Immuno-Microbial Signature of Vaccine-Induced Immunity against SARS-CoV-2
Abstract
1. Introduction
2. Materials and Methods
2.1. Participant Recruitment and Consent
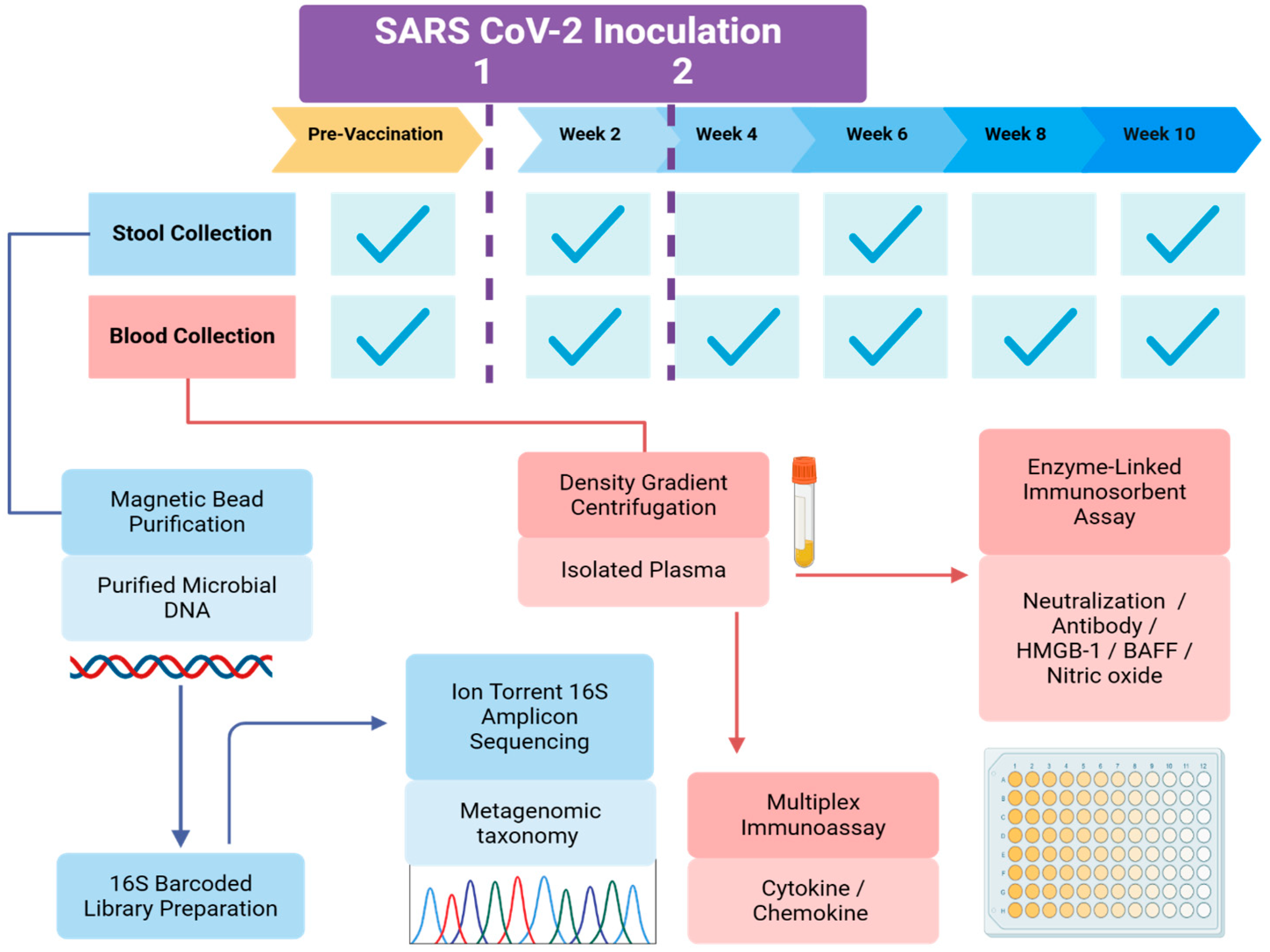
2.2. Sample and Data Collection
2.3. Antiviral Response: Anti-SARS-CoV-2 Neutralization Titer
2.4. Immune Response Profiling
2.5. Gut Microbiota Evaluation
2.6. Statistical Data Analysis
3. Results
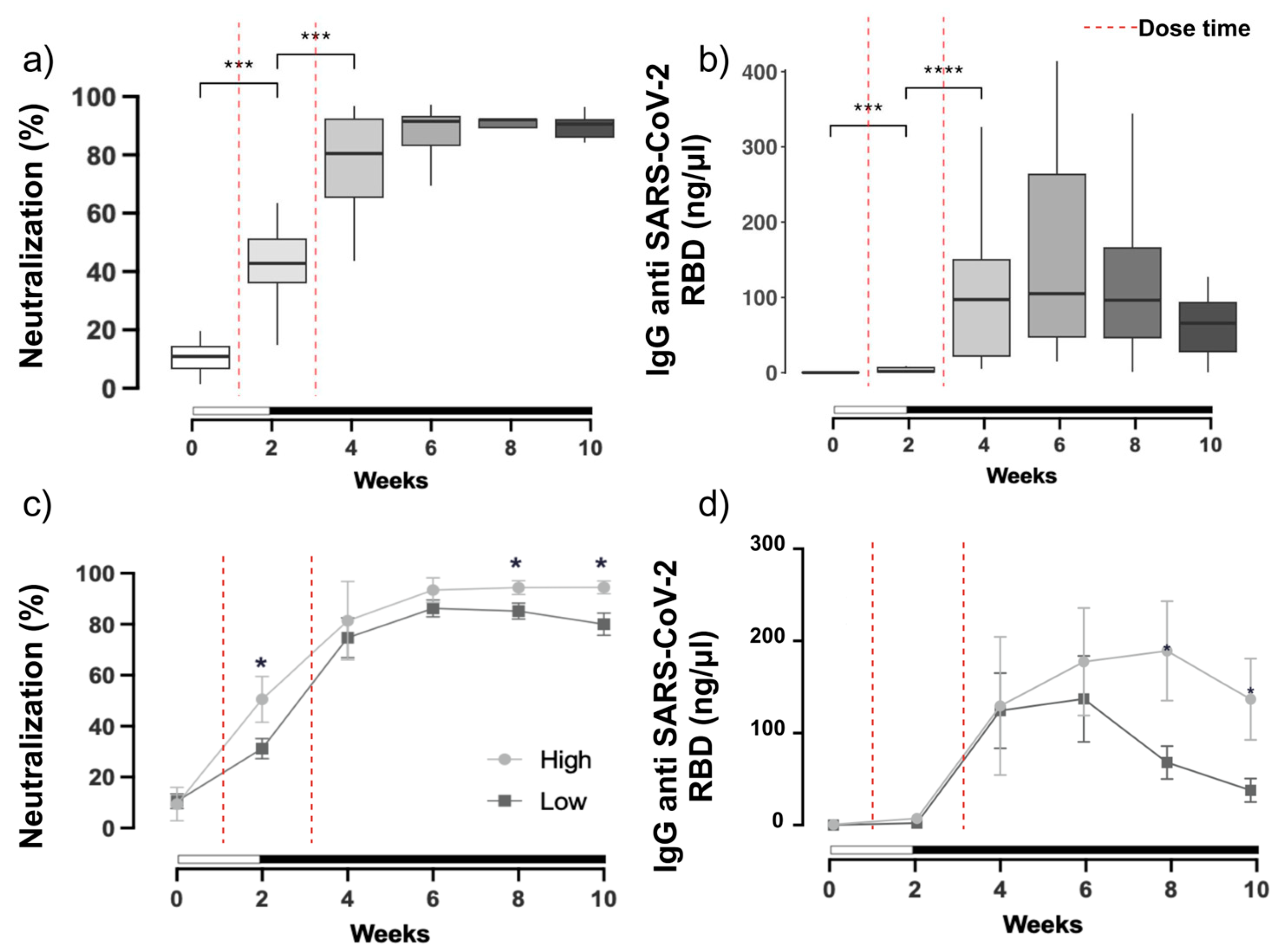
3.1. SARS-CoV-2 Neutralization and Antibody Production
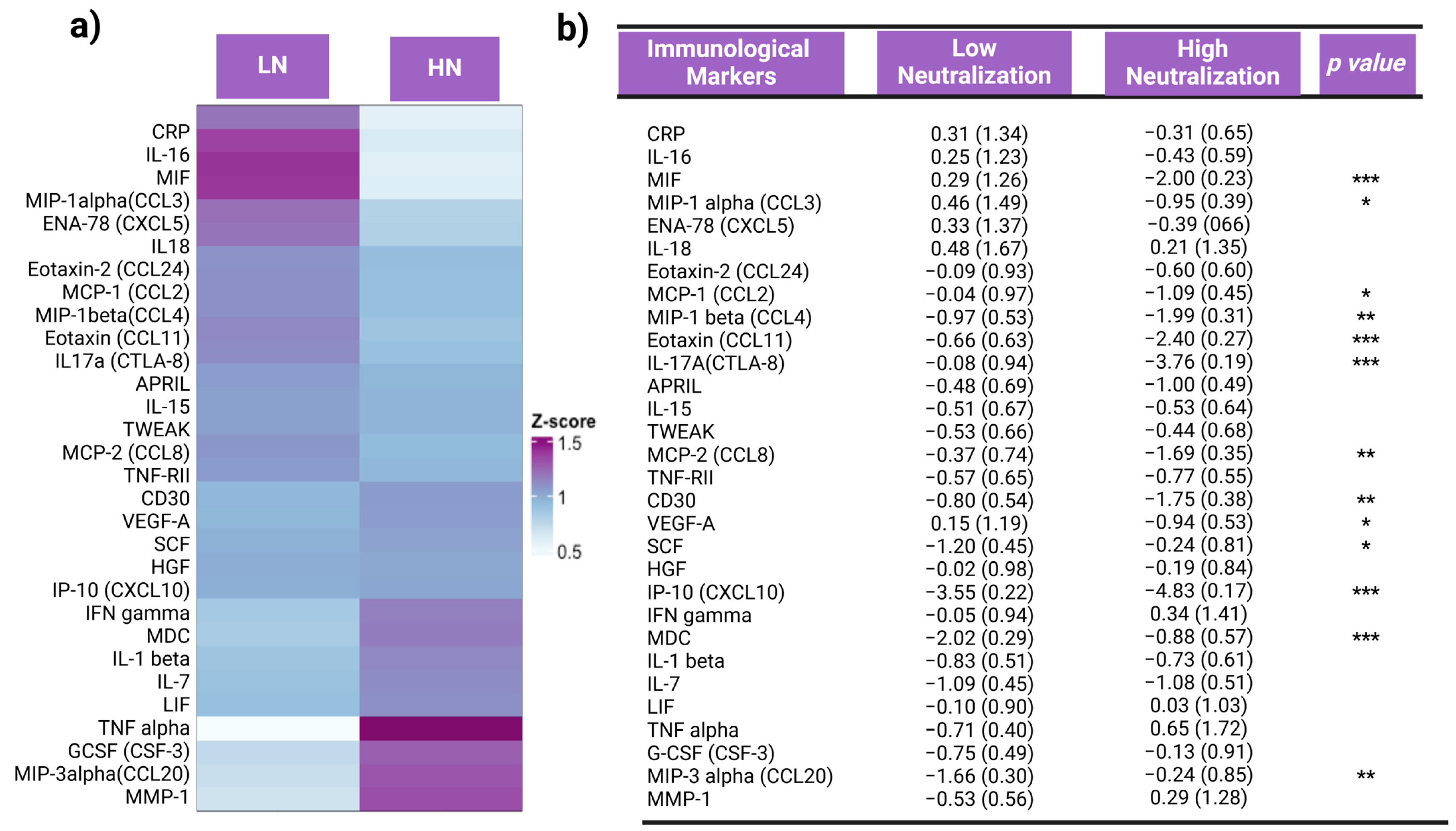
3.2. Inflammation Status and Inflammatory Responses
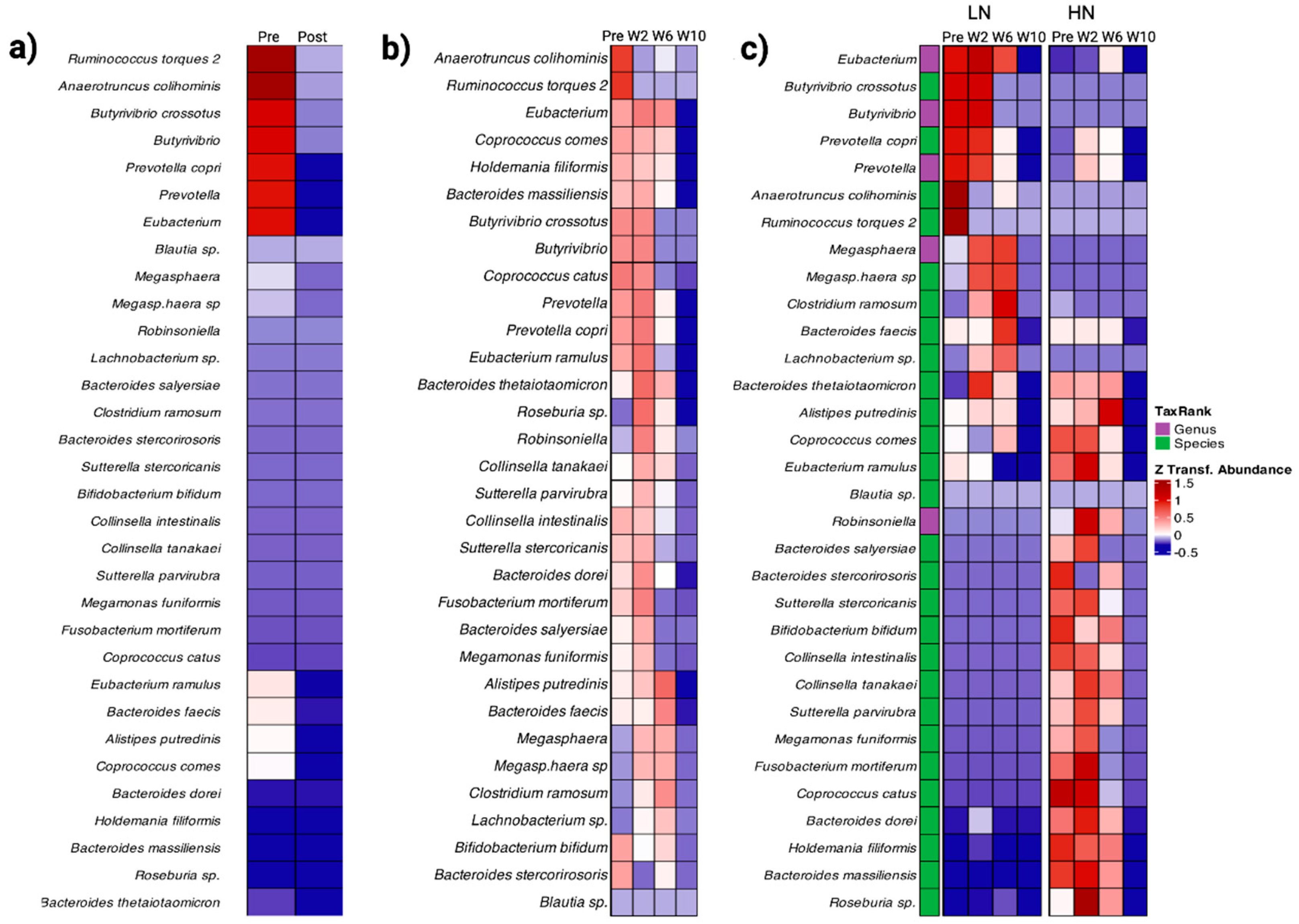
3.3. Gut Microbiome Composition
3.4. Inflammation Pathway Analysis
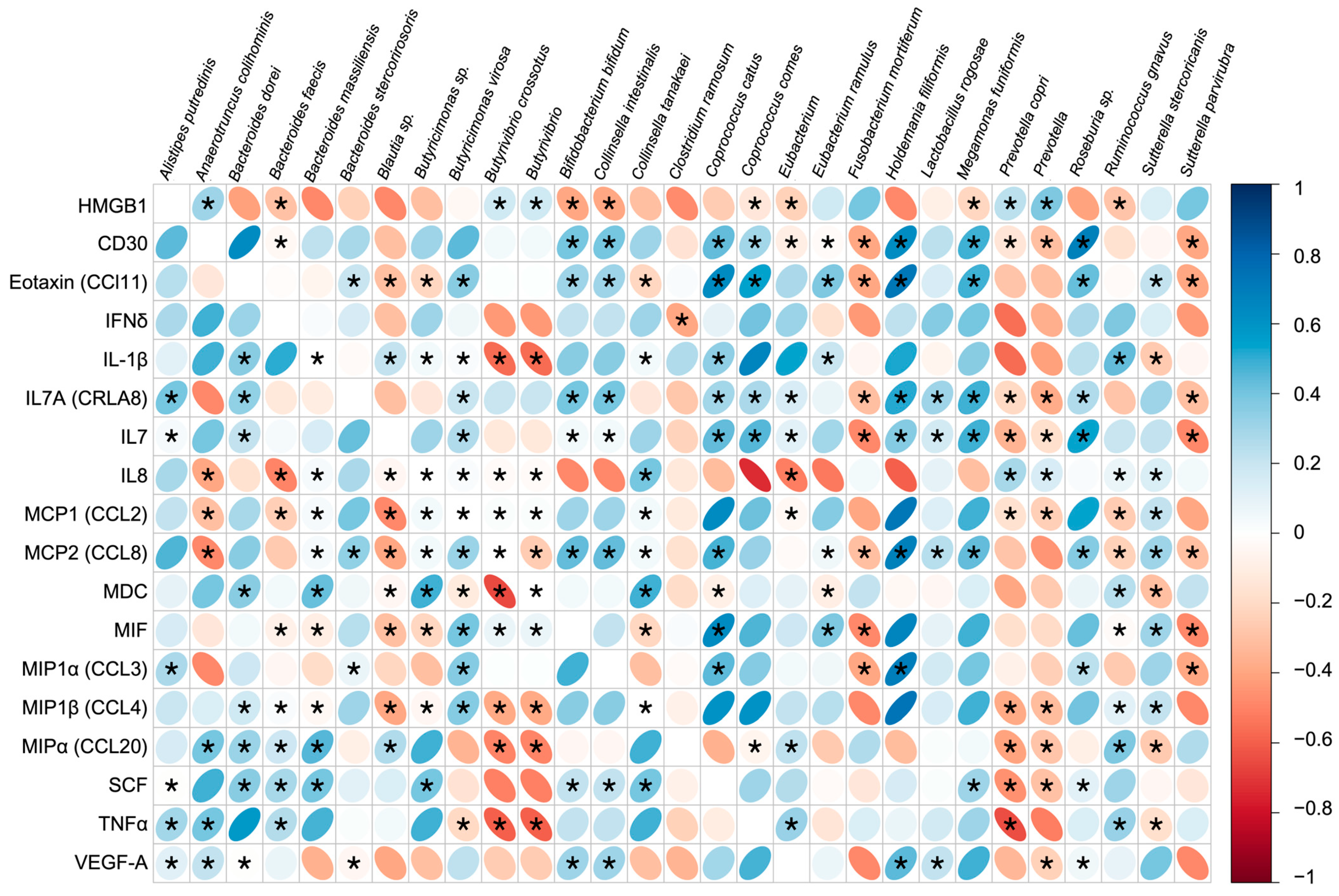
3.5. Immune Response and Gut Microbiota Composition
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shereen, M.A.; Khan, S.; Kazmi, A.; Bashir, N.; Siddique, R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020, 24, 91–98. [Google Scholar] [CrossRef] [PubMed]
- FDA Takes Key Action in Fight against COVID-19 by Issuing Emergency Use Authorization for First COVID-19 Vaccine. Available online: https://www.fda.gov/news-events/press-announcements/fda-takes-key-action-fight-against-covid-19-issuing-emergency-use-authorization-first-covid-19 (accessed on 20 December 2023).
- Barman, S.; Soni, D.; Brook, B.; Nanishi, E.; Dowling, D.J. Precision Vaccine Development: Cues from Natural Immunity. Front. Immunol. 2022, 12, 662218. [Google Scholar] [CrossRef] [PubMed]
- Wiertsema, S.P.; van Bergenhenegouwen, J.; Garssen, J.; Knippels, L.M.J. The Interplay between the Gut Microbiome and the Immune System in the Context of Infectious Diseases throughout Life and the Role of Nutrition in Optimizing Treatment Strategies. Nutrients 2021, 13, 886. [Google Scholar] [CrossRef] [PubMed]
- Wells, R.K.; Kunihiro, B.P.; Phankitnirundorn, K.; Peres, R.; McCracken, T.A.; Umeda, L.; Lee, R.H.; Kim, D.Y.; Juarez, R.; Maunakea, A.K. Gut microbial indicators of metabolic health underlie age-related differences in obesity and diabetes risk among Native Hawaiians and Pacific Islanders. Frontiers in cellular and infection microbiology. Front. Cell. Infect. Microbiol. 2022, 12, 1035641. [Google Scholar] [CrossRef] [PubMed]
- Becerra, C.Y.; Wells, R.K.; Kunihiro, B.P.; Lee, R.H.; Umeda, L.; Allan, N.P.; Rubas, N.C.; McCracken, T.A.; Nunokawa, C.K.L.; Lee, M.H.; et al. Examining the immunoepigenetic-gut microbiome axis in the context of self-esteem among Native Hawaiians and other Pacific Islanders. Front. Genet. 2023, 14, 1125217. [Google Scholar] [CrossRef] [PubMed]
- Lynn, D.J.; Benson, S.C.; Lynn, M.A.; Pulendran, B. Modulation of immune responses to vaccination by the microbiota: Implications and potential mechanisms. Nat. Rev. Immunol. 2022, 22, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Schwiertz, A.; Taras, D.; Schäfer, K.; Beijer, S.; Bos, N.A.; Donus, C.; Hardt, P.D. Microbiota and SCFA in lean and overweight healthy subjects. Obesity 2010, 18, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Christ, A.; Lauterbach, M.; Latz, E. Western Diet and the Immune System: An Inflammatory Connection. Immunity 2019, 51, 794–811. [Google Scholar] [CrossRef] [PubMed]
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef]
- Fourati, S.; Cristescu, R.; Loboda, A.; Talla, A.; Filali, A.; Railkar, R.; Schaeffer, A.K.; Favre, D.; Gagnon, D.; Peretz, Y.; et al. Pre-vaccination inflammation and B-cell signalling predict age-related hyporesponse to hepatitis B vaccination. Nat. Commun. 2016, 7, 10369. [Google Scholar] [CrossRef]
- De Heredia, F.P.; Gómez-Martínez, S.; Marcos, A. Obesity, inflammation and the immune system. Proc. Nutr. Soc. 2012, 71, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Dumitriu, I.E.; Baruah, P.; Manfredi, A.A.; Bianchi, M.E.; Rovere-Querini, P. HMGB1: An immmune odyssey. Discov. Med. 2005, 5, 388–392. [Google Scholar] [PubMed]
- Rubas, N.C.; Peres, R.; Kunihiro, B.P.; Allan, N.P.; Phankitnirundorn, K.; Wells, R.K.; McCraken, T.A.; Lee, R.H.; Umeda, L.; Conching, A.; et al. HMGB1 mediates microbiome-immune axis dysregulation underlying reduced neutralization capacity in obesity-related post-acute sequelae of SARS-CoV-2. Sci. Rep. 2024, 14, 355. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.L.P.; Minchin, P.; O’Hara, B.; Simpson, G.; Solymos, P.; Stevens, H.; Wagner, H. Vegan: Community Ecology Package; R Package Version 2.2-1, 2.1-2. Available online: https://github.com/vegandevs/vegan (accessed on 20 January 2023).
- Chamberlain, S.A.; Szöcs, E. Taxize: Taxonomic search and retrieval in R. F1000Research 2013, 2, 191. [Google Scholar] [CrossRef] [PubMed]
- Lahti, L.; Shetty, S. Tools for Microbiome Analysis in R. Microbiome R Package; Version 2012–2019. 2017. Available online: https://microbiome.github.io/tutorials/ (accessed on 20 January 2023).
- Kleanthous, H.; Silverman, J.M.; Makar, K.W.; Yoon, I.-K.; Jackson, N.; Vaughn, D.W. Scientific rationale for developing potent RBD-based vaccines targeting COVID-19. npj Vaccines 2021, 6, 128. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Wang, J.; Guo, C.; Cai, L.; Liao, C.; Yi, H.; Li, Q.; Hu, H.; Deng, Q.; Lu, Y.; Guo, Z.; et al. Pre-Existing Cross-Reactive Antibody Responses Do Not Significantly Impact Inactivated COVID-19 Vaccine-Induced Neutralization. Front. Immunol. 2021, 12, 772511. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Then, E.; Lucas, C.; Monteiro, V.S.; Miric, M.; Brache, V.; Cochon, L.; Vogels, C.B.F.; Malik, A.A.; De la Cruz, E.; Jorge, A.; et al. Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat. Med. 2022, 28, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Panova, N.; Allan, N.P.; Rubas, N.C.; Lee, R.H.; Kunihiro, B.P.; Umeda, L.; Peres, R.; Juarez, R.; Maunakea, A.K. Sequencing the SARS-CoV-2 Genome from Stool Samples of Post-acute Cases Implicates a Novel Mutation Associated with Reduced Antibody Neutralization. Eur. J. Biomed. Res. 2023, 2, 17–23. [Google Scholar] [CrossRef]
- Jiang, Y.; Rubin, L.; Peng, T.; Liu, L.; Xing, X.; Lazarovici, P.; Zheng, W. Cytokine storm in COVID-19: From viral infection to immune responses, diagnosis and therapy. Int. J. Biol. Sci. 2022, 18, 459–472. [Google Scholar] [CrossRef]
- Wulandari, S.; Hartono, W.T. The role of HMGB1 in COVID-19-induced cytokine storm and its potential therapeutic targets: A review. Immunology 2023, 169, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Street, M.E. HMGB1: A Possible Crucial Therapeutic Target for COVID-19? Horm. Res. Paediatr. 2020, 93, 73–75. [Google Scholar] [CrossRef] [PubMed]
- Guilloteau, P.; Martin, L.; Eeckhaut, V.; Ducatelle, R.; Zabielski, R.; Van Immerseel, F. From the gut to the peripheral tissues: The multiple effects of butyrate. Nutr. Res. Rev. 2010, 23, 366–384. [Google Scholar] [CrossRef]
- Liu, S.; Chen, H.-Z.; Xu, Z.-D.; Wang, F.; Fang, H.; Bellanfante, O.; Chen, X.-L. Sodium butyrate inhibits the production of HMGB1 and attenuates severe burn plus delayed resuscitation-induced intestine injury via the p38 signaling pathway. Burns 2019, 45, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.T.; Cresci, G.A.M. The Immunomodulatory Functions of Butyrate. J. Inflamm. Res. 2021, 14, 6025–6041. [Google Scholar]
- Liu, H.; Wang, J.; He, T.; Becker, S.; Zhang, G.; Li, D.; Ma, X. Butyrate: A Double-Edged Sword for Health? Adv. Nutr. 2018, 9, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Castaño, G.P.; Dorris, M.R.; Liu, X.; Bolling, B.W.; Acosta-Gonzalez, A.; Rey, F.E. Bacteroides thetaiotaomicron Starch Utilization Promotes Quercetin Degradation and Butyrate Production by Eubacterium ramulus. Front. Microbiol. 2019, 10, 1145. [Google Scholar] [CrossRef] [PubMed]
- Nie, K.; Ma, K.; Luo, W.; Shen, Z.; Yang, Z.; Xiao, M.; Tong, T.; Yang, Y.; Wang, X. Roseburia intestinalis: A Beneficial Gut Organism From the Discoveries in Genus and Species. Front. Cell Infect. Microbiol. 2021, 11, 757718. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.M. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 2017, 151, 363–374. [Google Scholar] [CrossRef]
- Precup, G.; Vodnar, D.C. Gut Prevotella as a possible biomarker of diet and its eubiotic versus dysbiotic roles: A comprehensive literature review. Br. J. Nutr. 2019, 122, 131–140. [Google Scholar] [CrossRef]
- Castrodeza-Sanz, J.; Sanz-Muñoz, I.; Eiros, J.M. Adjuvants for COVID-19 Vaccines. Vaccines 2023, 11, 902. [Google Scholar] [CrossRef] [PubMed]
- Magna, M.; Pisetsky, D.S. The role of HMGB1 in the pathogenesis of inflammatory and autoimmune diseases. Mol. Med. 2014, 20, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Svetkauskaite, D.; He, Q.; Kim, J.Y.; Strassheim, D.; Ishizaka, A.; Abraham, E. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J. Biol. Chem. 2004, 279, 7370–7377. [Google Scholar] [CrossRef] [PubMed]
- Candelli, M.; Franza, L.; Pignataro, G.; Ojetti, V.; Covino, M.; Piccioni, A.; Gasbarrini, A.; Franceschi, F. Interaction between Lipopolysaccharide and Gut Microbiota in Inflammatory Bowel Diseases. Int. J. Mol. Sci. 2021, 22, 6242. [Google Scholar] [CrossRef] [PubMed]
- An, L.; Wirth, U.; Koch, D.; Schirren, M.; Drefs, M.; Koliogiannis, D.; Nieß, H.; Andrassy, J.; Guba, M.; Bazhin, A.V.; et al. The Role of Gut-Derived Lipopolysaccharides and the Intestinal Barrier in Fatty Liver Diseases. J. Gastrointest. Surg. 2022, 26, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Si, J.; You, H.J.; Yu, J.; Sung, J.; Ko, G. Prevotella as a Hub for Vaginal Microbiota under the Influence of Host Genetics and Their Association with Obesity. Cell Host Microbe 2017, 21, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Vicentino, A.R.R.; Fraga-Junior, V.D.S.; Palazzo, M.; Tasmo, N.R.A.; Rodrigues, D.A.S.; Barroso, S.P.C.; Ferreira, S.N.; Neves-Borges, A.C.; Allonso, D.; Fantappié, M.R.; et al. High mobility group box 1, ATP, lipid mediators, and tissue factor are elevated in COVID-19 patients: HMGB1 as a biomarker of worst prognosis. Clin. Transl. Sci. 2023, 16, 631–646. [Google Scholar] [CrossRef]
- Chen, R.; Huang, Y.; Quan, J.; Liu, J.; Wang, H.; Billiar, T.R.; Lotze, M.T.; Zeh, H.J.; Kang, R.; Tang, D. HMGB1 as a potential biomarker and therapeutic target for severe COVID-19. Heliyon 2020, 6, e05672. [Google Scholar]

| Characteristics | Total | LN (n = 7) | HN (n = 7) | p-Value | |
|---|---|---|---|---|---|
| Age (±SD), age | 32.1 (±9.9) | 32.7 (±12.2) | 31.4 (±8.2) | 0.820 | |
| Residence time in Hawaii, years (±SD) | 14.1 (±10.3) | 14.2 (±10.4) | 14.0 (±10.9) | 0.981 | |
| Baseline cross-reactivity, pre-vaccine, % (±SD) | 10.0 (±7.0) | 10.6 (±7.9) | 9.5 (±6.6) | 0.768 | |
| BMI | kg/m2 (±SD) | 25.3 (±4.5) | 24.4 (±6.0) | 26.2 (±2.4) | 0.472 |
| Sex | Female | 7 (50%) | 4 (57%) | 3 (43%) | >0.999 |
| Male | 7 (50%) | 3 (43%) | 4 (57%) | ||
| Ethnicity | Asian | 8 (57%) | 4 (57%) | 4 (57%) | >0.999 |
| Non-Asian | 6 (43%) | 3 (43%) | 3 (43%) | ||
| Vaccine brand | Pfizer | 11 (50%) | 6 (86%) | 5 (72%) | 0.768 |
| Moderna | 3 (50%) | 1 (14%) | 2 (28%) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Umeda, L.; Torres, A.; Kunihiro, B.P.; Rubas, N.C.; Wells, R.K.; Phankitnirundorn, K.; Peres, R.; Juarez, R.; Maunakea, A.K. Immuno-Microbial Signature of Vaccine-Induced Immunity against SARS-CoV-2. Vaccines 2024, 12, 637. https://doi.org/10.3390/vaccines12060637
Umeda L, Torres A, Kunihiro BP, Rubas NC, Wells RK, Phankitnirundorn K, Peres R, Juarez R, Maunakea AK. Immuno-Microbial Signature of Vaccine-Induced Immunity against SARS-CoV-2. Vaccines. 2024; 12(6):637. https://doi.org/10.3390/vaccines12060637
Chicago/Turabian StyleUmeda, Lesley, Amada Torres, Braden P. Kunihiro, Noelle C. Rubas, Riley K. Wells, Krit Phankitnirundorn, Rafael Peres, Ruben Juarez, and Alika K. Maunakea. 2024. "Immuno-Microbial Signature of Vaccine-Induced Immunity against SARS-CoV-2" Vaccines 12, no. 6: 637. https://doi.org/10.3390/vaccines12060637
APA StyleUmeda, L., Torres, A., Kunihiro, B. P., Rubas, N. C., Wells, R. K., Phankitnirundorn, K., Peres, R., Juarez, R., & Maunakea, A. K. (2024). Immuno-Microbial Signature of Vaccine-Induced Immunity against SARS-CoV-2. Vaccines, 12(6), 637. https://doi.org/10.3390/vaccines12060637





