines as a third dose (booster) following two doses of ChAdOx1 nCov-19 or BNT162b2 in the UK (COV-BOOST): A blinded, multicentre, randomised, controlled, phase 2 trial. Lancet 2021, 398, 2258–2276. [Google Scholar] [CrossRef]
- Scheaffer, S.M.; Lee, D.; Whitener, B.; Ying, B.; Wu, K.; Liang, C.Y.; Jani, H.; Martin, P.; Amato, N.J.; Avena, L.E.; et al. Bivalent SARS-CoV-2 mRNA vaccines increase breadth of neutralization and protect against the BA.5 Omicron variant in mice. Nat. Med. 2023, 29, 247–257. [Google Scholar] [CrossRef]
- Ying, B.; Whitener, B.; VanBlargan, L.A.; Hassan, A.O.; Shrihari, S.; Liang, C.Y.; Karl, C.E.; Mackin, S.; Chen, R.E.; Kafai, N.M.; et al. Protective activity of mRNA vaccines against ancestral and variant SARS-CoV-2 strains. Sci. Transl. Med. 2022, 14, eabm3302. [Google Scholar] [CrossRef]
- Ying, B.; Scheaffer, S.M.; Whitener, B.; Liang, C.Y.; Dmytrenko, O.; Mackin, S.; Wu, K.; Lee, D.; Avena, L.E.; Chong, Z.; et al. Boosting with variant-matched or historical mRNA vaccines protects against Omicron infection in mice. Cell 2022, 185, 1572–1587.e1511. [Google Scholar] [CrossRef]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schäfer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef]
- Creech, C.B.; Anderson, E.; Berthaud, V.; Yildirim, I.; Atz, A.M.; Melendez Baez, I.; Finkelstein, D.; Pickrell, P.; Kirstein, J.; Yut, C.; et al. Evaluation of mRNA-1273 COVID-19 vaccine in children 6 to 11 years of age. N. Engl. J. Med. 2022, 386, 2011–2023. [Google Scholar] [CrossRef] [PubMed]
- Andrews, N.; Stowe, J.; Kirsebom, F.; Toffa, S.; Rickeard, T.; Gallagher, E.; Gower, C.; Kall, M.; Groves, N.; O’Connell, A.M.; et al. COVID-19 vaccine effectiveness against the Omicron (B.1.1.529) variant. N. Engl. J. Med. 2022, 386, 1532–1546. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Chan, J.F.; Yuen, K.Y. Animal models in SARS-CoV-2 research. Nat. Methods 2022, 19, 392–394. [Google Scholar] [CrossRef] [PubMed]
- Imai, M.; Iwatsuki-Horimoto, K.; Hatta, M.; Loeber, S.; Halfmann, P.J.; Nakajima, N.; Watanabe, T.; Ujie, M.; Takahashi, K.; Ito, M.; et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA 2020, 117, 16587–16595. [Google Scholar] [CrossRef]
- Suzuki, R.; Yamasoba, D.; Kimura, I.; Wang, L.; Kishimoto, M.; Ito, J.; Morioka, Y.; Nao, N.; Nasser, H.; Uriu, K.; et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 2022, 603, 700–705. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, X.; Verma, A.K.; Wang, G.; Roy, A.; Perlman, S.; Du, L. A Unique mRNA Vaccine Elicits Protective Efficacy against the SARS-CoV-2 Omicron Variant and SARS-CoV. Vaccines 2024, 12, 605. https://doi.org/10.3390/vaccines12060605
Guan X, Verma AK, Wang G, Roy A, Perlman S, Du L. A Unique mRNA Vaccine Elicits Protective Efficacy against the SARS-CoV-2 Omicron Variant and SARS-CoV. Vaccines. 2024; 12(6):605. https://doi.org/10.3390/vaccines12060605
Chicago/Turabian StyleGuan, Xiaoqing, Abhishek K. Verma, Gang Wang, Abhijeet Roy, Stanley Perlman, and Lanying Du. 2024. "A Unique mRNA Vaccine Elicits Protective Efficacy against the SARS-CoV-2 Omicron Variant and SARS-CoV" Vaccines 12, no. 6: 605. https://doi.org/10.3390/vaccines12060605
APA StyleGuan, X., Verma, A. K., Wang, G., Roy, A., Perlman, S., & Du, L. (2024). A Unique mRNA Vaccine Elicits Protective Efficacy against the SARS-CoV-2 Omicron Variant and SARS-CoV. Vaccines, 12(6), 605. https://doi.org/10.3390/vaccines12060605





