Development of a Cell-Based Reporter Potency Assay for Live Virus Vaccines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Reporter Assay
2.3. Vaccine 1 Reporter Assay Qualification
2.4. Vaccine 1 Reporter Assay Robustness
2.5. Vaccine 1 Reporter Assay and Plaque Assay Correlation
3. Results
3.1. Vaccine 1 Reporter Assay Development
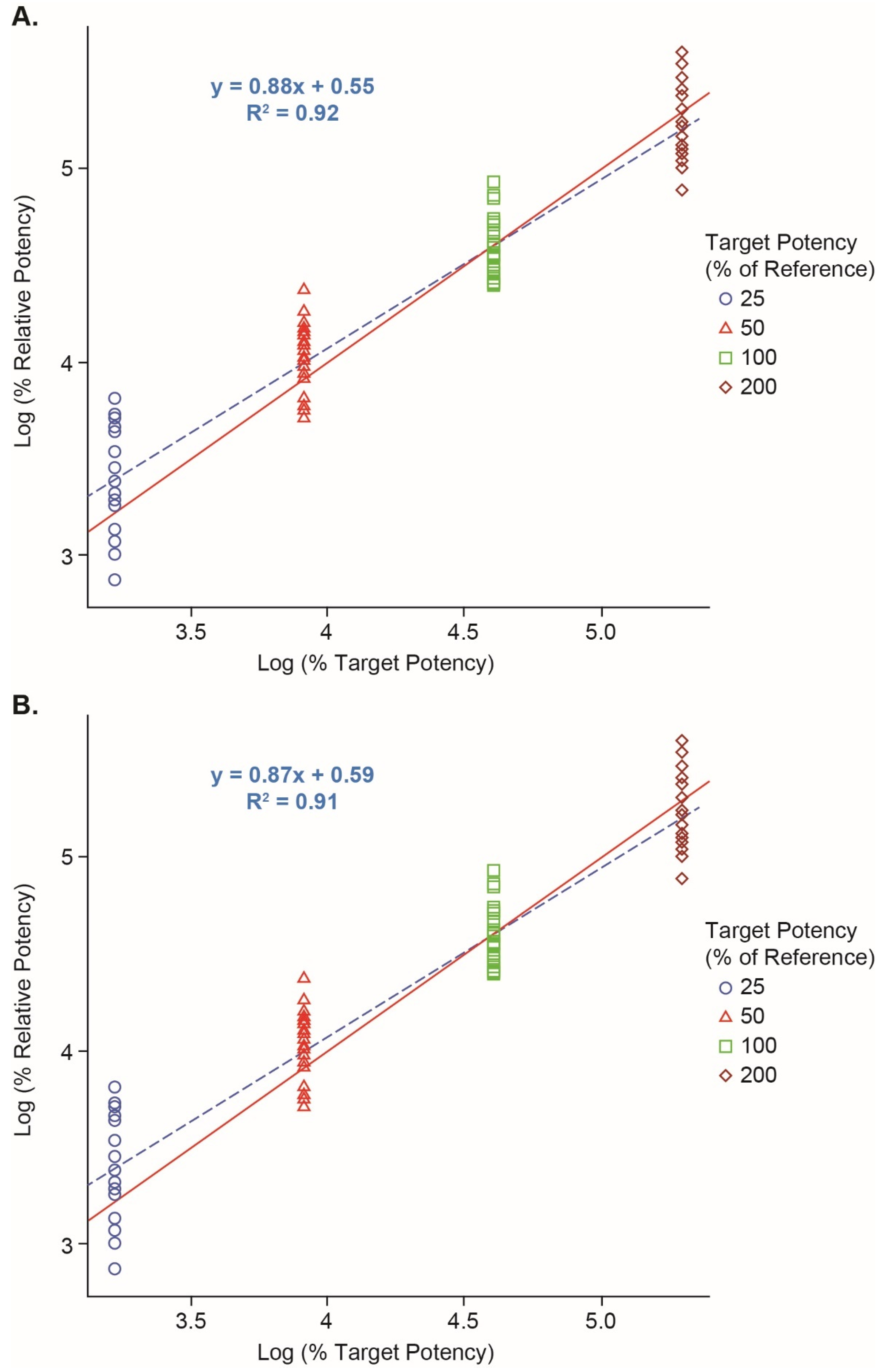
3.2. Vaccine 1 Reporter Assay Qualification
3.3. Vaccine 1 Reporter Assay Robustness Study
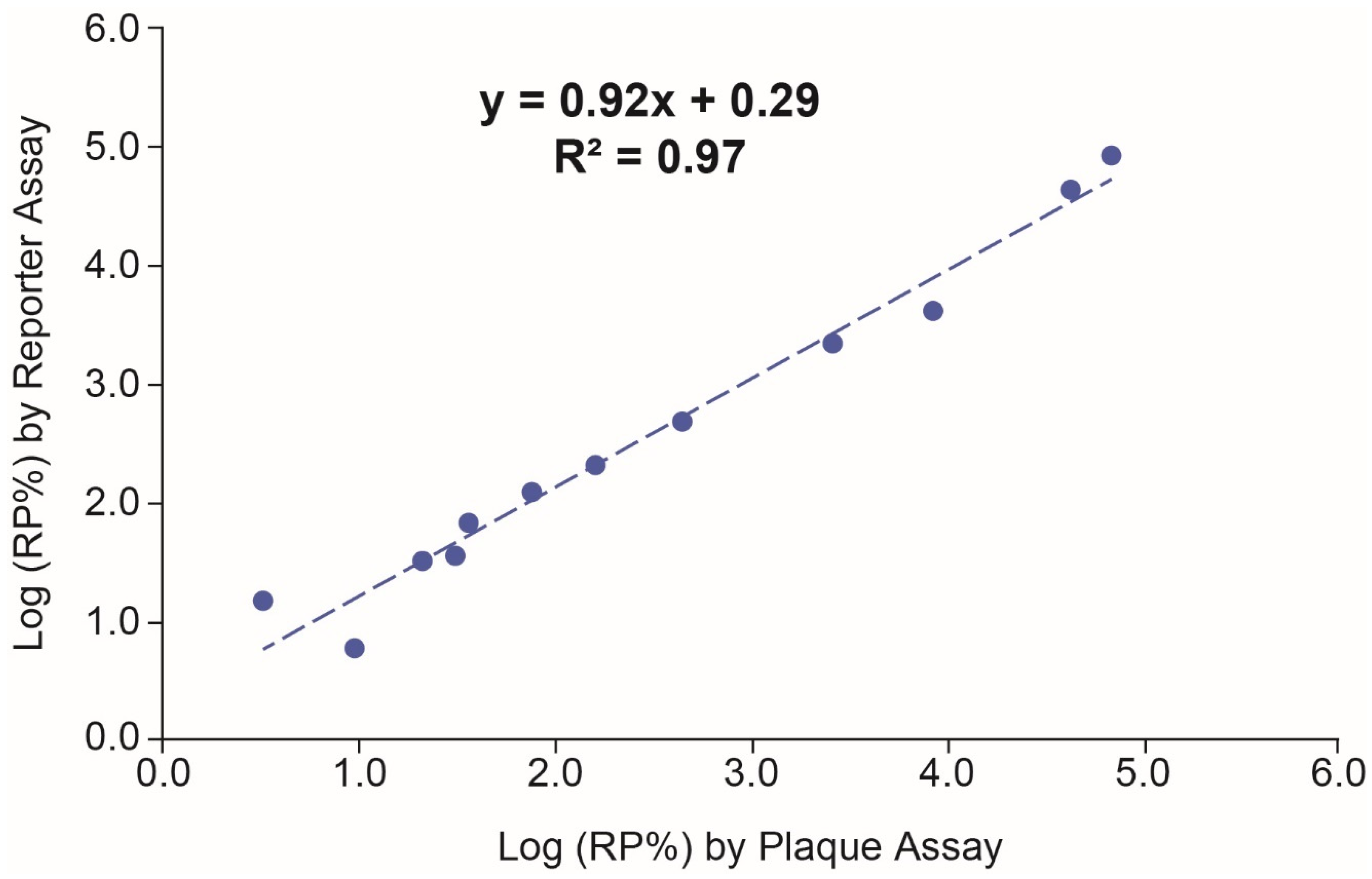
3.4. Correlation between the Reporter Assay and Plaque Assay
3.5. Vaccine 2 Reporter Assay
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sanyal, G. Development of functionally relevant potency assays for monovalent and multivalent vaccines delivered by evolving technologies. NPJ Vaccines 2022, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, G.; Särnefält, A.; Kumar, A. Considerations for bioanalytical characterization and batch release of COVID-19 vaccines. NPJ Vaccines 2021, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Verch, T.; Trausch, J.J.; Shank-Retzlaff, M. Principles of vaccine potency assays. Bioanalysis 2018, 10, 163–180. [Google Scholar] [CrossRef] [PubMed]
- Dulbecco, R.; Vogt, M. Some problems of animal virology as studied by the plaque technique. Cold Spring Harb. Symp. Quant. Biol. 1953, 18, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Baer, A.; Kehn-Hall, K. Viral concentration determination through plaque assays: Using traditional and novel overlay systems. J. Vis. Exp. 2014, 4, e52065. [Google Scholar]
- Juarez, D.; Long, K.C.; Aguilar, P.; Kochel, T.J.; Halsey, E.S. Assessment of plaque assay methods for alphaviruses. J. Virol. Methods. 2013, 187, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.L.; Loh, P.C. Comparison of the immunofluorescent-cell counting and plaque methods for the assay of vaccinia virus. Appl. Microbiol. 1972, 23, 214–217. [Google Scholar] [CrossRef] [PubMed]
- McClain, M.E.; Spendlove, R.S.; Lennette, E.H. Infectivity assay of Reoviruses: Comparison of immunofluorescent cell count and plaque methods. J. Immunol. 1967, 98, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Citron, M.; Bett, A.J.; Espeseth, A.S.; Vora, K.A.; Zhang, L.; DiStefano, D.J. Development and application of a higher throughput RSV plaque assay by immunofluorescent imaging. J. Virol. Methods 2019, 263, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Flint, S.J.; Enquist, W.; Racaniello, V.R.; Skalka, A.M. “Virological Methods”. In Principles of Virology; ASM Press: Washington, DC, USA, 2009; ISBN 978-1-55581-443-4. [Google Scholar]
- Smither, S.J.; Lear-Rooney, C.; Biggins, J.; Pettitt, J.; Lever, M.S.; Olinger, G.G., Jr. Comparison of the plaque assay and 50% tissue culture infectious dose assay as methods for measuring filovirus infectivity. J. Virol. Methods 2013, 193, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.T.; Tsai, H.Y.; Liu, C.P.; Yuan, T.T. Comparability of bovine virus titers obtained by TCID50/ml and FAID50/ml. J. Virol. Methods 2010, 165, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Schalk, J.A.; van den Elzen, C.; Ovelgönne, H.; Baas, C.; Jongen, P.M. Estimation of the number of infectious measles viruses in live virus vaccines using quantitative real-time PCR. J. Virol. Methods 2004, 117, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Ranheim, T.; Mathis, P.K.; Joelsson, D.B.; Smith, M.E.; Campbell, K.M.; Lucas, G.; Barmat, S.; Melissen, E.; Benz, R.; Lewis, J.A.; et al. Development and application of a quantitative RT-PCR potency assay for a pentavalent rotavirus vaccine (RotaTeq). J. Virol. Methods 2006, 131, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.; Bhambhani, A.; Blue, J. PubChem Patent Summary for US-2020181678-A1, Cell-Based Reporter Assay for Live Virus Vaccines. Available online: https://pubchem.ncbi.nlm.nih.gov/patent/US-2020181678-A1 (accessed on 25 June 2023).
- USP 1034; Analysis of Biological Assasys. The United States Pharmacopeial Convention: Frederick, MD, USA, 2010.
- Walhart, T.; Isaacson-Wechsler, E.; Ang, K.H.; Arkin, M.; Tugizov, S.; Palefsky, J.M. A Cell-Based Renilla Luminescence Reporter Plasmid Assay for High-Throughput Screening to Identify Novel FDA-Approved Drug Inhibitors of HPV-16 Infection. SLAS Discov. 2020, 25, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, Y.; Ohmiya, Y. Bioluminescence assays: Multicolor luciferase assay, secreted luciferase assay and imaging luciferase assay. Expert. Opin. Drug Discov. 2010, 5, 835–849. [Google Scholar] [CrossRef] [PubMed]
- England, C.G.; Ehlerding, E.B.; Cai, W. NanoLuc: A Small Luciferase Is Brightening Up the Field of Bioluminescence. Bioconjugate Chem. 2016, 27, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Yie, J.; Dey, M.; Su, J.; Sergi, J.; Zhang, Y.; Le, T.H.; Kashi, S.; Gurney, K. Development of a robust functional cell-based assay for replacing the rabbit blood sugar bioidentity test of insulin glargine drug substance. J. Pharm. Biomed. Anal. 2020, 186, 113328. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Hsu, A.; Quiroz, J.; He, X.; Whiteman, M.C.; Gurney, K.B.; Dellatore, S. Development and comparison of three cell-based potency assays for anti-respiratory syncytial virus monoclonal antibody. Biologicals 2021, 74, 1–9. [Google Scholar] [CrossRef] [PubMed]



| Parameter | Low | Set Point | High | |
|---|---|---|---|---|
| Cell seeding density (cell/well) | 8000 | 10,000 | 12,000 | |
| Cell seeding time (hours) | 20 | 24 | 26 | |
| Reference concentration (PFU/mL) | 8000 | 10,000 | 12,000 | |
| Infection time (hours) | 44 | 46 | 48 | |
| Cell passage | Low (≤25) | High (≥30) | ||
| Substrate incubation time (min) | 10 | 20 | ||
| Performance Characteristics | Results |
|---|---|
| Dilutional Linearity | The slope, and root mean square error (RMSE) of the regression analysis based on all 8 runs are estimated to be 0.94, 0.95 and 0.21, respectively.The %dilution bias based on the overall regression analysis is calculated to be −4.6%. |
| Accuracy | The largest %relative bias is 19.0% at the 50% potency level with the 90% CI being (3.0%, 37.5%). |
| Repeatability | The overall %RSD for repeatability is 16.8%. |
| Intermediate Precision | The overall %RSD for intermediate precision is 25.2%. |
| Substrate Incubation Time | Target RP% | n | Geomean | %Relative Bias | Repeatability | Intermediate Precision |
|---|---|---|---|---|---|---|
| 10 min | 25 | 32 | 29.2 | 16.6 | 29.4 | 29.4 |
| 50 | 32 | 54.9 | 9.8 | 16.8 | 17.3 | |
| 100 | 32 | 98.7 | −1.3 | 15.1 | 15.3 | |
| 200 | 32 | 183.0 | −8.5 | 17.3 | 17.9 | |
| Overall | 20.4 | 20.7 | ||||
| 20 min | 25 | 25 | 29.3 | 17.1 | 30.9 | 31.9 |
| 50 | 25 | 56.0 | 11.9 | 16.4 | 17.3 | |
| 100 | 25 | 99.6 | −0.4 | 14.7 | 15.1 | |
| 200 | 25 | 180.9 | −9.5 | 18.4 | 19.1 | |
| Overall | 21.0 | 21.9 | ||||
| Performance Characteristics | Results |
|---|---|
| Dilutional Linearity | The slope, and RMSE for the overall regression analysis are estimated to be 0.99, 0.98 and 0.14, respectively. The %dilution bias based on the overall regression analysis is calculated to be −0.8%. |
| Accuracy | The largest %relative bias is −10.6% at the 200% potency level with the 90% CI being (−16.0%, −4.8%). |
| Repeatability | The repeatability %RSD based on all samples is 14.0%. |
| Intermediate Precision | The intermediate precision %RSD based on all samples is 16.0%. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, D.; Meyer, B.K.; Deevi, D.S.; Mirza, A.; He, L.; Gruber, A.; Abbondanzo, S.J.; Benton, N.A.; Whiteman, M.C.; Capen, R.C.; et al. Development of a Cell-Based Reporter Potency Assay for Live Virus Vaccines. Vaccines 2024, 12, 769. https://doi.org/10.3390/vaccines12070769
Sun D, Meyer BK, Deevi DS, Mirza A, He L, Gruber A, Abbondanzo SJ, Benton NA, Whiteman MC, Capen RC, et al. Development of a Cell-Based Reporter Potency Assay for Live Virus Vaccines. Vaccines. 2024; 12(7):769. https://doi.org/10.3390/vaccines12070769
Chicago/Turabian StyleSun, Dengyun, Brian K. Meyer, Dhanvanthri S. Deevi, Asra Mirza, Li He, Ashley Gruber, Susan J. Abbondanzo, Noah A. Benton, Melissa C. Whiteman, Robert C. Capen, and et al. 2024. "Development of a Cell-Based Reporter Potency Assay for Live Virus Vaccines" Vaccines 12, no. 7: 769. https://doi.org/10.3390/vaccines12070769






