Influenza Vaccination Mediates SARS-CoV-2 Spike Protein Peptide-Induced Inflammatory Response via Modification of Histone Acetylation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Immunization
2.2. RBD-Antibody Enzyme-Linked Immunosorbent Assay (ELISA)
2.3. Hemagglutination-Inhibition (HI) Assay
2.4. Peptide Pools
2.5. Cell Isolation and In Vitro Stimulation
2.6. Cytokine Measurements
2.7. Total Histone Extraction and Histone-H3-Acetylation Detection
2.8. Flow-Cytometry Assays
2.9. Statistical Analysis
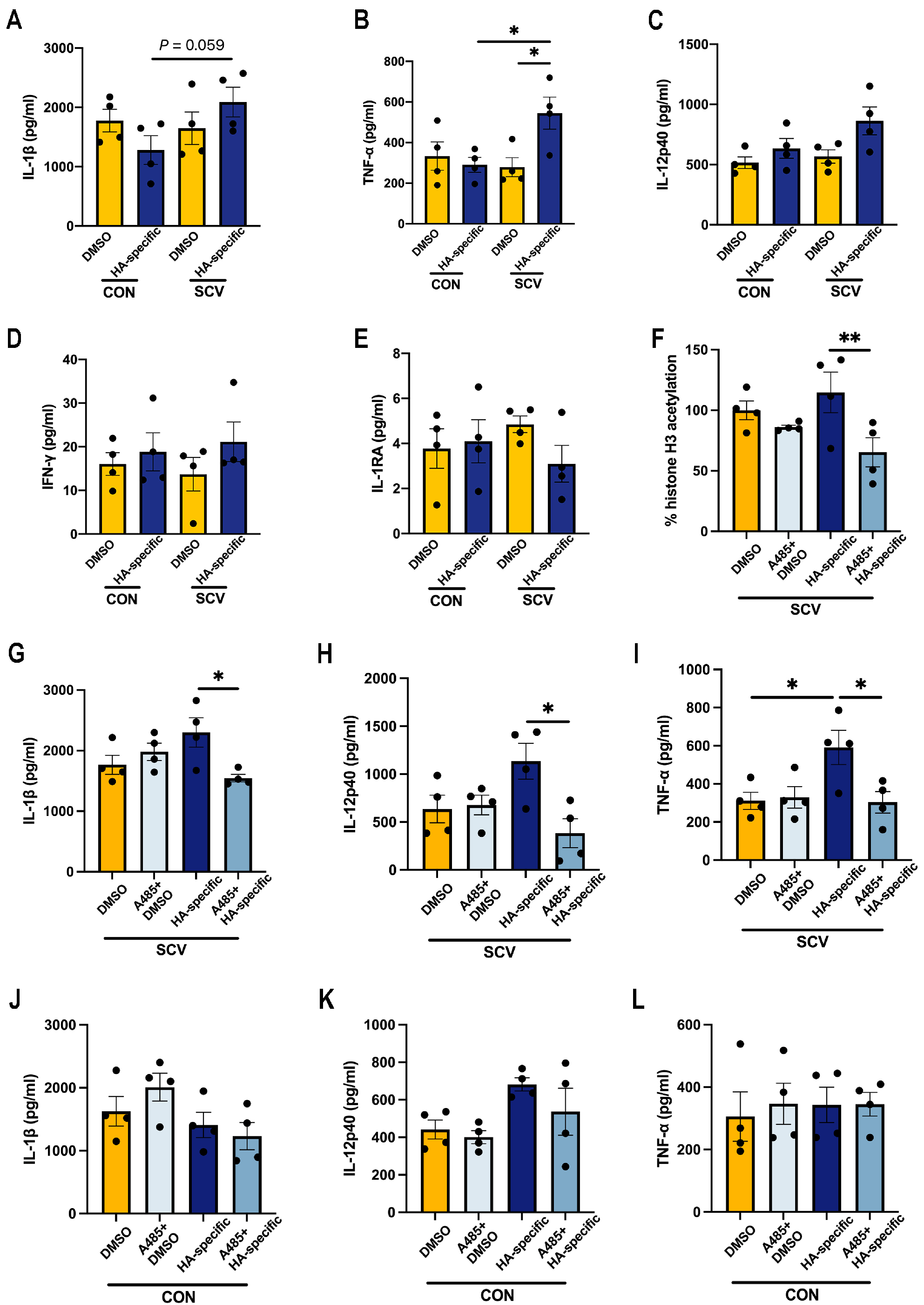
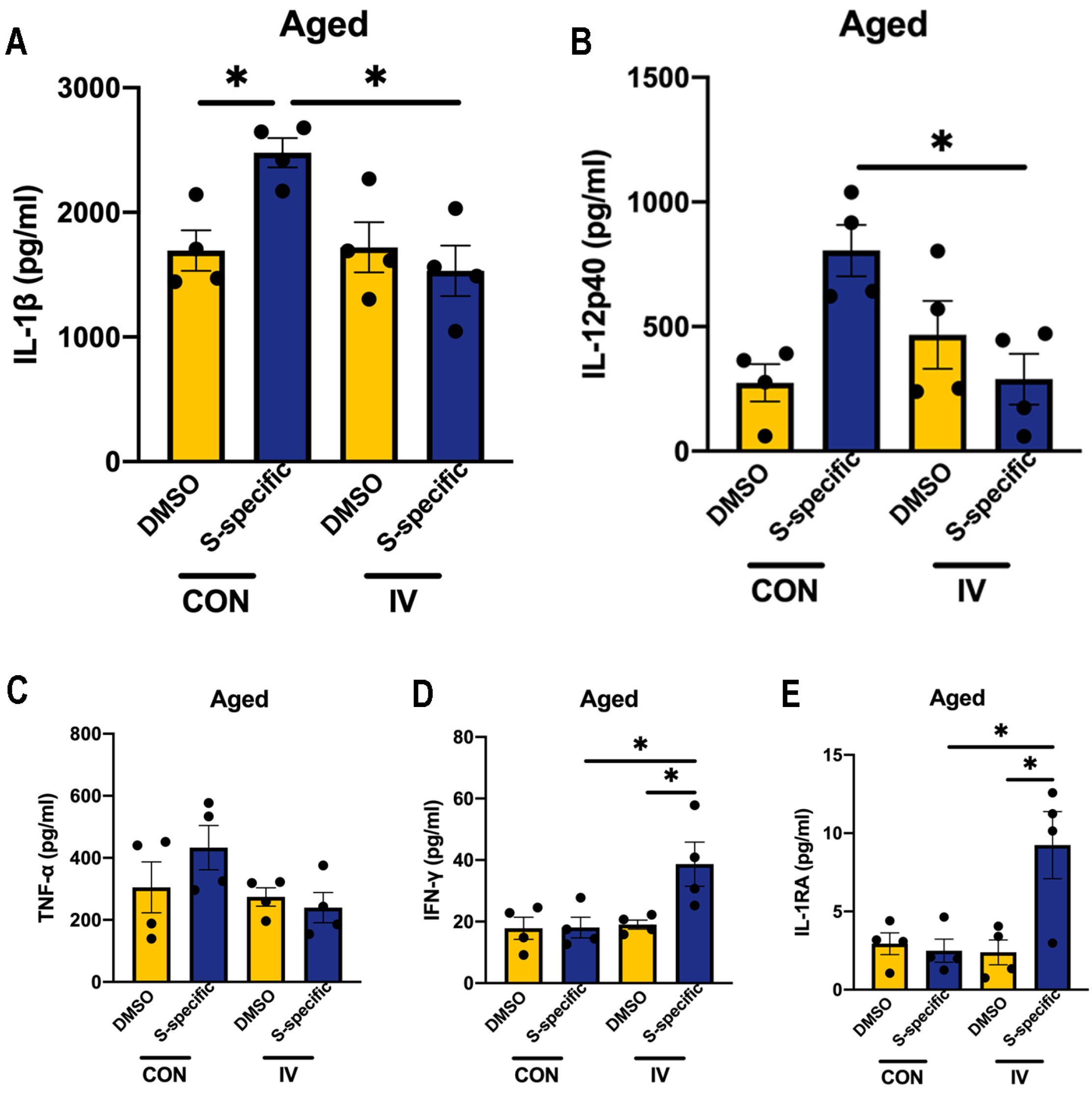
3. Results
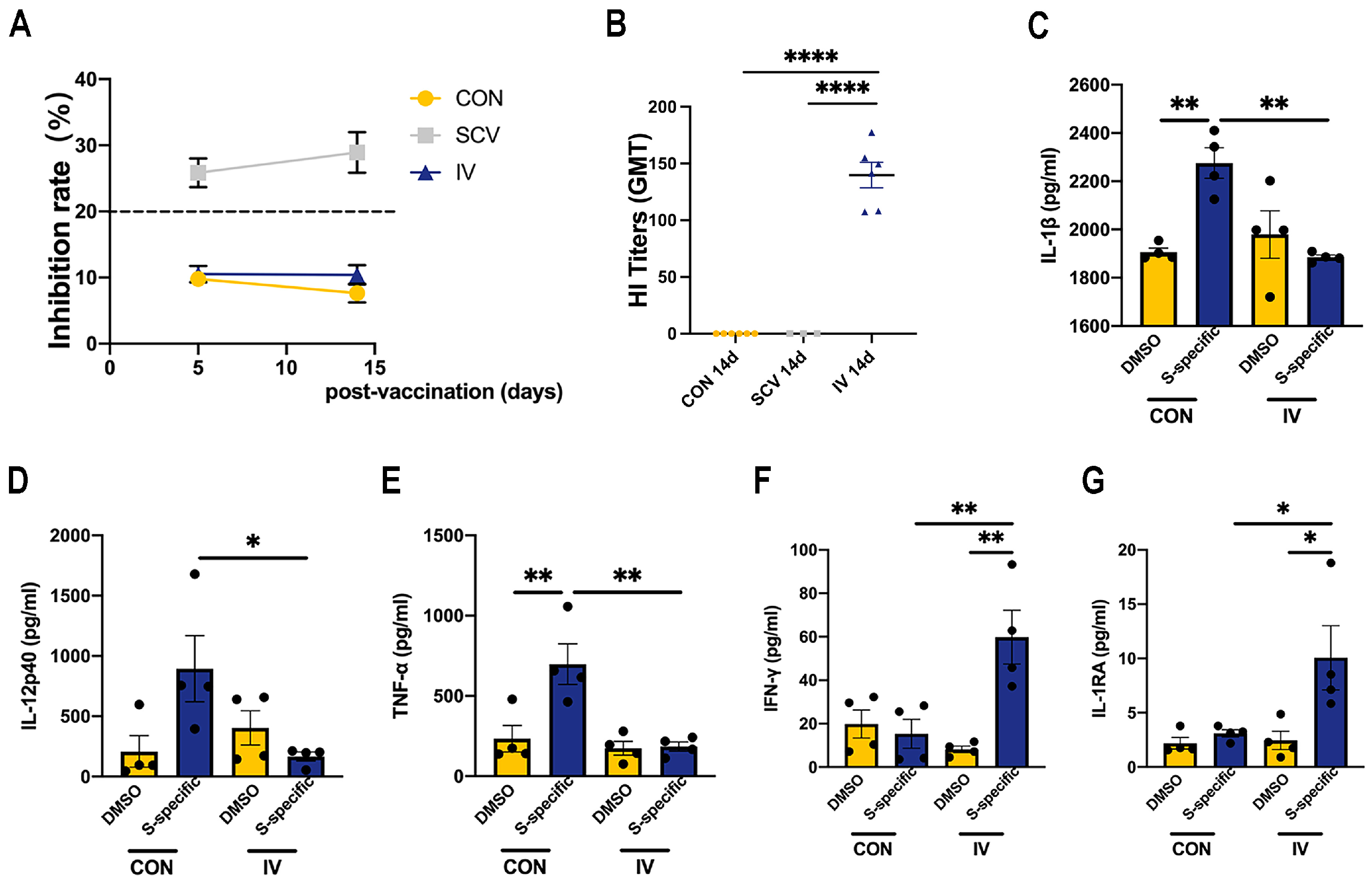
3.1. Influenza Vaccination Induces Changes in Cytokine Production Capacity in PBMCs after SARS-CoV-2 Spike Stimulation
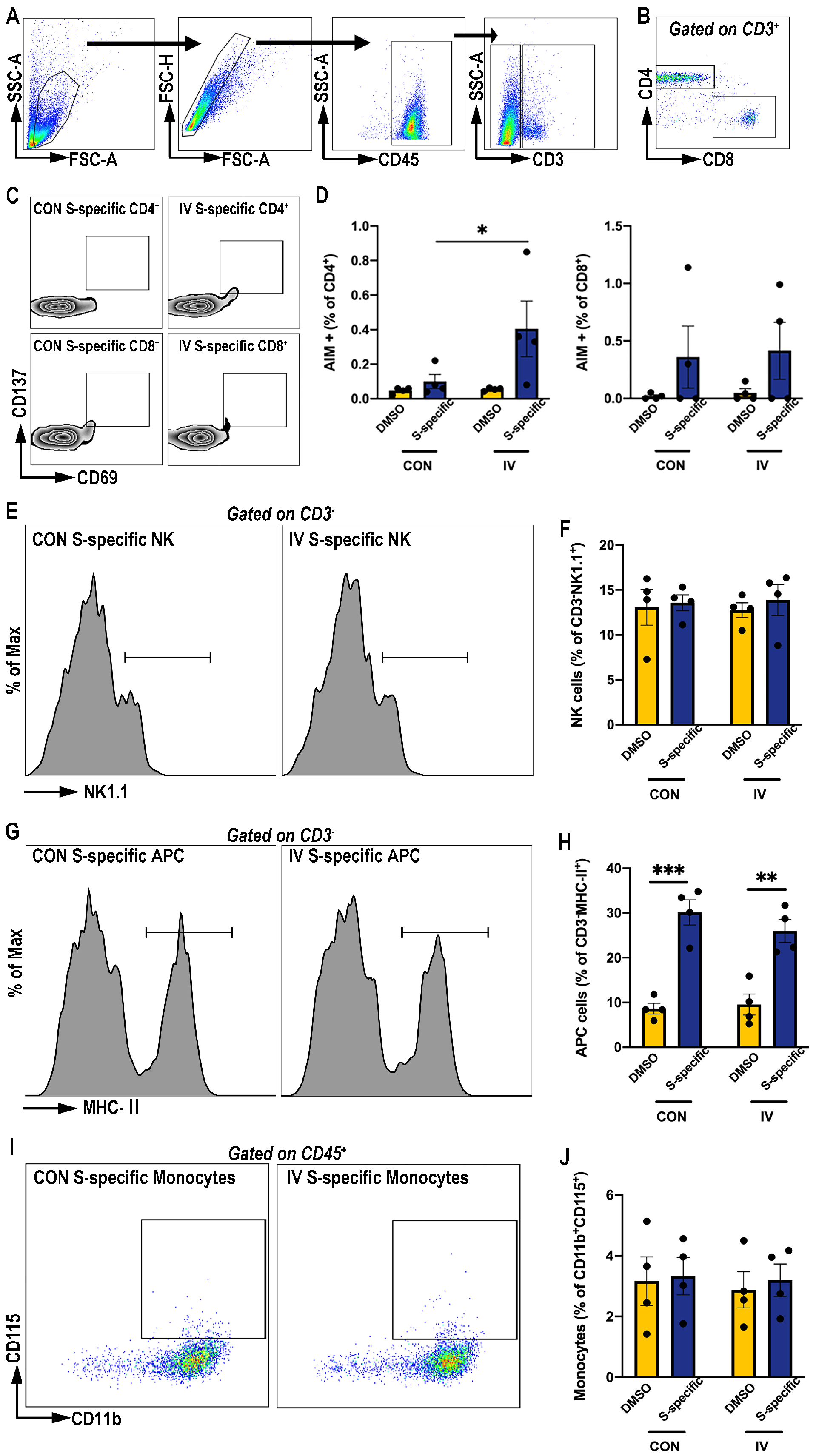
3.2. Influenza Vaccination Enhances Activation-Induced-Marker (AIM) Expression on CD4+ T Cells Post SARS-CoV-2 Spike Protein Peptide Pool Stimulation
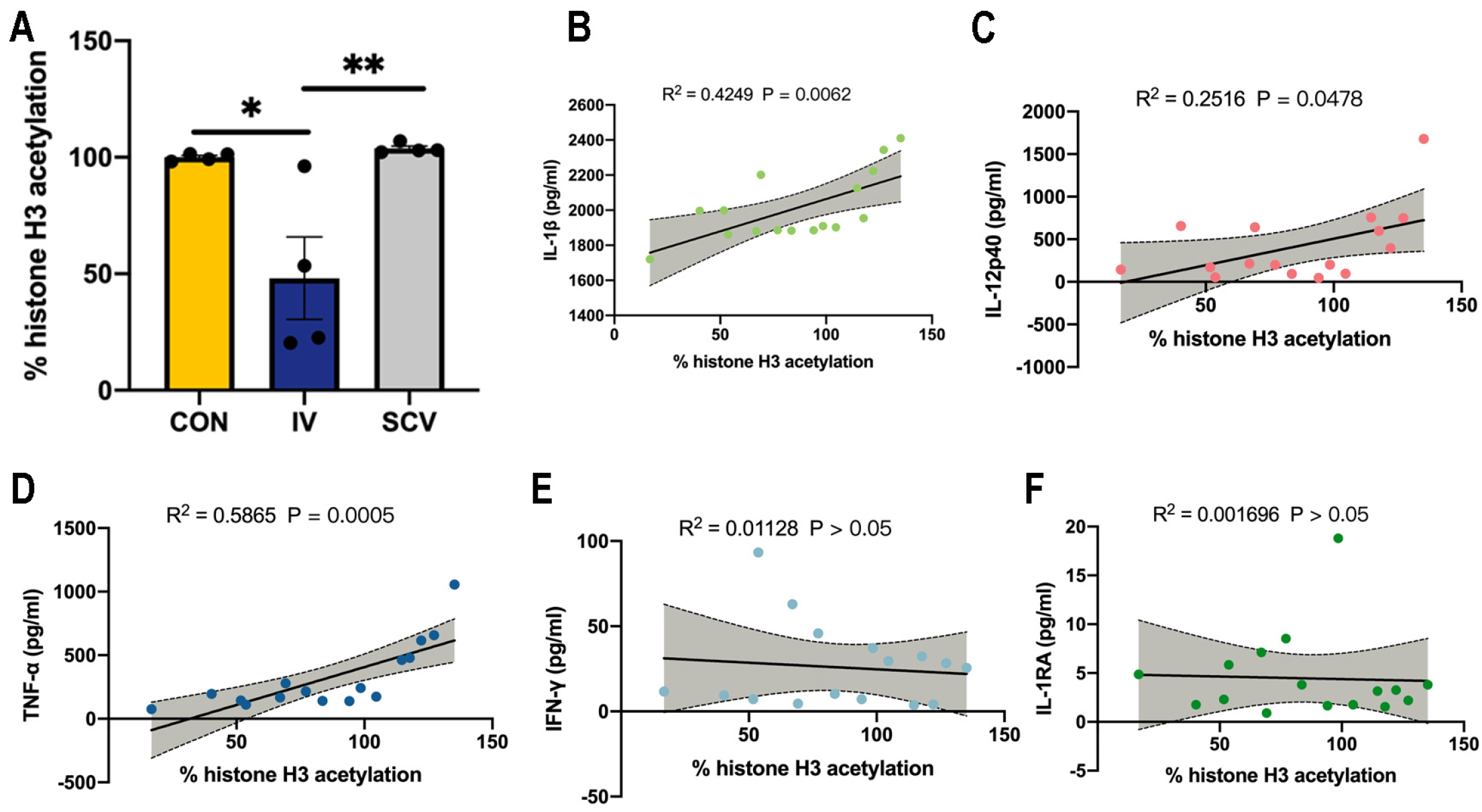
3.3. Influenza Vaccination Induces Histone-H3 Hypoacetylation in PBMCs
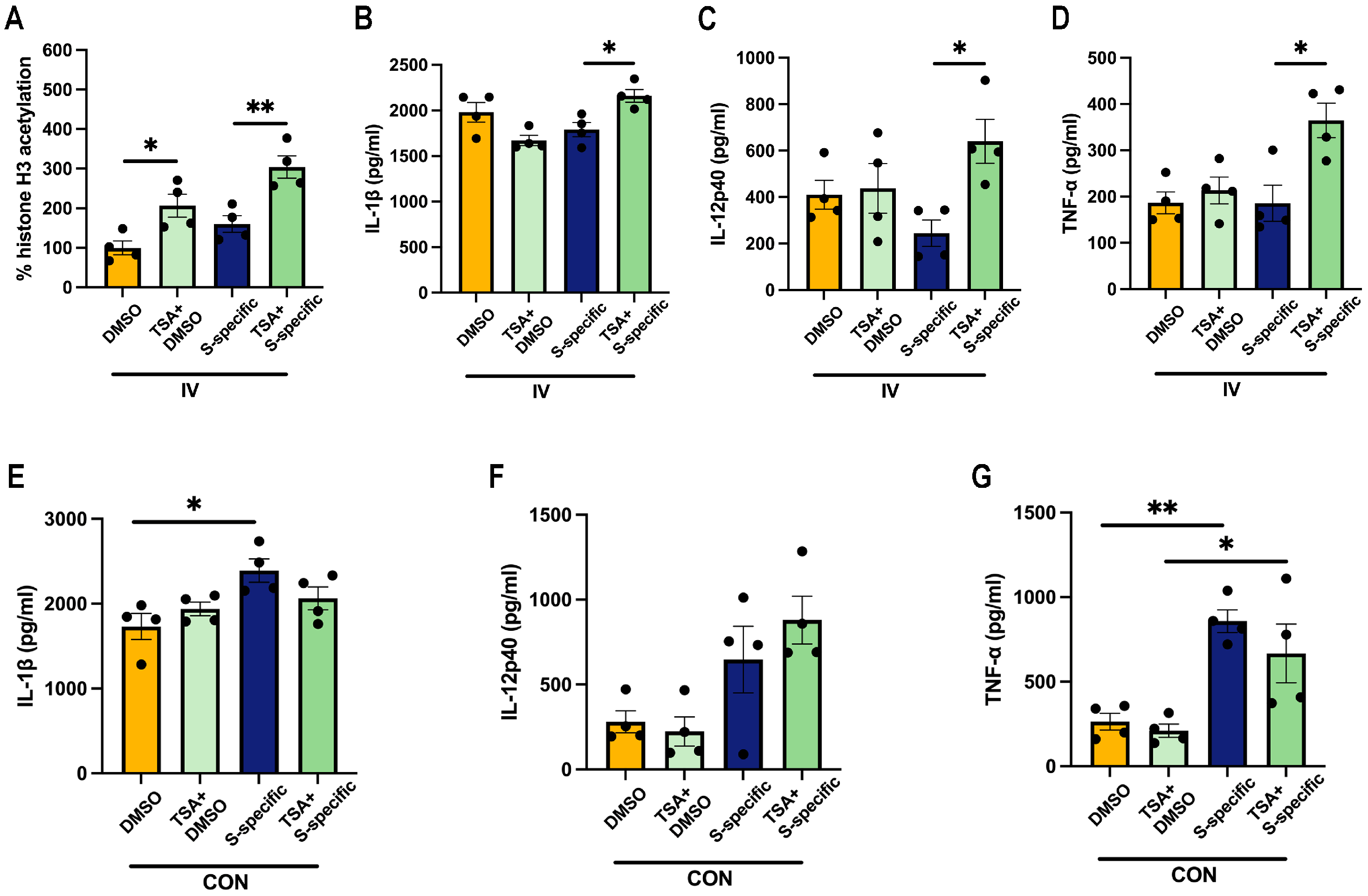
3.4. Prior SARS-CoV-2 Vaccination Does Not Rescue Proinflammatory Cytokine Production in HA-Peptide-Pool-Stimulated PBMCs
3.5. Influenza Vaccination Induces Similar Changes in Cytokine Production Capacity in PBMCs Isolated from Aged Mice
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tan, S.T.; Kwan, A.T.; Rodríguez-Barraquer, I.; Singer, B.J.; Park, H.J.; Lewnard, J.A.; Sears, D.; Lo, N.C. Infectiousness of SARS-CoV-2 breakthrough infections and reinfections during the Omicron wave. Nat. Med. 2023, 29, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Menegale, F.; Manica, M.; Zardini, A.; Guzzetta, G.; Marziano, V.; d’Andrea, V.; Trentini, F.; Ajelli, M.; Poletti, P.; Merler, S. Evaluation of waning of SARS-CoV-2 vaccine–induced immunity: A systematic review and meta-analysis. JAMA Netw. Open 2023, 6, e2310650. [Google Scholar] [CrossRef] [PubMed]
- Ghazy, R.M.; Ashmawy, R.; Hamdy, N.A.; Elhadi, Y.A.M.; Reyad, O.A.; Elmalawany, D.; Almaghraby, A.; Shaaban, R.; Taha, S.H.N. Efficacy and effectiveness of SARS-CoV-2 vaccines: A systematic review and meta-analysis. Vaccines 2022, 10, 350. [Google Scholar] [CrossRef] [PubMed]
- Murray, S.M.; Ansari, A.M.; Frater, J.; Klenerman, P.; Dunachie, S.; Barnes, E.; Ogbe, A. The impact of pre-existing cross-reactive immunity on SARS-CoV-2 infection and vaccine responses. Nat. Rev. Immunol. 2023, 23, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Conlon, A.; Ashur, C.; Washer, L.; Eagle, K.A.; Hofmann Bowman, M.A. Impact of the influenza vaccine on COVID-19 infection rates and severity. Am. J. Infect. Control 2021, 49, 694–700. [Google Scholar] [CrossRef]
- Jiang, B.; Huang, Q.; Jia, M.; Xue, X.; Wang, Q.; Yang, W.; Feng, L. Association between influenza vaccination and SARS-CoV-2 infection and its outcomes: Systematic review and meta-analysis. Chin. Med. J. 2022, 135, 2282–2293. [Google Scholar] [CrossRef] [PubMed]
- Fink, G.; Orlova-Fink, N.; Schindler, T.; Grisi, S.; Ferrer, A.P.S.; Daubenberger, C.; Brentani, A. Inactivated trivalent influenza vaccination is associated with lower mortality among patients with COVID-19 in Brazil. BMJ Evid.-Based Med. 2021, 26, 192–193. [Google Scholar] [CrossRef]
- Green, I.; Ashkenazi, S.; Merzon, E.; Vinker, S.; Golan-Cohen, A. The association of previous influenza vaccination and coronavirus disease-2019. Hum. Vaccin. Immunother. 2021, 17, 2169–2175. [Google Scholar] [CrossRef] [PubMed]
- Debisarun, P.A.; Gössling, K.L.; Bulut, O.; Kilic, G.; Zoodsma, M.; Liu, Z.; Oldenburg, M.; Rüchel, N.; Zhang, B.; Xu, C.-J. Induction of trained immunity by influenza vaccination-impact on COVID-19. PLoS Pathog. 2021, 17, e1009928. [Google Scholar] [CrossRef]
- Tayar, E.; Abdeen, S.; Alah, M.A.; Chemaitelly, H.; Bougmiza, I.; Ayoub, H.H.; Kaleeckal, A.H.; Latif, A.N.; Shaik, R.M.; Al-Romaihi, H.E. Effectiveness of influenza vaccination against SARS-CoV-2 infection among healthcare workers in Qatar. J. Infect. Public Health 2023, 16, 250–256. [Google Scholar] [CrossRef]
- Lopez, C.E.; Legge, K.L. Influenza A virus vaccination: Immunity, protection, and recent advances toward a universal vaccine. Vaccines 2020, 8, 434. [Google Scholar] [CrossRef] [PubMed]
- Wimmers, F.; Donato, M.; Kuo, A.; Ashuach, T.; Gupta, S.; Li, C.; Dvorak, M.; Foecke, M.H.; Chang, S.E.; Hagan, T. The single-cell epigenomic and transcriptional landscape of immunity to influenza vaccination. Cell 2021, 184, 3915–3935.e21. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Qi, F.; Zou, J.; Yang, J.; Yao, Z. Influenza vaccination during early pregnancy contributes to neurogenesis and behavioral function in offspring. Brain Behav. Immun. 2014, 42, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Babon, J.A.; Cruz, J.; Ennis, F.A.; Yin, L.; Terajima, M. A human CD4+ T cell epitope in the influenza hemagglutinin is cross-reactive to influenza A virus subtypes and to influenza B virus. J. Virol. 2012, 86, 9233–9243. [Google Scholar] [CrossRef] [PubMed]
- Chaisawangwong, W.; Wang, H.; Kouo, T.; Salathe, S.F.; Isser, A.; Bieler, J.G.; Zhang, M.L.; Livingston, N.K.; Li, S.; Horowitz, J.J. Cross-reactivity of SARS-CoV-2–and influenza A–specific T cells in individuals exposed to SARS-CoV-2. JCI Insight 2022, 7, e158308. [Google Scholar] [CrossRef]
- McElhaney, J.E.; Verschoor, C.P.; Andrew, M.K.; Haynes, L.; Kuchel, G.A.; Pawelec, G. The immune response to influenza in older humans: Beyond immune senescence. Immun. Ageing 2020, 17, 10. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Jing, X.; Li, X.; Lin, Z.; Plant, E.; Zoueva, O.; Yang, H.; Ye, Z. Immunogenicity and cross-reactivity of 2009-2010 inactivated seasonal influenza vaccine in US adults and elderly. PLoS ONE 2011, 6, e16650. [Google Scholar] [CrossRef]
- Castellino, F.; Germain, R.N. Cooperation between CD4+ and CD8+ T cells: When, where, and how. Annu. Rev. Immunol. 2006, 24, 519–540. [Google Scholar] [CrossRef] [PubMed]
- Doherty, P.C.; Turner, S.J.; Webby, R.G.; Thomas, P.G. Influenza and the challenge for immunology. Nat. Immunol. 2006, 7, 449–455. [Google Scholar] [CrossRef]
- Brown, D.M.; Dilzer, A.M.; Meents, D.L.; Swain, S.L. CD4 T cell-mediated protection from lethal influenza: Perforin and antibody-mediated mechanisms give a one-two punch. J. Immunol. 2006, 177, 2888–2898. [Google Scholar] [CrossRef]
- Strutt, T.M.; McKinstry, K.K.; Dibble, J.P.; Winchell, C.; Kuang, Y.; Curtis, J.D.; Huston, G.; Dutton, R.W.; Swain, S.L. Memory CD4+ T cells induce innate responses independently of pathogen. Nat. Med. 2010, 16, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Amato, M.; Werba, J.P.; Frigerio, B.; Coggi, D.; Sansaro, D.; Ravani, A.; Ferrante, P.; Veglia, F.; Tremoli, E.; Baldassarre, D. Relationship between influenza vaccination coverage rate and COVID-19 outbreak: An Italian ecological study. Vaccines 2020, 8, 535. [Google Scholar] [CrossRef]
- Marín-Hernández, D.; Schwartz, R.E.; Nixon, D.F. Epidemiological evidence for association between higher influenza vaccine uptake in the elderly and lower COVID-19 deaths in Italy. J. Med. Virol. 2021, 93, 64. [Google Scholar] [CrossRef] [PubMed]
- Zanettini, C.; Omar, M.; Dinalankara, W.; Imada, E.L.; Colantuoni, E.; Parmigiani, G.; Marchionni, L. Influenza vaccination and COVID-19 mortality in the USA: An ecological study. Vaccines 2021, 9, 427. [Google Scholar] [CrossRef] [PubMed]
- Al Mosawi, A.M.T.; Kadhim, H.M.; Hameed, H.M. The role of influenza vaccination in the COVID-19 infection: Impact on incidence and severity in Iraq. J. Appl. Pharm. Sci. 2022, 12, 130–136. [Google Scholar] [CrossRef]
- Paget, J.; Caini, S.; Cowling, B.; Esposito, S.; Falsey, A.R.; Gentile, A.; Kyncl, J.; MacIntyre, C.; Pitman, R.; Lina, B. The impact of influenza vaccination on the COVID-19 pandemic? Evidence and lessons for public health policies. Vaccine 2020, 38, 6485. [Google Scholar] [CrossRef] [PubMed]
- Hosseini-Moghaddam, S.M.; He, S.; Calzavara, A.; Campitelli, M.A.; Kwong, J.C. Association of influenza vaccination with SARS-CoV-2 infection and associated hospitalization and mortality among patients aged 66 years or older. JAMA Netw. Open 2022, 5, e2233730. [Google Scholar] [CrossRef] [PubMed]
- Pontiroli, A.E.; Scovenna, F.; Carlini, V.; Tagliabue, E.; Martin-Delgado, J.; La Sala, L.; Tanzi, E.; Zanoni, I. Vaccination against influenza viruses reduces infection, not hospitalization or death, from respiratory COVID-19: A systematic review and meta-analysis. J. Med. Virol. 2024, 96, e29343. [Google Scholar] [CrossRef] [PubMed]
- Shosha, S.H.; Ajlan, D.I.; Al-Ghatam, R. Does influenza vaccination help reduce incidence of COVID-19 infection among hospital employees? Medicine 2022, 101, e28479. [Google Scholar] [CrossRef]
- Su, W.; Wang, H.; Sun, C.; Li, N.; Guo, X.; Song, Q.; Liang, Q.; Liang, M.; Ding, X.; Sun, Y. The Association Between Previous Influenza Vaccination and COVID-19 Infection Risk and Severity: A Systematic Review and Meta-analysis. Am. J. Prev. Med. 2022, 63, 121–130. [Google Scholar] [CrossRef]
- Arokiaraj, M.C. Considering interim interventions to control COVID-19 associated morbidity and Mortality—Perspectives. Front. Public Health 2020, 8, 552576. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Hernández, J.G.; Quintanilla-Flores, D.L.; González-Galván, C.R.; Nuzzolo-Shihadeh, L.; Camacho-Ortiz, A.; Salinas-Martínez, R.; Morales-Delgado, R. Impact of influenza vaccination history in the clinical course of older adults hospitalized with COVID-19. Med. Clin. 2024, 162, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Lin, S.W.; Sheng, W.H.; Wang, C.C. Influenza vaccination and the risk of COVID-19 infection and severe illness in older adults in the United States. Sci. Rep. 2021, 11, 11025. [Google Scholar] [CrossRef] [PubMed]
- Veronese, N.; Smith, L.; Di Gennaro, F.; Bruyère, O.; Yang, L.; Demurtas, J.; Maggi, S.; Sabico, S.; Al-Daghri, N.M.; Barbagallo, M.; et al. Influenza Vaccination and COVID-19 Outcomes in People Older than 50 Years: Data from the Observational Longitudinal SHARE Study. Vaccines 2022, 10, 899. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zheng, X.; Ding, H.; Miao, T.; Zang, Y.; Shen, S.; Gao, Y. Association between combination COVID-19-influenza vaccination and long COVID in middle-aged and older Europeans: A cross-sectional study. Hum. Vaccin. Immunother. 2024, 20, 2345505. [Google Scholar] [CrossRef] [PubMed]
- Arts, R.J.; Moorlag, S.J.; Novakovic, B.; Li, Y.; Wang, S.-Y.; Oosting, M.; Kumar, V.; Xavier, R.J.; Wijmenga, C.; Joosten, L.A. BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe 2018, 23, 89–100.e5. [Google Scholar] [CrossRef] [PubMed]
- Gillard, J.; Blok, B.A.; Garza, D.R.; Venkatasubramanian, P.B.; Simonetti, E.; Eleveld, M.J.; Berbers, G.A.; van Gageldonk, P.G.; Joosten, I.; de Groot, R. BCG-induced trained immunity enhances acellular pertussis vaccination responses in an explorative randomized clinical trial. NPJ Vaccines 2022, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Counoupas, C.; Johansen, M.D.; Stella, A.O.; Nguyen, D.H.; Ferguson, A.L.; Aggarwal, A.; Bhattacharyya, N.D.; Grey, A.; Hutchings, O.; Patel, K. A single dose, BCG-adjuvanted COVID-19 vaccine provides sterilising immunity against SARS-CoV-2 infection. NPJ Vaccines 2021, 6, 143. [Google Scholar] [CrossRef] [PubMed]
- Fanucchi, S.; Domínguez-Andrés, J.; Joosten, L.A.; Netea, M.G.; Mhlanga, M.M. The intersection of epigenetics and metabolism in trained immunity. Immunity 2021, 54, 32–43. [Google Scholar] [CrossRef]
- Cheong, J.G.; Ravishankar, A.; Sharma, S.; Parkhurst, C.N.; Grassmann, S.A.; Wingert, C.K.; Laurent, P.; Ma, S.; Paddock, L.; Miranda, I.C.; et al. Epigenetic memory of coronavirus infection in innate immune cells and their progenitors. Cell 2023, 186, 3882–3902.e24. [Google Scholar] [CrossRef]
- Liu, Q.; Zhou, Y.-h.; Yang, Z.-q. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell. Mol. Immunol. 2016, 13, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Sinha, P.; Matthay, M.A.; Calfee, C.S. Is a “cytokine storm” relevant to COVID-19? JAMA Intern. Med. 2020, 180, 1152–1154. [Google Scholar] [CrossRef] [PubMed]
- Howlader, D.R.; Das, S.; Lu, T.; Mandal, R.S.; Hu, G.; Varisco, D.J.; Dietz, Z.K.; Ratnakaram, S.S.K.; Ernst, R.K.; Picking, W.D. A protein subunit vaccine elicits a balanced immune response that protects against Pseudomonas pulmonary infection. NPJ Vaccines 2023, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Kleinnijenhuis, J.; Quintin, J.; Preijers, F.; Joosten, L.A.; Ifrim, D.C.; Saeed, S.; Jacobs, C.; van Loenhout, J.; de Jong, D.; Stunnenberg, H.G. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. USA 2012, 109, 17537–17542. [Google Scholar] [CrossRef] [PubMed]
- Placek, K.; Schultze, J.L.; Aschenbrenner, A.C. Epigenetic reprogramming of immune cells in injury, repair, and resolution. J. Clin. Investig. 2019, 129, 2994–3005. [Google Scholar] [CrossRef] [PubMed]
- Jit, B.P.; Qazi, S.; Arya, R.; Srivastava, A.; Gupta, N.; Sharma, A. An immune epigenetic insight to COVID-19 infection. Epigenomics 2021, 13, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.; Vaishak, K.; Deka, D.; Radhakrishnan, A.K.; Paul, S.; Shanmugam, P.; Daniel, A.P.; Pathak, S.; Duttaroy, A.K.; Banerjee, A. Epigenetic perspectives associated with COVID-19 infection and related cytokine storm: An updated review. Infection 2023, 51, 1603–1618. [Google Scholar] [CrossRef]
- Takahashi, Y.; Hayakawa, A.; Sano, R.; Fukuda, H.; Harada, M.; Kubo, R.; Okawa, T.; Kominato, Y. Histone deacetylase inhibitors suppress ACE2 and ABO simultaneously, suggesting a preventive potential against COVID-19. Sci. Rep. 2021, 11, 3379. [Google Scholar] [CrossRef] [PubMed]
- Trionfetti, F.; Alonzi, T.; Bontempi, G.; Terri, M.; Battistelli, C.; Montaldo, C.; Repele, F.; Rotili, D.; Valente, S.; Zwergel, C.; et al. HDAC1-3 inhibition increases SARS-CoV-2 replication and productive infection in lung mesothelial and epithelial cells. Front. Cell Infect. Microbiol. 2023, 13, 1257683. [Google Scholar] [CrossRef]
- Achdout, H.; Vitner, E.B.; Politi, B.; Melamed, S.; Yahalom-Ronen, Y.; Tamir, H.; Erez, N.; Avraham, R.; Weiss, S.; Cherry, L. Increased lethality in influenza and SARS-CoV-2 coinfection is prevented by influenza immunity but not SARS-CoV-2 immunity. Nat. Commun. 2021, 12, 5819. [Google Scholar] [CrossRef]
- Skelly, D.T.; Harding, A.C.; Gilbert-Jaramillo, J.; Knight, M.L.; Longet, S.; Brown, A.; Adele, S.; Adland, E.; Brown, H. Two doses of SARS-CoV-2 vaccination induce robust immune responses to emerging SARS-CoV-2 variants of concern. Nat. Commun. 2021, 12, 5061. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Deng, W.; Huang, B.; Gao, H.; Liu, J.; Ren, L.; Wei, Q.; Yu, P.; Xu, Y.; Qi, F. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020, 583, 830–833. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Ji, K.; Tian, S.; Wang, F.; Huang, B.; Tong, Z.; Tan, S.; Hao, J.; Wang, Q.; Tan, W. A single-dose mRNA vaccine provides a long-term protection for hACE2 transgenic mice from SARS-CoV-2. Nat. Commun. 2021, 12, 776. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Sandgren, K.; Duette, G.; Stylianou, V.V.; Khanna, R.; Eden, J.S.; Blyth, E.; Gottlieb, D.; Cunningham, A.L.; Palmer, S. Identification of SARS-CoV-2 Nucleocapsid and Spike T-Cell Epitopes for Assessing T-Cell Immunity. J. Virol. 2021, 95, 10-1128. [Google Scholar] [CrossRef]
- Balz, K.; Kaushik, A.; Chen, M.; Cemic, F.; Heger, V.; Renz, H.; Nadeau, K.; Skevaki, C. Homologies between SARS-CoV-2 and allergen proteins may direct T cell-mediated heterologous immune responses. Sci. Rep. 2021, 11, 4792. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuo, Z.; Mu, Y.; Qi, F.; Zhang, H.; Li, Z.; Zhou, T.; Guo, W.; Guo, K.; Hu, X.; Yao, Z. Influenza Vaccination Mediates SARS-CoV-2 Spike Protein Peptide-Induced Inflammatory Response via Modification of Histone Acetylation. Vaccines 2024, 12, 731. https://doi.org/10.3390/vaccines12070731
Zuo Z, Mu Y, Qi F, Zhang H, Li Z, Zhou T, Guo W, Guo K, Hu X, Yao Z. Influenza Vaccination Mediates SARS-CoV-2 Spike Protein Peptide-Induced Inflammatory Response via Modification of Histone Acetylation. Vaccines. 2024; 12(7):731. https://doi.org/10.3390/vaccines12070731
Chicago/Turabian StyleZuo, Zejie, Yating Mu, Fangfang Qi, Hongyang Zhang, Zhihui Li, Tuo Zhou, Wenhai Guo, Kaihua Guo, Xiquan Hu, and Zhibin Yao. 2024. "Influenza Vaccination Mediates SARS-CoV-2 Spike Protein Peptide-Induced Inflammatory Response via Modification of Histone Acetylation" Vaccines 12, no. 7: 731. https://doi.org/10.3390/vaccines12070731
APA StyleZuo, Z., Mu, Y., Qi, F., Zhang, H., Li, Z., Zhou, T., Guo, W., Guo, K., Hu, X., & Yao, Z. (2024). Influenza Vaccination Mediates SARS-CoV-2 Spike Protein Peptide-Induced Inflammatory Response via Modification of Histone Acetylation. Vaccines, 12(7), 731. https://doi.org/10.3390/vaccines12070731





