Evolution of the Antigenic Landscape in Children and Young Adults with COVID-19 and MIS-C
Abstract
:1. Introduction
2. Methods
2.1. Ethics Statement
2.2. Study Design
2.3. Lentivirus Pseudovirion Neutralization Assay (PsVNA)
2.4. Antibody Binding Kinetics to SARS-CoV-2 RBD by Surface Plasmon Resonance (SPR)
2.5. Statistical Analysis
2.6. Antigenic Landscape and Cartography
2.7. Code Availability
3. Results
3.1. Study Cohort
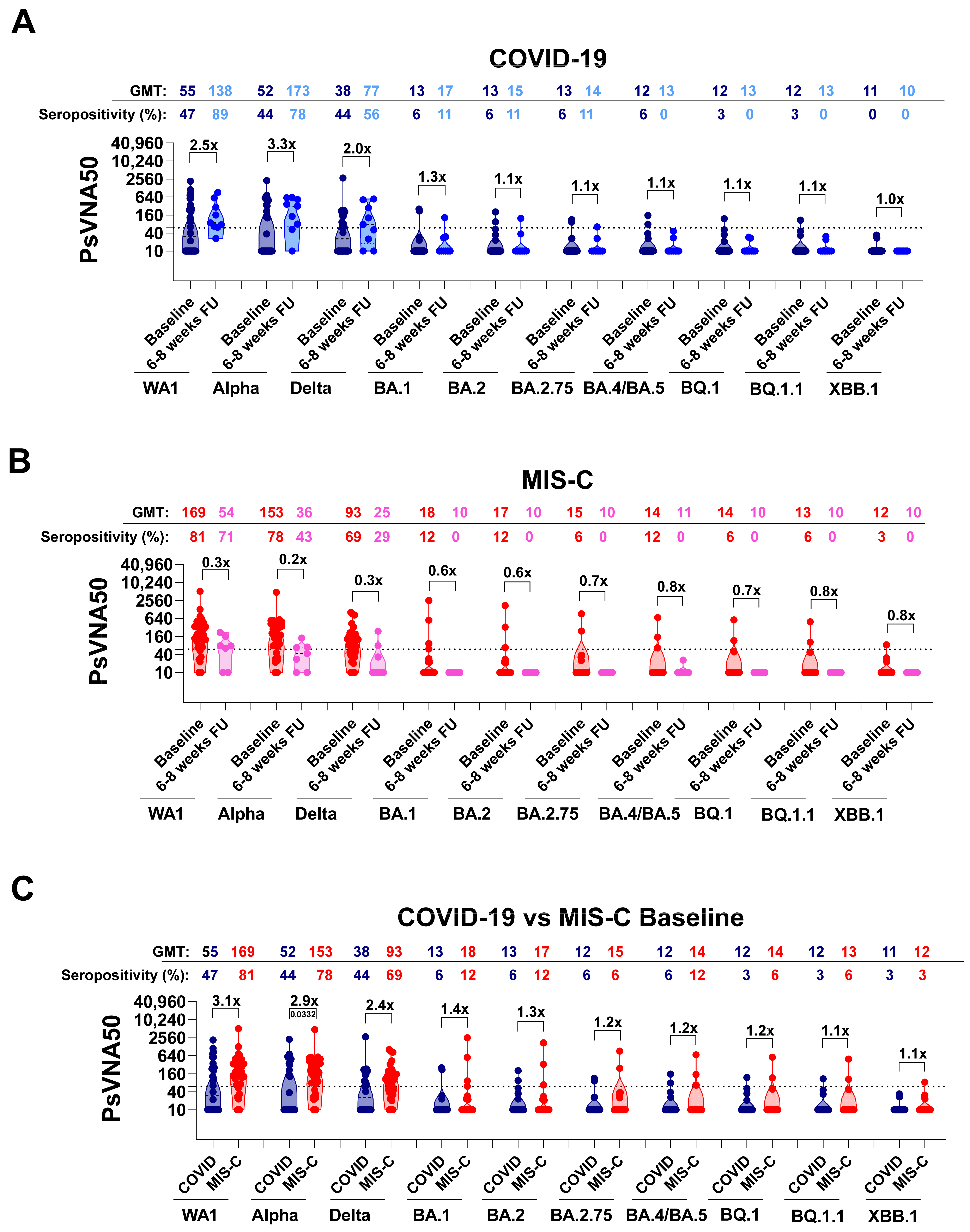
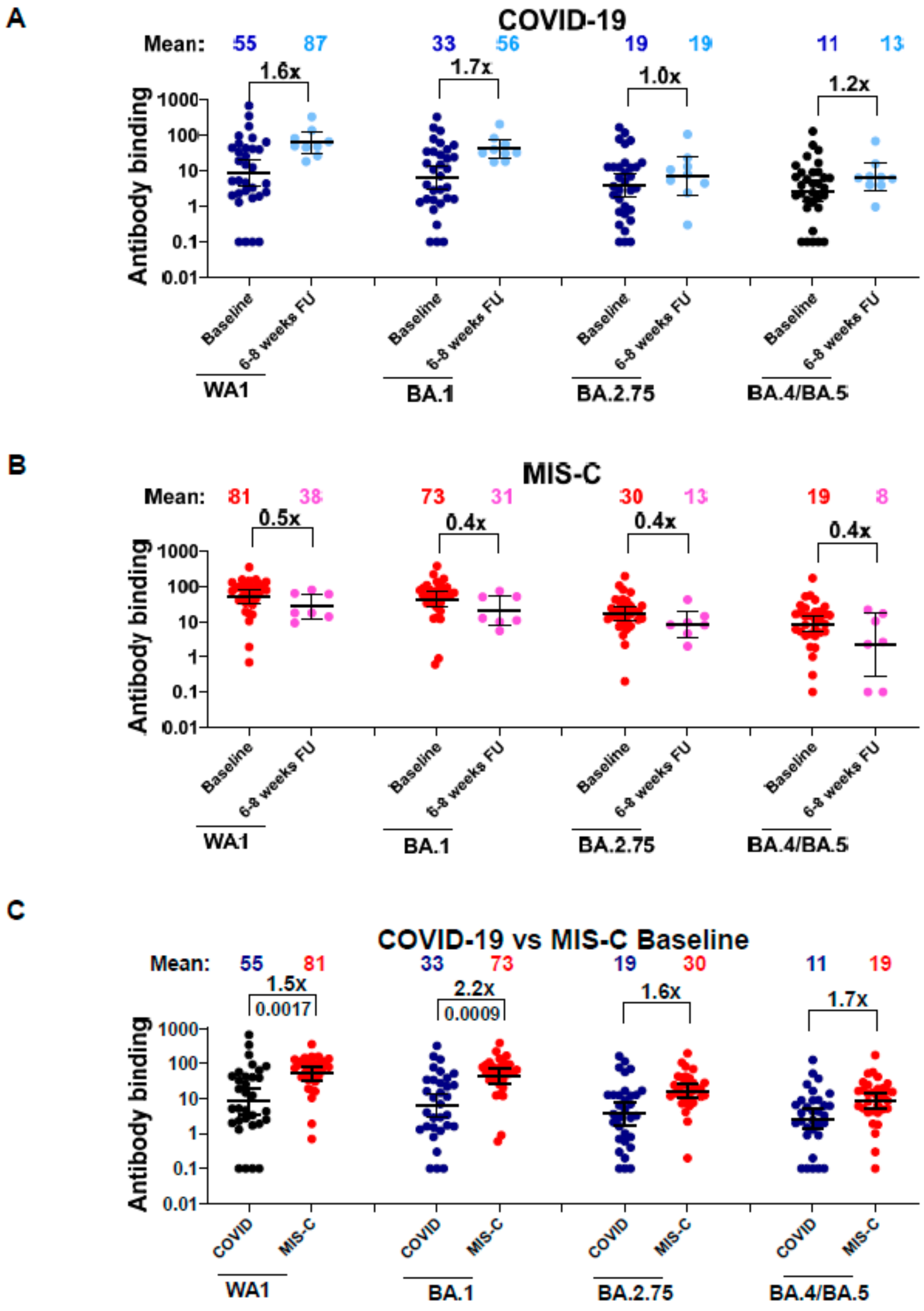
3.2. Neutralizing Antibodies in Children with COVID-19 vs. MIS-C against Circulating SARS-CoV-2 Variants
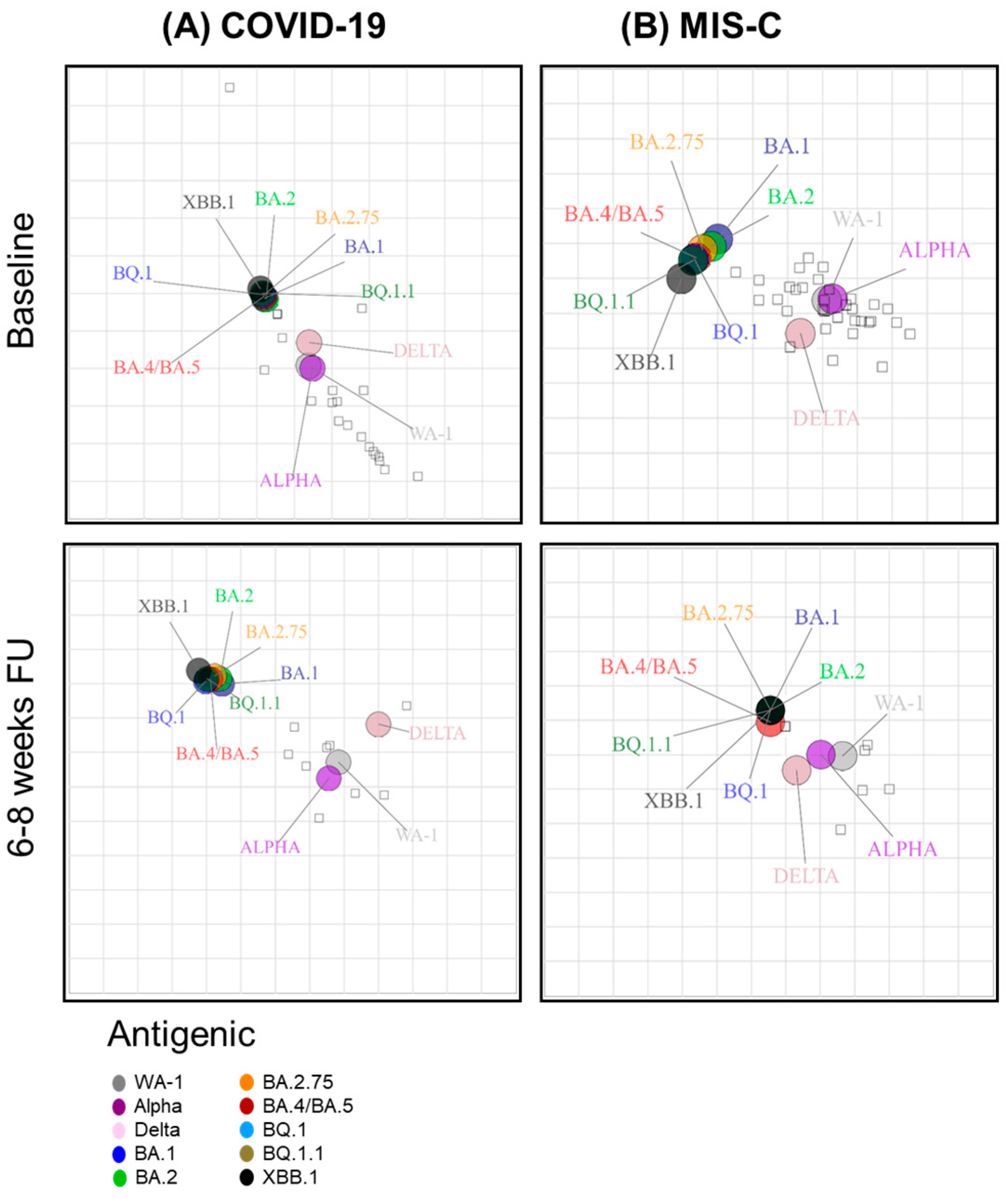
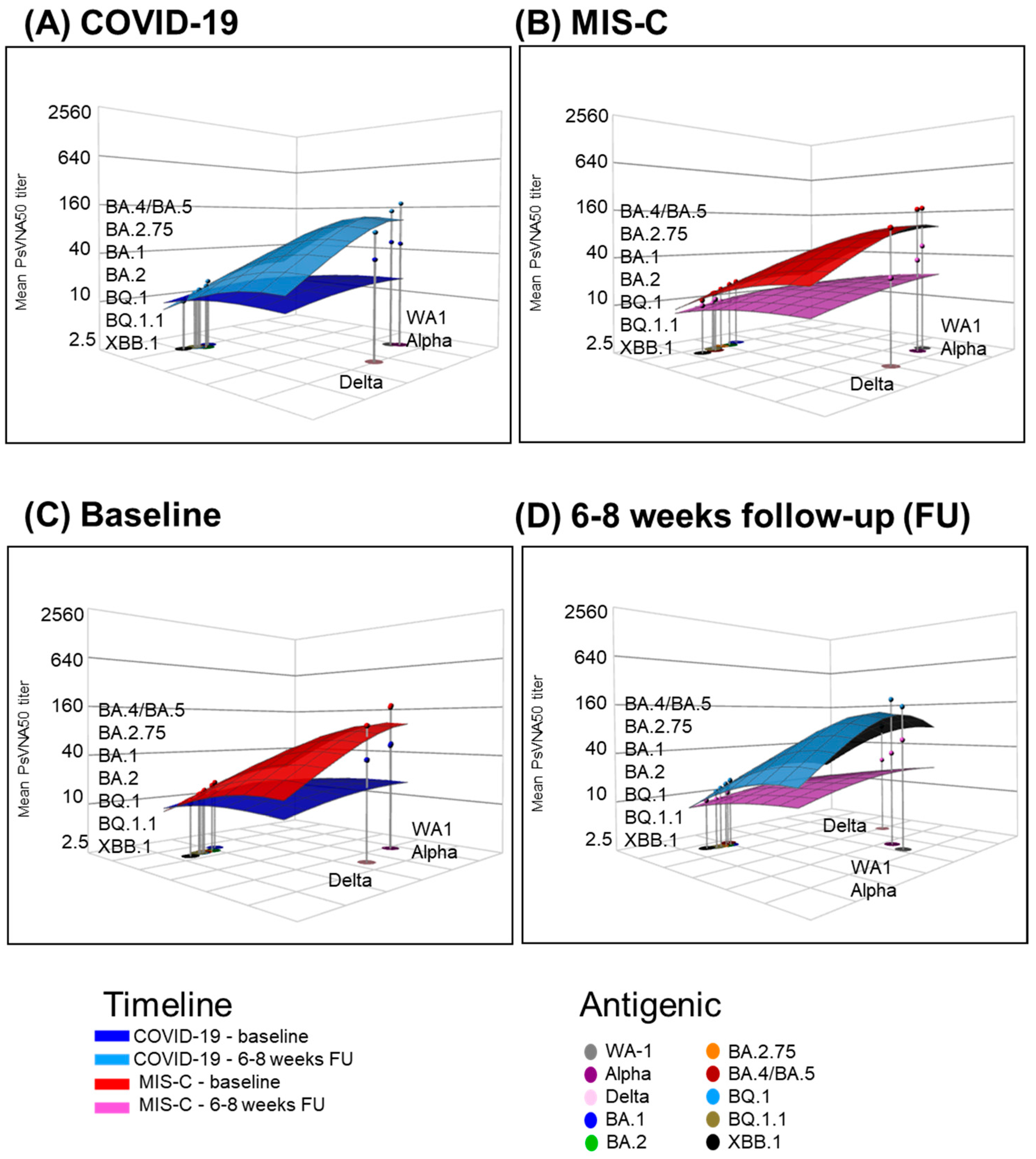
3.3. Neutralizing Antibody Landscapes in Children with COVID-19 vs. MIS-C
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- UN News. Available online: https://news.un.org/en/story/2023/05/1136367#:~:text=The%20head%20of%20the%20UN,no%20longer%20a%20global%20threat (accessed on 2 March 2024).
- CDC. COVID Data Tracker. Available online: https://covid.cdc.gov/covid-data-tracker/#datatracker-home (accessed on 2 December 2023).
- Liu, L.; Iketani, S.; Guo, Y.; Chan, J.F.; Wang, M.; Liu, L.; Luo, Y.; Chu, H.; Huang, Y.; Nair, M.S.; et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 2022, 602, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Saunders, N.; Maes, P.; Guivel-Benhassine, F.; Planchais, C.; Buchrieser, J.; Bolland, W.H.; Porrot, F.; Staropoli, I.; Lemoine, F.; et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2022, 602, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Kurhade, C.; Zou, J.; Xia, H.; Liu, M.; Chang, H.C.; Ren, P.; Xie, X.; Shi, P.Y. Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1, and XBB.1 by parental mRNA vaccine or a BA.5-bivalent booster. Nat. Med. 2022, 29, 344–347. [Google Scholar] [CrossRef] [PubMed]
- Miller, N.L.; Clark, T.; Raman, R.; Sasisekharan, R. Insights on the mutational landscape of the SARS-CoV-2 Omicron variant receptor-binding domain. Cell Rep. Med. 2022, 3, 100527. [Google Scholar] [CrossRef] [PubMed]
- Uraki, R.; Ito, M.; Furusawa, Y.; Yamayoshi, S.; Iwatsuki-Horimoto, K.; Adachi, E.; Saito, M.; Koga, M.; Tsutsumi, T.; Yamamoto, S.; et al. Humoral immune evasion of the omicron subvariants BQ. 1.1 and XBB. Lancet Infect. Dis. 2023, 23, 30–32. [Google Scholar] [CrossRef] [PubMed]
- Bi, Q.; Wu, Y.; Mei, S.; Ye, C.; Zou, X.; Zhang, Z.; Liu, X.; Wei, L.; Truelove, S.A.; Zhang, T.; et al. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: A retrospective cohort study. Lancet Infect. Dis. 2020, 20, 911–919. [Google Scholar] [CrossRef] [PubMed]
- CDC. COVID Data Tracker: Multisystem Inflammatory Syndrome in Children (MIS-C). Available online: https://www.cdc.gov/mmwr/volumes/73/wr/mm7310a2.htm#:~:text=Top-,Multisystem%20inflammatory%20syndrome%20in%20children%20(MIS%2DC)%20is%20a,involvement%20(1%2C2 (accessed on 16 March 2024).
- Nakra, N.A.; Blumberg, D.A.; Herrera-Guerra, A.; Lakshminrusimha, S. Multi-system inflammatory syndrome in children (MIS-C) following SARS-CoV-2 infection: Review of clinical presentation, hypothetical pathogenesis, and proposed management. Children 2020, 7, 69. [Google Scholar] [CrossRef] [PubMed]
- Godfred-Cato, S.; Bryant, B.; Leung, J.; Oster, M.E.; Conklin, L.; Abrams, J.; Roguski, K.; Wallace, B.; Prezzato, E.; Koumans, E.H.; et al. COVID-19–associated multisystem inflammatory syndrome in children—United States, March–July 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 1074. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention 2024. Available online: https://www.cdc.gov/ncird/whats-new/covid-19-vaccine-effectiveness.html#:~:text=Updated%20COVID%2D19%20vaccine%20protects%20against%20many%20variants&text=People%20who%20received%20the%20updated,1%20variant (accessed on 1 February 2024).
- Hill, H.A.; Yankey, D.; Elam-Evans, L.D.; Chen, M.; Singleton, J.A. Vaccination Coverage by Age 24 Months among Children Born in 2019 and 2020—National Immunization Survey-Child, United States, 2020–2022. MMWR Morb. Mortal. Wkly. Rep. 2023, 72, 1190–1196. [Google Scholar] [CrossRef]
- Lu, P.J.; Zhou, T.; Santibanez, T.A.; Jain, A.; Black, C.L.; Srivastav, A.; Hung, M.C.; Kriss, J.L.; Schorpp, S.; Yankey, D.; et al. COVID-19 Bivalent Booster Vaccination Coverage and Intent to Receive Booster Vaccination Among Adolescents and Adults—United States, November-December 2022. MMWR Morb. Mortal. Wkly. Rep. 2023, 72, 190–198. [Google Scholar] [CrossRef]
- Pingali, C.; Yankey, D.; Elam-Evans, L.D.; Markowitz, L.E.; Valier, M.R.; Fredua, B.; Crowe, S.J.; DeSisto, C.L.; Stokley, S.; Singleton, J.A. Vaccination Coverage Among Adolescents Aged 13–17 Years—National Immunization Survey-Teen, United States, 2022. MMWR Morb. Mortal. Wkly. Rep. 2023, 72, 912–919. [Google Scholar] [CrossRef] [PubMed]
- Santibanez, T.A.; Black, C.L.; Vogt, T.M.; Chatham-Stephens, K.; Zhou, T.; Lendon, J.P.; Singleton, J.A. Where are children ages 5-17 years receiving their COVID-19 vaccinations? Variations over time and by sociodemographic characteristics, United States. Vaccine 2022, 40, 6917–6923. [Google Scholar] [CrossRef] [PubMed]
- Santibanez, T.A.; Zhou, T.; Black, C.L.; Vogt, T.M.; Murthy, B.P.; Pineau, V.; Singleton, J.A. Sociodemographic Variation in Early Uptake of COVID-19 Vaccine and Parental Intent and Attitudes Toward Vaccination of Children Aged 6 Months-4 Years—United States, July 1-29, 2022. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 1479–1484. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Lee, Y.; Ravichandran, S.; Grubbs, G.; Huang, C.; Stauft, C.B.; Wang, T.; Golding, B.; Golding, H.; Khurana, S. Epitope diversity of SARS-CoV-2 hyperimmune intravenous human immunoglobulins and neutralization of variants of concern. iScience 2021, 24, 103006. [Google Scholar] [CrossRef]
- Ravichandran, S.; Coyle, E.M.; Klenow, L.; Tang, J.; Grubbs, G.; Liu, S.; Wang, T.; Golding, H.; Khurana, S. Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits. Sci. Transl. Med. 2020, 12, eabc3539. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, S.; Tang, J.; Grubbs, G.; Lee, Y.; Pourhashemi, S.; Hussaini, L.; Lapp, S.A.; Jerris, R.C.; Singh, V.; Chahroudi, A.; et al. SARS-CoV-2 immune repertoire in MIS-C and pediatric COVID-19. Nat. Immunol. 2021, 22, 1452–1464. [Google Scholar] [CrossRef]
- Tang, J.; Novak, T.; Hecker, J.; Grubbs, G.; Zahra, F.T.; Bellusci, L.; Pourhashemi, S.; Chou, J.; Moffitt, K.; Halasa, N.B.; et al. Cross-reactive immunity against the SARS-CoV-2 Omicron variant is low in pediatric patients with prior COVID-19 or MIS-C. Nat. Commun. 2022, 13, 2979. [Google Scholar] [CrossRef] [PubMed]
- Neerukonda, S.N.; Vassell, R.; Herrup, R.; Liu, S.; Wang, T.; Takeda, K.; Yang, Y.; Lin, T.L.; Wang, W.; Weiss, C.D. Establishment of a well-characterized SARS-CoV-2 lentiviral pseudovirus neutralization assay using 293T cells with stable expression of ACE2 and TMPRSS2. PLoS ONE 2021, 16, e0248348. [Google Scholar] [CrossRef]
- Roessler, A.; Netzl, A.; Knabl, L.; Schaefer, H.; Wilks, S.H.; Bante, D.; Falkensammer, B.; Borena, W.; von Laer, D.; Smith, D.J.; et al. BA.2 and BA.5 omicron differ immunologically from both BA.1 omicron and pre-omicron variants. Nat. Commun. 2022, 13, 7701. [Google Scholar] [CrossRef]
- Fonville, J.M.; Fraaij, P.L.A.; de Mutsert, G.; Wilks, S.H.; van Beek, R.; Fouchier, R.A.M.; Rimmelzwaan, G.F. Antigenic Maps of Influenza A(H3N2) Produced With Human Antisera Obtained After Primary Infection. J. Infect. Dis. 2016, 213, 31–38. [Google Scholar] [CrossRef]
- Wilks, S.H.; Mühlemann, B.; Shen, X.; Türeli, S.; LeGresley, E.B.; Netzl, A.; Caniza, M.A.; Chacaltana-Huarcaya, J.N.; Corman, V.M.; Daniell, X.; et al. Mapping SARS-CoV-2 antigenic relationships and serological responses. Science 2023, 382, eadj0070. [Google Scholar] [CrossRef] [PubMed]
- Bellusci, L.; Grubbs, G.; Sait, S.; Yonker, L.M.; Randolph, A.G.; Novak, T.; Kobayashi, T.; Overcoming COVID-19 Investigators; Khurana, S. Neutralization of SARS-CoV-2 Omicron BQ. 1, BQ. 1.1 and XBB. 1 variants following SARS-CoV-2 infection or vaccination in children. Nat. Commun. 2023, 14, 7952. [Google Scholar] [CrossRef] [PubMed]
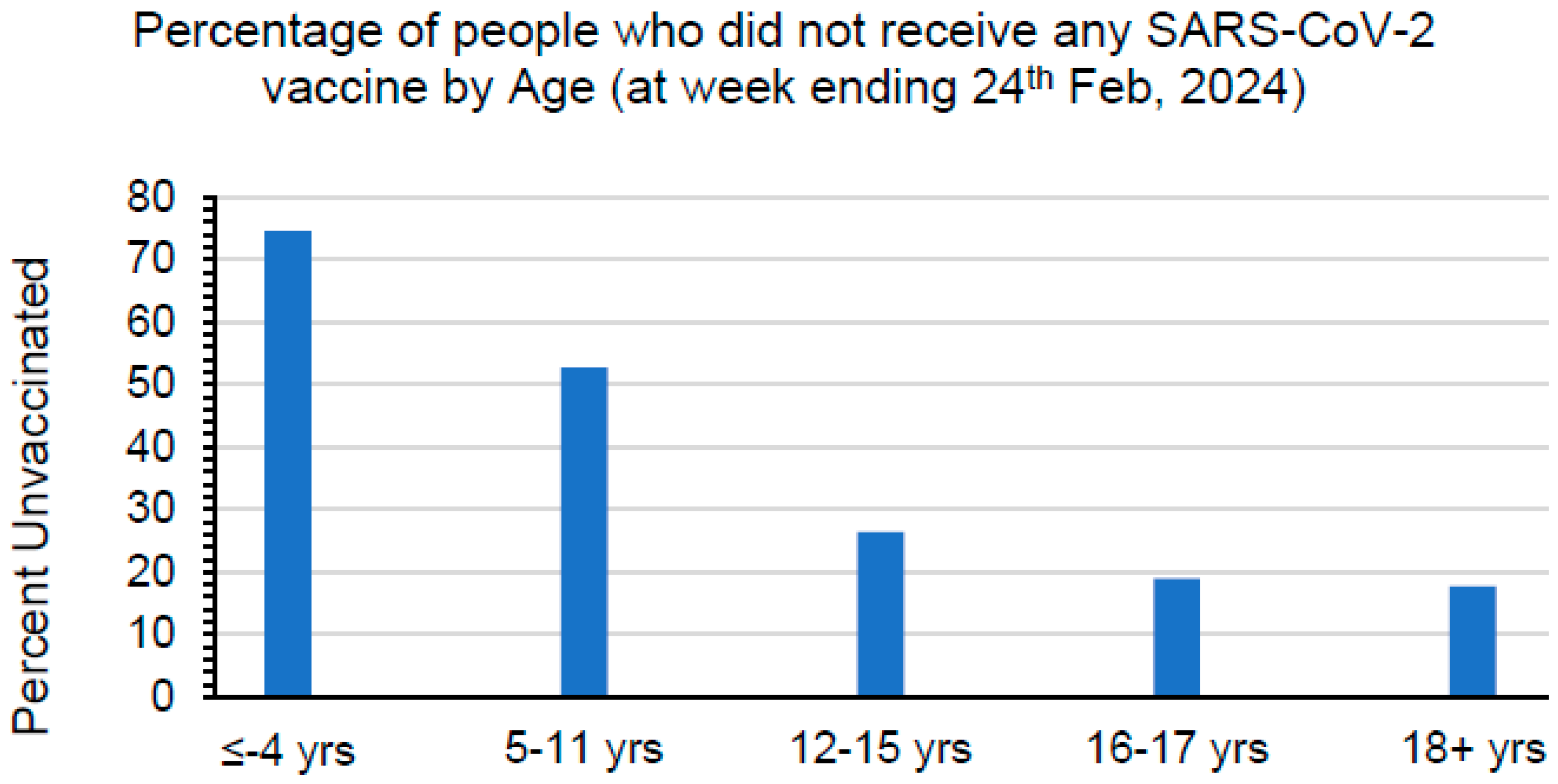

| Characteristic | COVID-19 (n = 32) | MIS-C (n = 32) | p-Value |
|---|---|---|---|
| Age (months) | 113 (44–198) | 116 (69–154) | 0.50 |
| Male | 17 (53%) | 19 (59%) | 0.80 |
| Race | 0.15 | ||
| Black | 5 (16%) | 12 (38%) | |
| White | 13 (41%) | 5 (16%) | |
| Other Race | 11 (34%) | 12 (37%) | |
| >1 Race | 1 (3%) | 3 (9%) | |
| Prefer not to answer | 2 (6%) | - | |
| Hispanic | 11 (34%) | 12 (37%) | 1.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellusci, L.; Grubbs, G.; Sait, S.; Herbst, K.W.; Salazar, J.C.; Khurana, S.; The Connecticut Children’s COVID Collaborative. Evolution of the Antigenic Landscape in Children and Young Adults with COVID-19 and MIS-C. Vaccines 2024, 12, 638. https://doi.org/10.3390/vaccines12060638
Bellusci L, Grubbs G, Sait S, Herbst KW, Salazar JC, Khurana S, The Connecticut Children’s COVID Collaborative. Evolution of the Antigenic Landscape in Children and Young Adults with COVID-19 and MIS-C. Vaccines. 2024; 12(6):638. https://doi.org/10.3390/vaccines12060638
Chicago/Turabian StyleBellusci, Lorenza, Gabrielle Grubbs, Shaimaa Sait, Katherine W. Herbst, Juan C. Salazar, Surender Khurana, and The Connecticut Children’s COVID Collaborative. 2024. "Evolution of the Antigenic Landscape in Children and Young Adults with COVID-19 and MIS-C" Vaccines 12, no. 6: 638. https://doi.org/10.3390/vaccines12060638







