The Human Genetic Differences in the Outcomes of mRNA Vaccination against COVID-19: A Prospective Cohort Study
Abstract
:1. Introduction
2. Materials and Methods
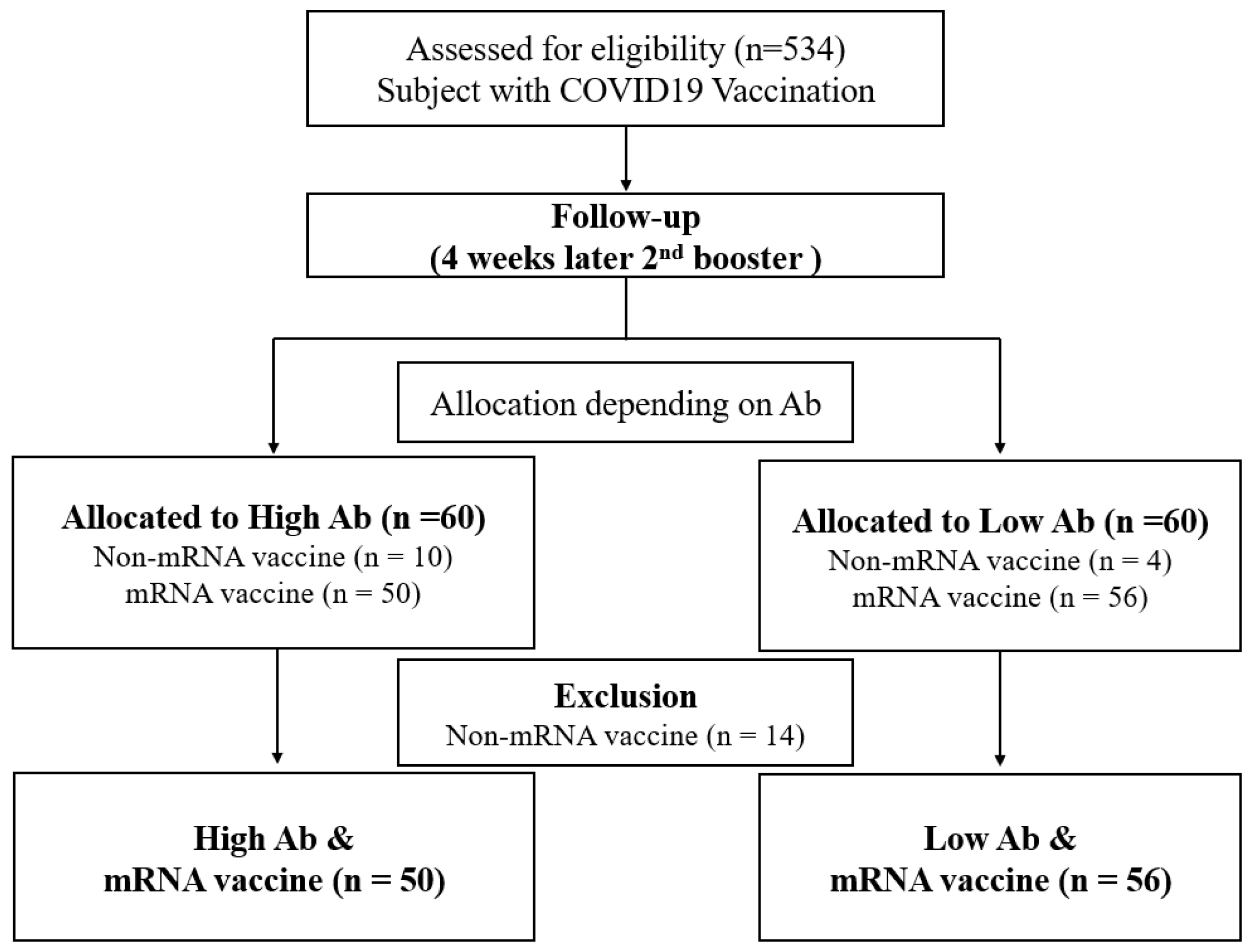
2.1. Study Design and Participants
2.2. Measurement of Anthropometric and Biochemical Parameters
2.3. Detection of Virus-Specific Antibodies
2.4. Genome-Wide Association Study
2.5. Real-Time PCR
2.6. Statistical Analysis
2.7. Power Calculation
3. Results
3.1. Population Characteristics
3.2. GWAS Results
3.3. Sample Distribution and Allele Frequency by Genotype of HDAC9 SNP (rs7795433)
3.4. Real-Time PCR Analysis of HDAC9 SNP (rs7795433)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, H.; Paulson, K.R.; Pease, S.A.; Watson, S.; Comfort, H.; Zheng, P.; Aravkin, A.Y.; Bisignano, C.; Barber, R.M.; Alam, T. Estimating excess mortality due to the COVID-19 pandemic: A systematic analysis of COVID-19-related mortality, 2020–2021. Lancet 2022, 399, 1513–1536. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B. Efficacy and safety of the mrna-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C. Safety and efficacy of the bnt162b2 mrna COVID-19 vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Emergency, C.; Group, C.C.; Graña, C.; Ghosn, L.; Evrenoglou, T.; Jarde, A.; Minozzi, S.; Bergman, H.; Buckley, B.S.; Probyn, K.; et al. Efficacy and safety of COVID-19 vaccines. Cochrane Database Syst. Rev. 2023, 2023, CD015477. [Google Scholar]
- Yan, Z.-P.; Yang, M.; Lai, C.-L. COVID-19 vaccines: A review of the safety and efficacy of current clinical trials. Pharmaceuticals 2021, 14, 406. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam, P.S.; Scott, M.K.; Hagan, T.; Li, C.; Feng, Y.; Wimmers, F.; Grigoryan, L.; Trisal, M.; Edara, V.V.; Lai, L. Systems vaccinology of the bnt162b2 mrna vaccine in humans. Nature 2021, 596, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A. Phase i/ii study of COVID-19 rna vaccine bnt162b1 in adults. Nature 2020, 586, 589–593. [Google Scholar] [CrossRef]
- Reusch, J.; Wagenhäuser, I.; Gabel, A.; Eggestein, A.; Höhn, A.; Lâm, T.T.; Frey, A.; Schubert-Unkmeir, A.; Dölken, L.; Frantz, S. Influencing factors of anti-SARS-CoV-2-spike-igg antibody titers in healthcare workers: A cross-section study. J. Med. Virol. 2023, 95, e28300. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, T.; Ikeda, K.; Tanaka, S.; Taniguchi, T.; Igari, H.; Onouchi, Y.; Kaneda, A.; Matsushita, K.; Hanaoka, H.; Nakada, T.-A. Antibody responses to bnt162b2 mrna COVID-19 vaccine and their predictors among healthcare workers in a tertiary referral hospital in Japan. Clin. Microbiol. Infect. 2021, 27, 1861.e1–1861.e5. [Google Scholar] [CrossRef]
- COVID-19 Host Genetics Initiative. Mapping the human genetic architecture of COVID-19. Nature 2021, 600, 472–477. [Google Scholar] [CrossRef]
- Čiučiulkaitė, I.; Siffert, W.; Elsner, C.; Dittmer, U.; Wichert, M.; Wagner, B.; Volbracht, L.; Mosel, F.; Möhlendick, B. Influence of the single nucleotide polymorphisms rs12252 and rs34481144 in ifitm3 on the antibody response after vaccination against COVID-19. Vaccines 2023, 11, 1257. [Google Scholar] [CrossRef] [PubMed]
- Kodde, C.; Tafelski, S.; Balamitsa, E.; Nachtigall, I.; Bonsignore, M. Factors influencing antibody response to SARS-CoV-2 vaccination. Vaccines 2023, 11, 451. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, R.B.; Ovsyannikova, I.G.; Haralambieva, I.H.; Lambert, N.D.; Pankratz, V.S.; Poland, G.A. Genome-wide snp associations with rubella-specific cytokine responses in measles-mumps-rubella vaccine recipients. Immunogenetics 2014, 66, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Hennig, B.J.; Fielding, K.; Broxholme, J.; Diatta, M.; Mendy, M.; Moore, C.; Pollard, A.J.; Rayco-Solon, P.; Sirugo, G.; van der Sande, M.A. Host genetic factors and vaccine-induced immunity to hepatitis b virus infection. PLoS ONE 2008, 3, e1898. [Google Scholar] [CrossRef] [PubMed]
- Gelder, C.M.; Lambkin, R.; Hart, K.W.; Fleming, D.; Williams, O.M.; Bunce, M.; Welsh, K.I.; Marshall, S.E.; Oxford, J. Associations between human leukocyte antigens and nonresponsiveness to influenza vaccine. J. Infect. Dis. 2002, 185, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Franco, L.M.; Bucasas, K.L.; Wells, J.M.; Niño, D.; Wang, X.; Zapata, G.E.; Arden, N.; Renwick, A.; Yu, P.; Quarles, J.M. Integrative genomic analysis of the human immune response to influenza vaccination. Elife 2013, 2, e00299. [Google Scholar] [CrossRef]
- Wolday, D.; Fung, C.Y.J.; Morgan, G.; Casalino, S.; Frangione, E.; Taher, J.; Lerner-Ellis, J.P. Hla variation and SARS-CoV-2 specific antibody response. Viruses 2023, 15, 906. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.H.; Gregersen, P.K. Genomics and the multifactorial nature of human autoimmune disease. N. Engl. J. Med. 2011, 365, 1612–1623. [Google Scholar] [CrossRef] [PubMed]
- Weiner, J.; Suwalski, P.; Holtgrewe, M.; Rakitko, A.; Thibeault, C.; Mueller, M.; Patriki, D.; Quedenau, C.; Krueger, U.; Ilinsky, V. Increased risk of severe clinical course of COVID-19 in carriers of hla-c* 04: 01. EClinicalMedicine 2021, 40, 101099. [Google Scholar] [CrossRef]
- Langton, D.J.; Bourke, S.C.; Lie, B.A.; Reiff, G.; Natu, S.; Darlay, R.; Burn, J.; Echevarria, C. The influence of hla genotype on the severity of COVID-19 infection. Hla 2021, 98, 14–22. [Google Scholar] [CrossRef]
- Nguyen, A.; David, J.K.; Maden, S.K.; Wood, M.A.; Weeder, B.R.; Nellore, A.; Thompson, R.F. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J. Virol. 2020, 94, e00510-20. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Althaus, T.; Tan, C.W.; Costantini, A.; Chia, W.N.; Chau, N.V.V.; Mattiuzzo, G.; Rose, N.J.; Voiglio, E.; Wang, L.-F. Who international standard for SARS-CoV-2 antibodies to determine markers of protection. Lancet Microbe 2022, 3, e81–e82. [Google Scholar] [CrossRef] [PubMed]
- Pruim, R.J.; Welch, R.P.; Sanna, S.; Teslovich, T.M.; Chines, P.S.; Gliedt, T.P.; Boehnke, M.; Abecasis, G.R.; Willer, C.J. Locuszoom: Regional visualization of genome-wide association scan results. Bioinformatics 2010, 26, 2336–2337. [Google Scholar] [CrossRef]
- Moore, C.M.; Jacobson, S.A.; Fingerlin, T.E. Power and sample size calculations for genetic association studies in the presence of genetic model misspecification. Hum. Hered. 2020, 84, 256–271. [Google Scholar] [CrossRef] [PubMed]
- Gauderman, W.J. Sample size requirements for association studies of gene-gene interaction. Am. J. Epidemiol. 2002, 155, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Gauderman, W.J. Sample size requirements for matched case-control studies of gene–environment interaction. Stat. Med. 2002, 21, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Scola, L.; Ferraro, D.; Sanfilippo, G.L.; De Grazia, S.; Lio, D.; Giammanco, G.M. Age and cytokine gene variants modulate the immunogenicity and protective effect of SARS-CoV-2 mrna-based vaccination. Vaccines 2023, 11, 413. [Google Scholar] [CrossRef] [PubMed]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Mai, A.; Massa, S.; Rotili, D.; Cerbara, I.; Valente, S.; Pezzi, R.; Simeoni, S.; Ragno, R. Histone deacetylation in epigenetics: An attractive target for anticancer therapy. Med. Res. Rev. 2005, 25, 261–309. [Google Scholar] [CrossRef] [PubMed]
- Haberland, M.; Montgomery, R.L.; Olson, E.N. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat. Rev. Genet. 2009, 10, 32–42. [Google Scholar] [CrossRef]
- Noureen, N.; Rashid, H.; Kalsoom, S. Identification of type-specific anticancer histone deacetylase inhibitors: Road to success. Cancer Chemother. Pharmacol. 2010, 66, 625–633. [Google Scholar] [CrossRef]
- Li, Y.; Seto, E. Hdacs and hdac inhibitors in cancer development and therapy. Cold Spring Harb. Perspect. Med. 2016, 6, a026831. [Google Scholar] [CrossRef] [PubMed]
- Sanaei, M.; Kavoosi, F. Histone deacetylases and histone deacetylase inhibitors: Molecular mechanisms of action in various cancers. Adv. Biomed. Res. 2019, 8, 63. [Google Scholar]
- Chen, I.-C.; Sethy, B.; Liou, J.-P. Recent update of hdac inhibitors in lymphoma. Front. Cell Dev. Biol. 2020, 8, 576391. [Google Scholar] [CrossRef] [PubMed]
- Fournel, M.; Bonfils, C.; Hou, Y.; Yan, P.T.; Trachy-Bourget, M.-C.; Kalita, A.; Liu, J.; Lu, A.-H.; Zhou, N.Z.; Robert, M.-F. Mgcd0103, a novel isotype-selective histone deacetylase inhibitor, has broad spectrum antitumor activity in vitro and in vivo. Mol. Cancer Ther. 2008, 7, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Deroanne, C.F.; Bonjean, K.; Servotte, S.; Devy, L.; Colige, A.; Clausse, N.; Blacher, S.; Verdin, E.; Foidart, J.-M.; Nusgens, B.V. Histone deacetylases inhibitors as anti-angiogenic agents altering vascular endothelial growth factor signaling. Oncogene 2002, 21, 427–436. [Google Scholar] [CrossRef] [PubMed]
- McClure, J.J.; Li, X.; Chou, C.J. Advances and challenges of hdac inhibitors in cancer therapeutics. Adv. Cancer Res. 2018, 138, 183–211. [Google Scholar]
- Hu, Z.; Wei, F.; Su, Y.; Wang, Y.; Shen, Y.; Fang, Y.; Ding, J.; Chen, Y. Histone deacetylase inhibitors promote breast cancer metastasis by elevating nedd9 expression. Signal Transduct. Target. Ther. 2023, 8, 11. [Google Scholar] [CrossRef]
- Hull, E.E.; Montgomery, M.R.; Leyva, K.J. Hdac inhibitors as epigenetic regulators of the immune system: Impacts on cancer therapy and inflammatory diseases. BioMed Res. Int. 2016, 2016, 8797206. [Google Scholar] [CrossRef] [PubMed]
- Adcock, I. Hdac inhibitors as anti-inflammatory agents. Br. J. Pharmacol. 2007, 150, 829–831. [Google Scholar] [CrossRef]
- Karin, M. Nuclear factor-κb in cancer development and progression. Nature 2006, 441, 431–436. [Google Scholar] [CrossRef]
- Petrie, K.; Guidez, F.; Howell, L.; Healy, L.; Waxman, S.; Greaves, M.; Zelent, A. The histone deacetylase 9 gene encodes multiple protein isoforms. J. Biol. Chem. 2003, 278, 16059–16072. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Croteau, S.; Hardy, P. Histone deacetylase (hdac) 9: Versatile biological functions and emerging roles in human cancer. Cell. Oncol. 2021, 44, 997–1017. [Google Scholar] [CrossRef] [PubMed]
- Conte, M.; De Palma, R.; Altucci, L. Hdac inhibitors as epigenetic regulators for cancer immunotherapy. Int. J. Biochem. Cell Biol. 2018, 98, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Cho, E.-H.; Lee, J.-Y. Histone deacetylase 9: Its role in the pathogenesis of diabetes and other chronic diseases. Diabetes Metab. J. 2020, 44, 234–244. [Google Scholar] [CrossRef]
- Han, Z.; Dong, X.; Zhang, C.; Wu, Y.; Yuan, Z.; Wang, X. Polymorphism of hdac9 gene is associated with increased risk of acute coronary syndrome in chinese han population. BioMed Res. Int. 2016, 2016, 3746276. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Q.; Ding, Y.; Liu, Y.; Zhao, D.; Zhao, K.; Shen, Q.; Liu, X.; Zhu, X.; Li, N. Methyltransferase dnmt3a upregulates hdac9 to deacetylate the kinase tbk1 for activation of antiviral innate immunity. Nat. Immunol. 2016, 17, 806–815. [Google Scholar] [CrossRef] [PubMed]
- Ripamonti, C.; Spadotto, V.; Pozzi, P.; Stevenazzi, A.; Vergani, B.; Marchini, M.; Sandrone, G.; Bonetti, E.; Mazzarella, L.; Minucci, S. Hdac inhibition as potential therapeutic strategy to restore the deregulated immune response in severe COVID-19. Front. Immunol. 2022, 13, 841716. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.-C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020, 181, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
- Trionfetti, F.; Alonzi, T.; Bontempi, G.; Terri, M.; Battistelli, C.; Montaldo, C.; Repele, F.; Rotili, D.; Valente, S.; Zwergel, C. Hdac1-3 inhibition increases SARS-CoV-2 replication and productive infection in lung mesothelial and epithelial cells. Front. Cell. Infect. Microbiol. 2023, 13, 1257683. [Google Scholar] [CrossRef]
- Husain, M.; Cheung, C.-Y. Histone deacetylase 6 inhibits influenza a virus release by downregulating the trafficking of viral components to the plasma membrane via its substrate, acetylated microtubules. J. Virol. 2014, 88, 11229–11239. [Google Scholar] [CrossRef]
- Milazzo, G.; Mercatelli, D.; Di Muzio, G.; Triboli, L.; De Rosa, P.; Perini, G.; Giorgi, F.M. Histone deacetylases (hdacs): Evolution, specificity, role in transcriptional complexes, and pharmacological actionability. Genes 2020, 11, 556. [Google Scholar] [CrossRef] [PubMed]
- Bondarev, A.D.; Attwood, M.M.; Jonsson, J.; Chubarev, V.N.; Tarasov, V.V.; Schiöth, H.B. Recent developments of hdac inhibitors: Emerging indications and novel molecules. Br. J. Clin. Pharmacol. 2021, 87, 4577–4597. [Google Scholar] [CrossRef] [PubMed]
- Gatla, H.R.; Muniraj, N.; Thevkar, P.; Yavvari, S.; Sukhavasi, S.; Makena, M.R. Regulation of chemokines and cytokines by histone deacetylases and an update on histone decetylase inhibitors in human diseases. Int. J. Mol. Sci. 2019, 20, 1110. [Google Scholar] [CrossRef] [PubMed]
| High Ab | Low Ab | p-Value | ||
|---|---|---|---|---|
| N | 50 | 56 | ||
| Age, years | 45.9 ± 8.1 | 46.9 ± 7.5 | 0.489 | |
| Sex (male, %) | 24 (48.0) | 33 (58.9) | 0.035 | |
| Body mass index, kg/m2 | 26.4 ± 4.3 | 23.9 ± 3.1 | 0.001 | |
| Glucose, mg/dL | 95.6 ± 19.1 | 99.5 ± 15.9 | 0.256 | |
| Total cholesterol, mg/dL | 182.4 ± 48.7 | 187.0 ± 40.0 | 0.597 | |
| Triglyceride, mg/dL | 118.5 (80.2–203.7) | 127.5 (66.7–196.5) | 0.482 | |
| HDL cholesterol, mg/dL | 47.6 ± 15.6 | 54.2 ± 14.4 | 0.024 | |
| LDL cholesterol, mg/dL | 119.1 ± 42.0 | 119.1 ± 38.6 | 0.990 | |
| Ig E | 69.4 (26.1–170.7) | 47.0 (18.9–102.0) | 0.055 | |
| Vitamin D | 21.4 (16.5–26.9) | 19.8 (13.9–23.3) | 0.430 | |
| WBC ( L) | 6.24 ± 1.62 | 5.65 ± 1.31 | 0.040 | |
| AST (IU/L) | 28.9 ± 10.2 | 22.5 ± 9.7 | 0.166 | |
| ALT (IU/L) | 26.7 ± 18.0 | 24.2 ± 16.8 | 0.463 | |
| Comorbid condition, n (%) | ||||
| Hypertension, (%) | 6 (12.0) | 5 (8.9) | 0.604 | |
| Diabetes, (%) | 4 (8) | 4 (7.1) | 0.867 | |
| Neutralizing Ab Titer | ||||
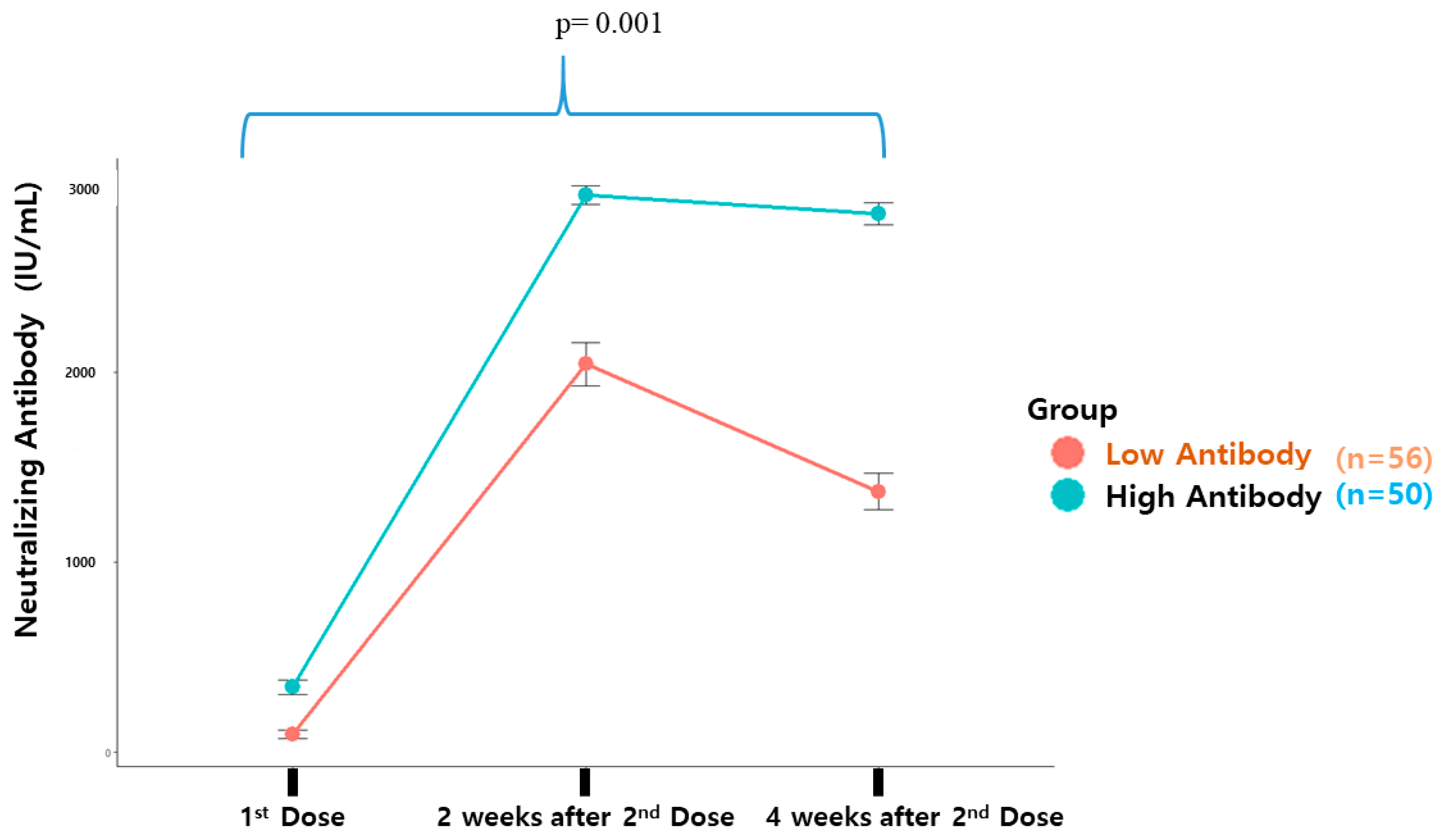
| First vaccination | 342.3 ± 269.0 | 92.7 ± 168.7 | 0.001 | |
| Second vaccination | 2935.2 ± 334.3 | 2044.0 ± 857.9 | 0.001 | |
| 4 Weeks later | 2837.9 ± 410.3 | 1373.3 ± 707.1 | 0.001 | |
| Vaccination, n (%) | ||||
| First dose | Second dose | 0.001 | ||
| Moderna | Moderna | 29 (58.0) | 12 (21.4) | |
| Pfizer | Pfizer | 21 (42.0) | 44 (78.6) | |
| Model 1 | Model 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Chr a | Gene | BP b | SNP c | Minor Allele | MAF | OR (L95–U95) d | p-Value | OR (L95–U95) d | p-Value |
| 7 | HDAC9 | 18833779 | rs7795433 | G | 0.48 | 5.63 (2.51–12.65) | 3 × 10−5 | 6.99 (2.74–17.87) | 5 × 10−5 |
| 8 | MSC-AS1 | 71979513 | rs10111413 | A | 0.50 | 0.24 (0.12–0.47) | 5 × 10−5 | 0.21 (0.09–0.47) | 1 × 10−4 |
| 7 | HDAC9 | 18828330 | rs6951522 | C | 0.47 | 5.18 (2.37–11.36) | 4 × 10−5 | 6.76 (2.69–16.96) | 5 × 10−5 |
| 7 | HDAC9 | 18838251 | rs2073963 | G | 0.48 | 5.14 (2.35–11.24) | 4 × 10−5 | 6.24 (2.53–15.37) | 7 × 10−5 |
| 4 | NDST4/MIR1973 | 115308340 | rs118002192 | C | 0.13 | 0.12 (0.04–0.38) | 4 × 10−4 | 0.17 (0.05–0.60) | 6 × 10−3 |
| 11 | HTR3A | 113984620 | rs1176717 | A | 0.21 | 0.23 (0.10–0.54) | 7 × 10−4 | 0.31 (0.12–0.79) | 1 × 10−2 |
| 7 | AUTS2 | 70660723 | rs17141963 | T | 0.47 | 0.20 (0.17–0.63) | 8 × 10−4 | 0.26 (0.12–0.59) | 1 × 10−3 |
| 7 | HDAC9 | 18837993 | rs957958 | G | 0.50 | 4.42 (2.12–9.22) | 8 × 10−5 | 5.04 (2.20–11.56) | 1 × 10−4 |
| 6 | LOC105377862 | 3491893 | rs7742726 | C | 0.36 | 3.73 (1.69–8.25) | 1 × 10−3 | 3.58 (1.52–8.44) | 4 × 10−3 |
| 7 | HDAC9 | 18837785 | rs957960 | A | 0.47 | 5.02 (2.29–11.00) | 6 × 10−5 | 6.23 (2.51–15.45) | 8 × 10−5 |
| Antibody Production | rs7795433 Genotype | rs7795433 Allele | |||||
|---|---|---|---|---|---|---|---|
| AA | GA | GG | Total (n) | A | G | Total (n) | |
| High (%) | Group A | Group B | Group C | Effect Alle | Non-Effect Alle | ||
| 16 (32%) | 26 (52%) | 8 (16%) | 50 | 58 (58%) | 42 (42%) | 100 | |
| Low (%) | Group D | Group E | Group F | ||||
| 12 (21%) | 30 (54%) | 14 (25%) | 56 | 54 (48%) | 58 (52%) | 112 | |
| Average Delta ct per Sample (Hdac9–GapDH) | ||||||
|---|---|---|---|---|---|---|
| AA | GA | GG | ||||
| MM | Mm | mm | ||||
| Group A | Group B | Group C | ||||
| R374 | 4.088497 | R082 | 5.982103 | R035 | NA | |
| R434 | 4.125577 | R362 | 4.662499 | R391 | NA | |
| R503 | 3.808289 | R436 | 5.774843 | R498 | 4.644077 | |
| Group D | Group E | Group F | ||||
| R501 | 5.253894 | R038 | 6.569895 | R360 | 4.760575 | |
| R383 | 5.220209 | R386 | 6.568871 | R365 | 4.2658 | |
| R471 | 4.61415 | R415 | 6.731386 | R504 | 4.924133 | |
| Average Delta ct for Each Group (Hdac9–GapDH) | ||||||
| MM | Mm | mm | ||||
| Group A | Group B | Group C | ||||
| Mean and SD | 4.007454 | 0.173476 | 5.473148 | 0.70965 | 4.644077 | 0 |
| Group D | Group E | Group F | ||||
| Mean and SD | 5.029417 | 0.360026 | 6.623384 | 0.093534 | 4.650169 | 0.342772 |
| Relative Quantitative Comparison between Each Group | ||||||
| A vs. D | B vs. E | C vs. F | ||||
| MM | Mn | mm | ||||
| Delta delta CT | −1.02196 | −1.15024 | −0.00609 | |||
| RQ Ct (relative quantification) | 2.03068 | 2.219501 | 1.004232 | |||
| t-test p-value | 0.011429 | 0.04965 | 0.09891 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ryu, H.-E.; Yoon, J.; Choi, J.-E.; Heo, S.-J.; Hong, K.-W.; Jung, D.-H. The Human Genetic Differences in the Outcomes of mRNA Vaccination against COVID-19: A Prospective Cohort Study. Vaccines 2024, 12, 626. https://doi.org/10.3390/vaccines12060626
Ryu H-E, Yoon J, Choi J-E, Heo S-J, Hong K-W, Jung D-H. The Human Genetic Differences in the Outcomes of mRNA Vaccination against COVID-19: A Prospective Cohort Study. Vaccines. 2024; 12(6):626. https://doi.org/10.3390/vaccines12060626
Chicago/Turabian StyleRyu, Ha-Eun, Jihyun Yoon, Ja-Eun Choi, Seok-Jae Heo, Kyung-Won Hong, and Dong-Hyuk Jung. 2024. "The Human Genetic Differences in the Outcomes of mRNA Vaccination against COVID-19: A Prospective Cohort Study" Vaccines 12, no. 6: 626. https://doi.org/10.3390/vaccines12060626







