Subsequent Waves of Convergent Evolution in SARS-CoV-2 Genes and Proteins
Abstract
:1. Introduction
2. Convergent Evolution in SARS-CoV-2 Structural Proteins (SP)
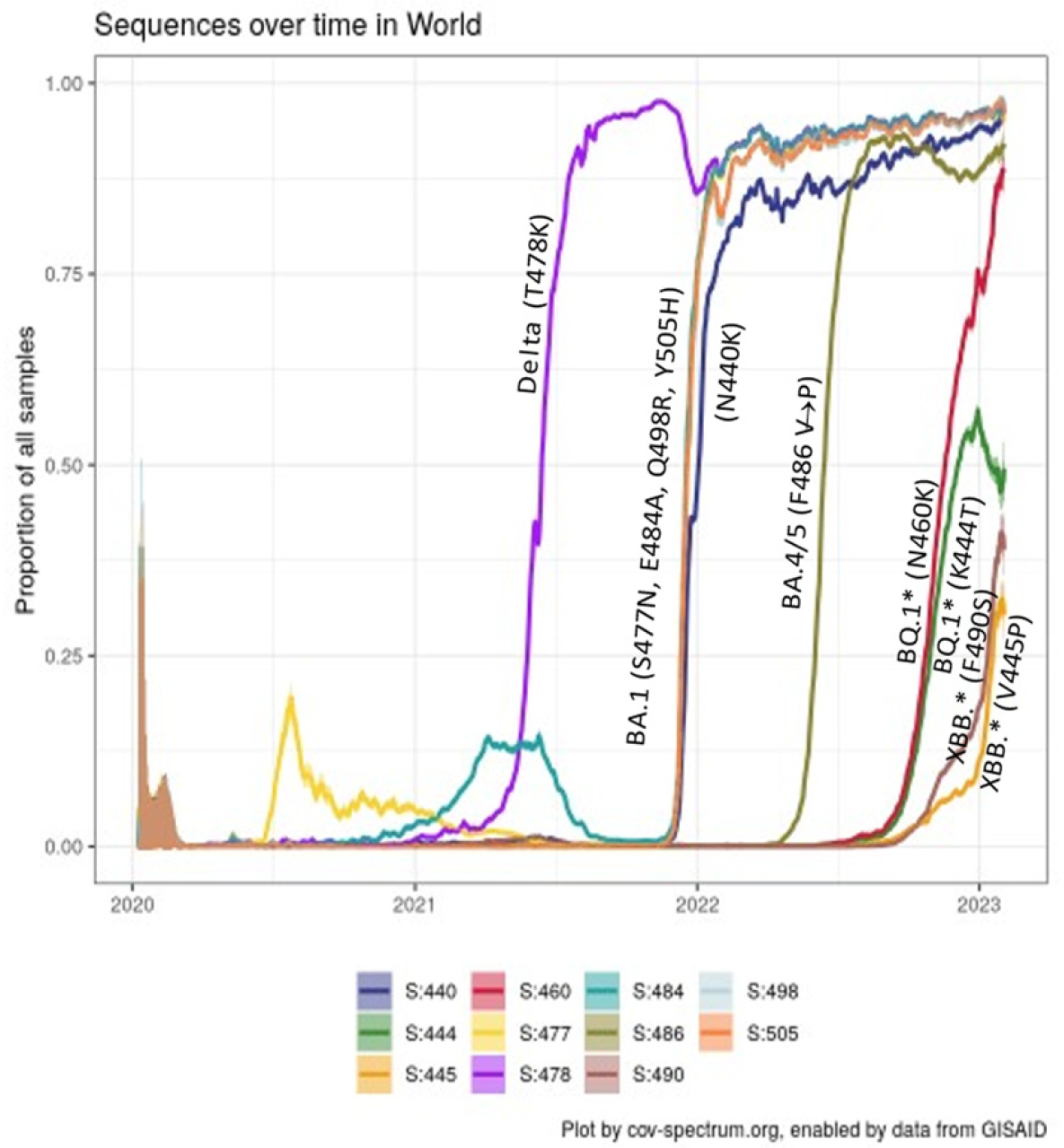
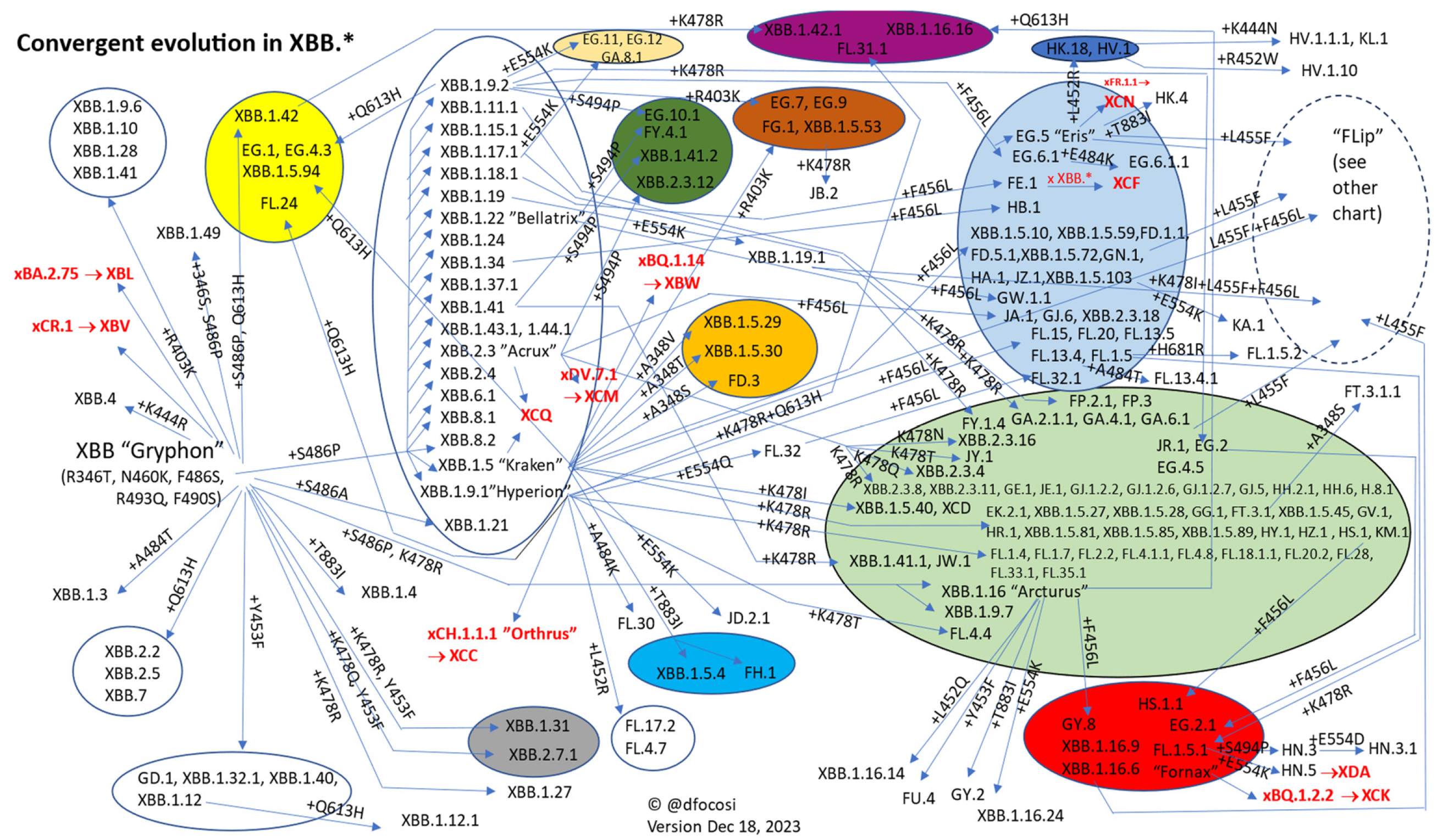
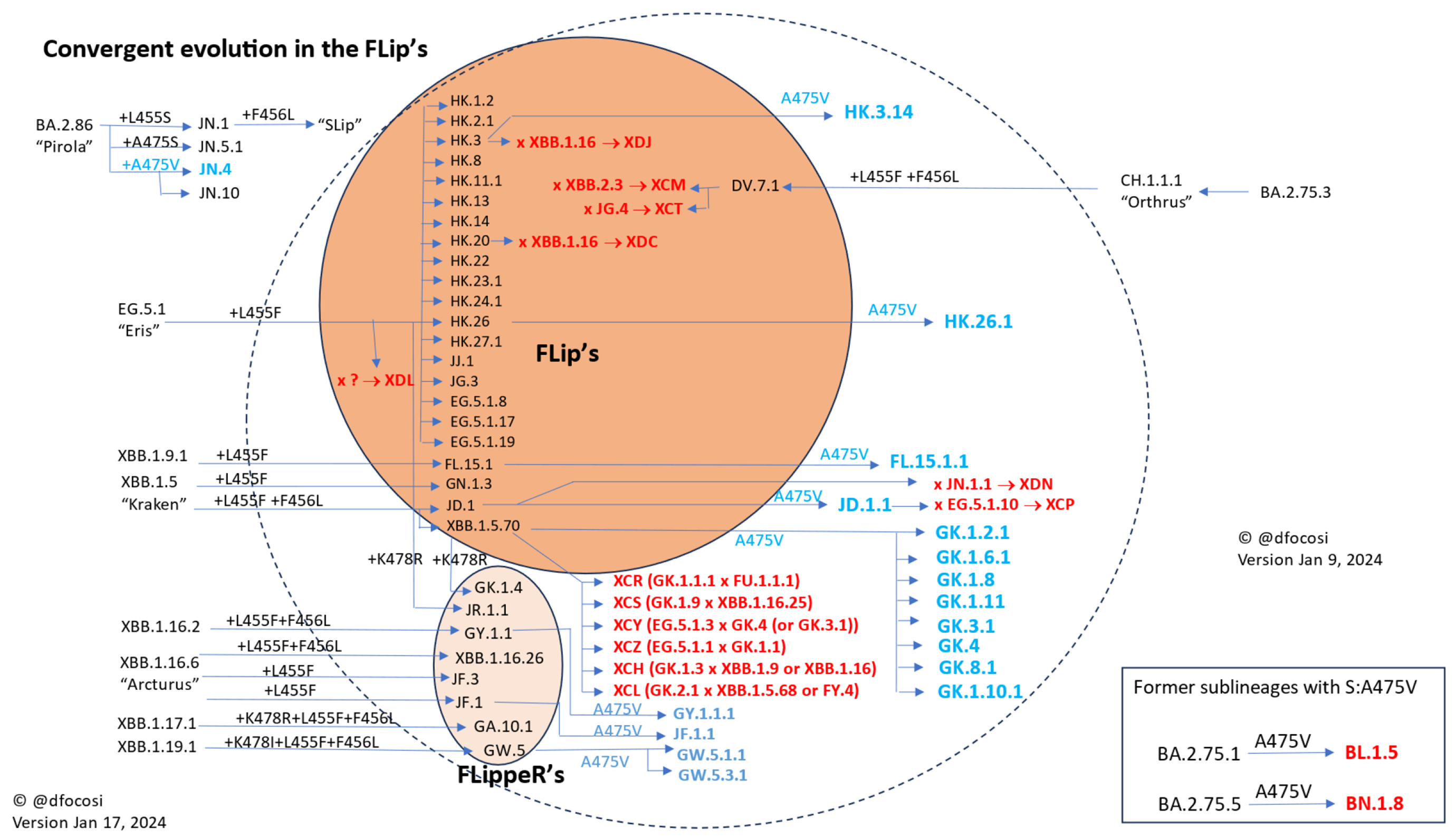
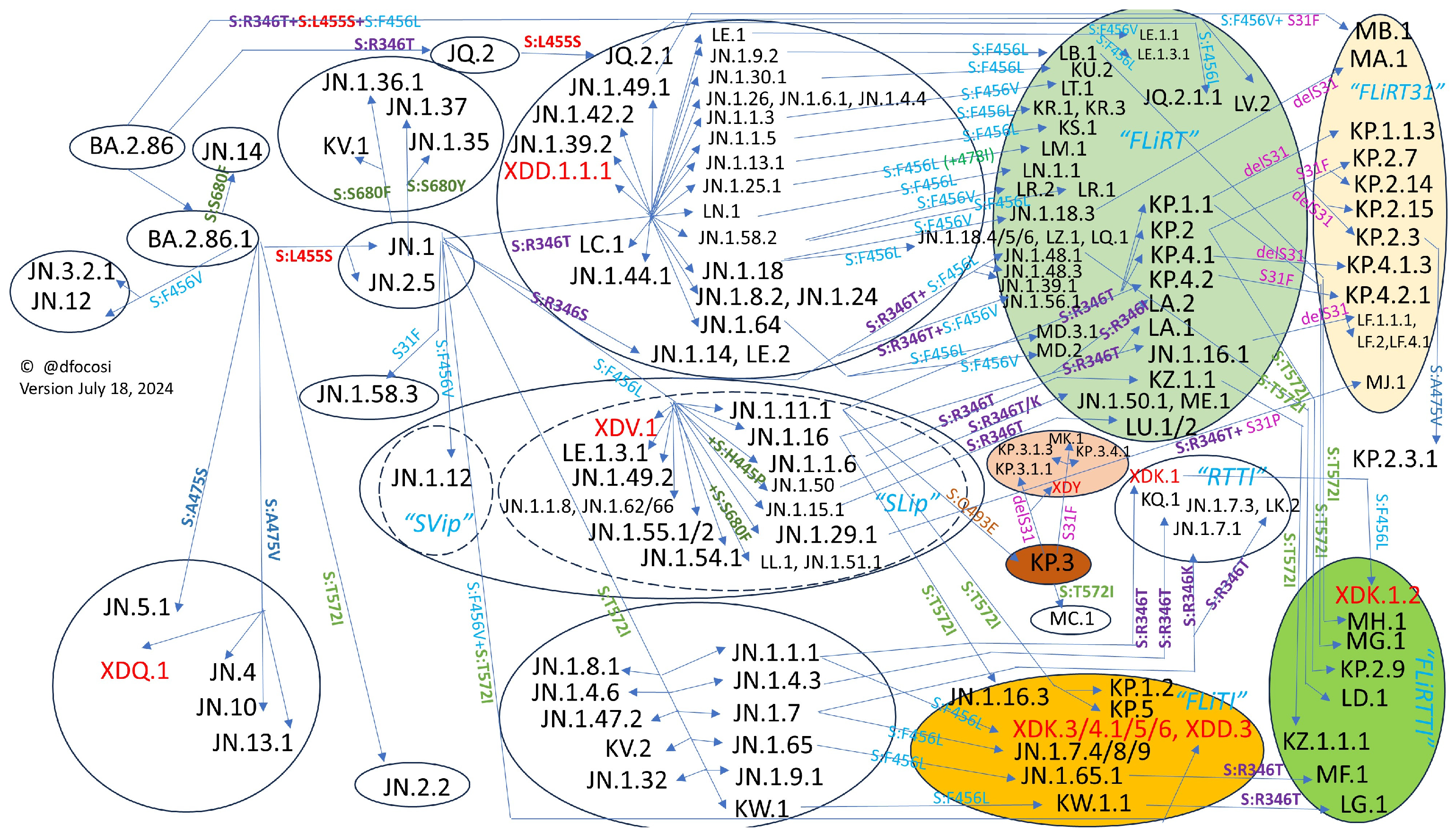
2.1. Convergence in the Spike Protein
2.2. Convergence in the RBD
2.3. Epistatic Interactions among Spike Mutations
2.4. “Yo-Yo” and Fixated Mutations
2.5. Disappearance of the Furin-Cleavage Site (FCS)
2.6. Saltation Variants
2.7. Alterations at N-Glycosylation Sites
- •
- S:F32S (convergent in chronics) leads to a N-glycosylation site at N30
- •
- S:R190T (found in Gamma and KP.3.1.1) leads to a N-glycosylation site at N188
- •
- S:H245N (found in BA.2.86) leads to a N-glycosylation site at N245
- •
- S:Y248N (found in BA.2.76 and BA.2.86) leads to a N-glycosylation site at 250
- •
- S:K356T (e.g., in BA.2.86*) leads to a N-glycosylation site at N354. Glycosylation at N354 is a regulatory element for the RBD conformation that lessens viral infectivity.
2.8. NTD Deletions and Their Functional Consequences
- •
- Alpha and BA.1.1;
- •
- •
- C.1.2 from Africa similarly had both P9L and C136F;
- •
- B.1.427 (VOI Epsilon) from US West had S13I, but a new disulfide bond with C136 was introduced by W152C, causing total rearrangement and immune evasion [70];
- •
- BA.2.87 had Δ14-22 and Δ135-146.
2.9. Reversion to SARS-CoV-1 and/or bat-CoV Residues
2.10. Truncation and Mutations of Nucleocapsid
3. Convergent Evolution in Nonstructural Proteins (NSPs)
4. Convergent Evolution in Accessory Proteins (AP)
4.1. ORF6 Mutations
4.2. ORF7a Loss
4.3. ORF7b Stop Codons Create New Proteins
4.4. ORF8 Deletion
4.5. ORF9b Increased Translation
5. Shared Mutations in Persistent Infections
6. Forecasting SARS-CoV-2 Evolution
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pawlica, P.; Yario, T.A.; White, S.; Wang, J.; Moss, W.N.; Hui, P.; Vinetz, J.M.; Steitz, J.A. SARS-CoV-2 expresses a microRNA-like small RNA able to selectively repress host genes. Proc. Natl. Acad. Sci. USA 2021, 118, e2116668118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Bisht, P.; Flamier, A.; Barrasa, M.I.; Friesen, M.; Richards, A.; Hughes, S.H.; Jaenisch, R. LINE1-Mediated Reverse Transcription and Genomic Integration of SARS-CoV-2 mRNA Detected in Virus-Infected but Not in Viral mRNA-Transfected Cells. Viruses 2023, 15, 629. [Google Scholar] [CrossRef] [PubMed]
- Farjo, M.; Koelle, K.; Martin, M.A.; Gibson, L.L.; Walden, K.K.O.; Rendon, G.; Fields, C.J.; Alnaji, F.G.; Gallagher, N.; Luo, C.H.; et al. Within-host evolutionary dynamics and tissue compartmentalization during acute SARS-CoV-2 infection. bioRxiv 2022. [Google Scholar] [CrossRef] [PubMed]
- Normandin, E.; Rudy, M.; Barkas, N.; Schaffner, S.F.; Levine, Z.; Padera, R.F.; Babadi, M.; Mukerji, S.S.; Park, D.J.; MacInnis, B.L.; et al. High-depth sequencing characterization of viral dynamics across tissues in fatal COVID-19 reveals compartmentalized infection. Nat. Commun. 2023, 14, 574. [Google Scholar] [CrossRef] [PubMed]
- Swank, Z.; Senussi, Y.; Manickas-Hill, Z.; Yu, X.G.; Li, J.Z.; Alter, G.; Walt, D.R. Persistent Circulating Severe Acute Respiratory Syndrome Coronavirus 2 Spike Is Associated With Post-acute Coronavirus Disease 2019 Sequelae. Clin. Infect. Dis. 2023, 76, e487–e490. [Google Scholar] [CrossRef] [PubMed]
- Stein, S.R.; Ramelli, S.C.; Grazioli, A.; Chung, J.-Y.; Singh, M.; Yinda, C.K.; Winkler, C.W.; Sun, J.; Dickey, J.M.; Ylaya, K.; et al. SARS-CoV-2 infection and persistence in the human body and brain at autopsy. Nature 2022, 612, 758–763. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, A.; Zlitni, S.; Brooks, E.F.; Vance, S.E.; Dahlen, A.; Hedlin, H.; Park, R.M.; Han, A.; Schmidtke, D.T.; Verma, R.; et al. Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA suggest prolonged gastrointestinal infection. Med 2022, 3, 371–387.e9. [Google Scholar] [CrossRef]
- Tomasicchio, M.; Jaumdally, S.; Pooran, A.; Esmail, A.; Wilson, L.; Kotze, A.; Semple, L.; Meier, S.; Pillay, K.; Roberts, R.; et al. SARS-CoV-2 viral replication persists in the human lung for several weeks after symptom onset in mechanically ventilated patients with severe COVID-19: A prospective tissue-sampling study. medRxiv 2023. [Google Scholar] [CrossRef]
- Yonker, L.M.; Gilboa, T.; Ogata, A.F.; Senussi, Y.; Lazarovits, R.; Boribong, B.P.; Bartsch, Y.C.; Loiselle, M.; Rivas, M.N.; Porritt, R.A.; et al. Multisystem inflammatory syndrome in children is driven by zonulin-dependent loss of gut mucosal barrier. J. Clin. Investig. 2021, 131, e149633. [Google Scholar] [CrossRef]
- Su, Y.; Yuan, D.; Chen, D.G.; Ng, R.H.; Wang, K.; Choi, J.; Li, S.; Hong, S.; Zhang, R.; Xie, J.; et al. Multiple early factors anticipate post-acute COVID-19 sequelae. Cell 2022, 185, 881–895.e20. [Google Scholar] [CrossRef]
- Wang, Y.; Ye, M.; Zhang, F.; Freeman, Z.T.; Yu, H.; Ye, X.; He, Y. Taxonomical and ontological analysis of verified natural and laboratory human coronavirus hosts. bioRxiv 2023. [Google Scholar] [CrossRef]
- Oude Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Pickering, B.; Lung, O.; Maguire, F.; Kruczkiewicz, P.; Kotwa, J.D.; Buchanan, T.; Gagnier, M.; Guthrie, J.L.; Jardine, C.M.; Marchand-Austin, A.; et al. Divergent SARS-CoV-2 variant emerges in white-tailed deer with deer-to-human transmission. Nat. Microbiol. 2022, 7, 2011–2024. [Google Scholar] [CrossRef] [PubMed]
- Fischhoff, I.R.; Castellanos, A.A.; Rodrigues, J.; Varsani, A.; Han, B.A. Predicting the zoonotic capacity of mammals to transmit SARS-CoV-2. Proceedings. Biol. Sci. 2021, 288, 20211651. [Google Scholar] [CrossRef] [PubMed]
- Caserta, L.C.; Martins, M.; Butt, S.L.; Hollingshead, N.A.; Covaleda, L.M.; Ahmed, S.; Everts, M.R.R.; Schuler, K.L.; Diel, D.G. White-tailed deer (Odocoileus virginianus) may serve as a wildlife reservoir for nearly extinct SARS-CoV-2 variants of concern. Proc. Natl. Acad. Sci. USA 2023, 120, e2215067120. [Google Scholar] [CrossRef] [PubMed]
- Domańska-Blicharz, K.; Oude Munnink, B.B.; Orłowska, A.; Smreczak, M.; Opolska, J.; Lisowska, A.; Trębas, P.; Socha, W.; Giza, A.; Bomba, A.; et al. Cryptic SARS-CoV-2 lineage identified on two mink farms as a possible result of long-term undetected circulation in an unknown animal reservoir, Poland, November 2022 to January 2023. Eurosurveillance Bull. Eur. Mal. Transm. Eur. Commun. Dis. Bull. 2023, 28, 2300188. [Google Scholar] [CrossRef]
- Cov-Lineages/Pango-Designation. Available online: https://github.com/cov-lineages/pango-designation/blob/7651872d633c8e94437830f792b272712d4a22fc/lineage_notes.txt (accessed on 13 February 2024).
- WHO. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 13 February 2024).
- Focosi, D.; Maggi, F. How SARS-CoV-2 Big Data Are Challenging Viral Taxonomy Rules. Viruses 2023, 15, 715. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; McConnell, S.; Casadevall, A. The Omicron variant of concern: Diversification and convergent evolution in spike protein, and escape from anti-Spike monoclonal antibodies. Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer Chemother. 2022, 65, 100882. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; Maggi, F.; McConnell, S.; Casadevall, A. Spike mutations in SARS-CoV-2 AY Sublineages of Delta Variant of Concern: Implications for the future of the pandemic. Future Microbiol. 2022, 17, 219–221. [Google Scholar] [CrossRef]
- Focosi, D.; Maggi, F. Second-Generation SARS-CoV-2 Recombinants: Lessons from Other Viruses. Viruses 2023, 15, 1063. [Google Scholar] [CrossRef]
- Variant Report 2023-06-29. Available online: https://github.com/neherlab/SARS-CoV-2_variant-reports/blob/main/reports/variant_report_latest_draft.md (accessed on 3 July 2023).
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef] [PubMed]
- Carabelli, A.M.; Peacock, T.P.; Thorne, L.G.; Harvey, W.T.; Hughes, J.; de Silva, T.I.; Peacock, S.J.; Barclay, W.S.; de Silva, T.I.; Towers, G.J.; et al. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D. SARS-CoV-2 Spike Protein Convergent Evolution; Springer: Cham, Switzerland, 2021. [Google Scholar]
- Zhang, J.; Xiao, T.; Cai, Y.; Chen, B. Structure of SARS-CoV-2 spike protein. Curr. Opin. Virol. 2021, 50, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nature reviews. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Paredes, M.I.; Lunn, S.M.; Famulare, M.; Frisbie, L.A.; Painter, I.; Burstein, R.; Roychoudhury, P.; Xie, H.; Mohamed Bakhash, S.A.; Perez, R.; et al. Associations Between Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Variants and Risk of Coronavirus Disease 2019 (COVID-19) Hospitalization Among Confirmed Cases in Washington State: A Retrospective Cohort Study. Clin. Infect. Dis. 2022, 75, e536–e544. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Abdullahi, A.; Ferreira, I.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Hui, K.P.Y.; Ho, J.C.W.; Cheung, M.C.; Ng, K.C.; Ching, R.H.H.; Lai, K.L.; Kam, T.T.; Gu, H.; Sit, K.Y.; Hsin, M.K.Y.; et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 2022, 603, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Yamasoba, D.; Kimura, I.; Wang, L.; Kishimoto, M.; Ito, J.; Morioka, Y.; Nao, N.; Nasser, H.; Uriu, K.; et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 2022, 603, 700–705. [Google Scholar] [CrossRef] [PubMed]
- Willett, B.J.; Grove, J.; MacLean, O.A.; Wilkie, C.; De Lorenzo, G.; Furnon, W.; Cantoni, D.; Scott, S.; Logan, N.; Ashraf, S.; et al. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat. Microbiol. 2022, 7, 1161–1179. [Google Scholar] [CrossRef]
- Qing, E.; Gallagher, T. Adaptive variations in SARS-CoV-2 spike proteins: Effects on distinct virus-cell entry stages. mBio 2023, 14, e0017123. [Google Scholar] [CrossRef]
- Peacock, T.P.; Brown, J.C.; Zhou, J.; Thakur, N.; Sukhova, K.; Newman, J.; Kugathasan, R.; Yan, A.W.C.; Furnon, W.; Lorenzo, G.D.; et al. The altered entry pathway and antigenic distance of the SARS-CoV-2 Omicron variant map to separate domains of spike protein. bioRxiv 2022. [Google Scholar] [CrossRef]
- Gartner, M.J.; Lee, L.Y.Y.; Mordant, F.L.; Suryadinata, R.; Chen, J.; Robinson, P.; Polo, J.M.; Subbarao, K. Ancestral, Delta, and Omicron (BA.1) SARS-CoV-2 strains are dependent on serine proteases for entry throughout the human respiratory tract. Med 2023, 4, 944–955.e7. [Google Scholar] [CrossRef] [PubMed]
- Mykytyn, A.Z.; Breugem, T.I.; Geurts, M.H.; Beumer, J.; Schipper, D.; van Acker, R.; van den Doel, P.B.; van Royen, M.E.; Zhang, J.; Clevers, H.; et al. SARS-CoV-2 Omicron entry is type II transmembrane serine protease-mediated in human airway and intestinal organoid models. J. Virol. 2023, 97, e0085123. [Google Scholar] [CrossRef] [PubMed]
- Metzdorf, K.; Jacobsen, H.; Greweling-Pils, M.C.; Hoffmann, M.; Lüddecke, T.; Miller, F.; Melcher, L.; Kempf, A.M.; Nehlmeier, I.; Bruder, D.; et al. TMPRSS2 Is Essential for SARS-CoV-2 Beta and Omicron Infection. Viruses 2023, 15, 271. [Google Scholar] [CrossRef] [PubMed]
- Heurich, A.; Hofmann-Winkler, H.; Gierer, S.; Liepold, T.; Jahn, O.; Pöhlmann, S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014, 88, 1293–1307. [Google Scholar] [CrossRef] [PubMed]
- Shulla, A.; Heald-Sargent, T.; Subramanya, G.; Zhao, J.; Perlman, S.; Gallagher, T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011, 85, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, A.; Fichter, C.; Milogiannakis, V.; Akerman, A.; Ison, T.; Silva, M.R.; Esneau, C.; Bartlett, N.; Burrell, L.; Patel, S.; et al. TMPRSS2 activation of Omicron lineage Spike glycoproteins is regulated by TMPRSS2 cleavage of ACE2. bioRxiv 2023. [Google Scholar] [CrossRef]
- Stevens, B.R. TMPRSS2 and ADAM17 interactions with ACE2 complexed with SARS-CoV-2 and B0AT1 putatively in intestine, cardiomyocytes, and kidney. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kistler, K.E.; Bedford, T. Evidence for adaptive evolution in the receptor-binding domain of seasonal coronaviruses OC43 and 229e. eLife 2021, 10, e64509. [Google Scholar] [CrossRef] [PubMed]
- Foster, C.S.P.; Walker, G.J.; Jean, T.; Wong, M.; Brassil, L.; Isaacs, S.; Lu, Y.; Turville, S.G.; Kelleher, A.; Rawlinson, W.D. Long-term serial passaging of SARS-CoV-2 reveals signatures of convergent evolution. bioRxiv 2023. [Google Scholar] [CrossRef]
- Focosi, D.; Quiroga, R.; McConnell, S.; Johnson, M.C.; Casadevall, A. Convergent evolution in SARS-CoV-2 Spike creates a variant soup from which new COVID-19 waves emerge. Int. J. Mol. Sci. 2023, 24, 2264. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; Spezia, P.G.; Gueli, F.; Maggi, F. The Era of the FLips: How Spike Mutations L455F and F456L (and A475V) Are Shaping SARS-CoV-2 Evolution. Viruses 2024, 16, 3. [Google Scholar] [CrossRef]
- Jian, F.; Feng, L.; Yang, S.; Yu, Y.; Wang, L.; Song, W.; Yisimayi, A.; Chen, X.; Xu, Y.; Wang, P.; et al. Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455–456 synergistically enhances antibody evasion and ACE2 binding. PLoS Pathog. 2023, 19, e1011868. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; Spezia, P.G.; Maggi, F. SARS-CoV-2 BA.2.86 (“Pirola”): Is it Pi or Just Another Omicron Sublineage? Vaccines 2023, 11, 1634. [Google Scholar] [CrossRef] [PubMed]
- Kaku, Y.; Uriu, K.; Kosugi, Y.; Okumura, K.; Yamasoba, D.; Uwamino, Y.; Kuramochi, J.; Sadamasu, K.; Yoshimura, K.; Asakura, H.; et al. Virological characteristics of the SARS-CoV-2 KP.2 variant. bioRxiv 2024. [Google Scholar] [CrossRef]
- Jian, F.; Yisimayi, A.; Song, W.; Wang, J.; Xu, Y.; Chen, X.; Yu, Y.; Niu, X.; Yang, S.; Wang, P.; et al. Humoral immunogenicity comparison of XBB and JN.1 in human infections. bioRxiv 2024. [Google Scholar] [CrossRef]
- Leist, S.R.; Dinnon, K.H., 3rd; Schäfer, A.; Tse, L.V.; Okuda, K.; Hou, Y.J.; West, A.; Edwards, C.E.; Sanders, W.; Fritch, E.J.; et al. A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell 2020, 183, 1070–1085.e12. [Google Scholar] [CrossRef]
- Liu, Z.; Zheng, H.; Lin, H.; Li, M.; Yuan, R.; Peng, J.; Xiong, Q.; Sun, J.; Li, B.; Wu, J.; et al. Identification of Common Deletions in the Spike Protein of Severe Acute Respiratory Syndrome Coronavirus 2. J. Virol. 2020, 94, 10-1128. [Google Scholar] [CrossRef]
- Heo, G.; Kang, Y.; An, S.; Kim, Y.; Cha, R.; Jang, Y. Concurrent Infection with Clade 2.3.4.4b Highly Pathogenic Avian Influenza H5N6 and H5N1 Viruses, South Korea, 2023. Emerg. Infect. Dis. 2024, 30, 1223–1227. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Hannon, W.W.; Loes, A.N.; Hauser, K.; Dillen, J.R.; Ferri, E.; Farrell, A.G.; Dadonaite, B.; McCallum, M.; et al. Shifting mutational constraints in the SARS-CoV-2 receptor-binding domain during viral evolution. Science 2022, 377, 420–424. [Google Scholar] [CrossRef]
- Moulana, A.; Dupic, T.; Phillips, A.M.; Chang, J.; Nieves, S.; Roffler, A.A.; Greaney, A.J.; Starr, T.N.; Bloom, J.D.; Desai, M.M. Compensatory epistasis maintains ACE2 affinity in SARS-CoV-2 Omicron BA.1. Nat. Commun. 2022, 13, 7011. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Mizuma, K.; Nasser, H.; Deguchi, S.; Padilla-Blanco, M.; Oda, Y.; Uriu, K.; Tolentino, J.E.M.; Tsujino, S.; Suzuki, R.; et al. Virological characteristics of the SARS-CoV-2 BA.2.86 variant. Cell Host Microbe 2024, 32, 170–180.e12. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; Spezia, P.G.; Maggi, F. Fixation and reversion of mutations in the receptor-binding domain of SARS-CoV-2 Spike protein. Diagn. Microbiol. Infect. Dis. 2023, 108, 116104. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.-J.; Lee, Y.-h.; Khalid, M.M.; Chen, I.P.; Zhang, Y.; Ott, M.; Verdin, E. SARS-CoV-2 protein ORF8 limits expression levels of Spike antigen and facilitates immune evasion of infected host cells. J. Biol. Chem. 2023, 299, 104955. [Google Scholar] [CrossRef] [PubMed]
- South African National HIV Prevalence, Incidence, Behaviour and Communication Survey. 2017. Available online: https://repository.hsrc.ac.za/bitstream/handle/20.500.11910/15052/11091.pdf#page=32 (accessed on 3 February 2024).
- Liu, P.; Yue, C.; Meng, B.; Xiao, T.; Yang, S.; Liu, S.; Jian, F.; Zhu, Q.; Yu, Y.; Ren, Y.; et al. Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through multiple mechanisms. bioRxiv 2024. [Google Scholar] [CrossRef]
- Cantoni, D.; Murray, M.J.; Kalemera, M.D.; Dicken, S.J.; Stejskal, L.; Brown, G.; Lytras, S.; Coey, J.D.; McKenna, J.; Bridgett, S.; et al. Evolutionary remodelling of N-terminal domain loops fine-tunes SARS-CoV-2 spike. EMBO Rep. 2022, 23, e54322. [Google Scholar] [CrossRef] [PubMed]
- Qing, E.; Li, P.; Cooper, L.; Schulz, S.; Jäck, H.M.; Rong, L.; Perlman, S.; Gallagher, T. Inter-domain communication in SARS-CoV-2 spike proteins controls protease-triggered cell entry. Cell Rep. 2022, 39, 110786. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Go, E.P.; Ding, H.; Anang, S.; Kappes, J.C.; Desaire, H.; Sodroski, J.G. Analysis of Glycosylation and Disulfide Bonding of Wild-Type SARS-CoV-2 Spike Glycoprotein. J. Virol. 2022, 96, e0162621. [Google Scholar] [CrossRef]
- McCarthy, K.R.; Rennick, L.J.; Nambulli, S.; Robinson-McCarthy, L.R.; Bain, W.G.; Haidar, G.; Duprex, W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 2021, 371, 1139–1142. [Google Scholar] [CrossRef]
- Meng, B.; Kemp, S.A.; Papa, G.; Datir, R.; Ferreira, I.; Marelli, S.; Harvey, W.T.; Lytras, S.; Mohamed, A.; Gallo, G.; et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021, 35, 109292. [Google Scholar] [CrossRef] [PubMed]
- Qing, E.; Kicmal, T.; Kumar, B.; Hawkins, G.M.; Timm, E.; Perlman, S.; Gallagher, T. Dynamics of SARS-CoV-2 Spike Proteins in Cell Entry: Control Elements in the Amino-Terminal Domains. mBio 2021, 12, e0159021. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Juraszek, J.; Rutten, L.; Bakkers, M.J.G.; Blokland, S.; Melchers, J.M.; van den Broek, N.J.F.; Verwilligen, A.Y.W.; Abeywickrema, P.; Vingerhoets, J.; et al. Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike. PLoS Pathog. 2023, 19, e1011308. [Google Scholar] [CrossRef] [PubMed]
- Bolland, W.; Michel, V.; Planas, D.; Hubert, M.; Staropoli, I.; Guivel-Benhassine, F.; Porrot, F.; N’Debi, M.; Rodriguez, C.; Fourati, S.; et al. High fusion and cytopathy of SARS-CoV-2 variant B.1.640.1. J. Virol. 2024, 98, e0135123. [Google Scholar] [CrossRef] [PubMed]
- McCallum, M.; Bassi, J.; De Marco, A.; Chen, A.; Walls, A.C.; Di Iulio, J.; Tortorici, M.A.; Navarro, M.-J.; Silacci-Fregni, C.; Saliba, C.; et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science 2021, 373, 648–654. [Google Scholar] [CrossRef]
- Zhang, L.; Kempf, A.; Nehlmeier, I.; Cossmann, A.; Richter, A.; Bdeir, N.; Graichen, L.; Moldenhauer, A.S.; Dopfer-Jablonka, A.; Stankov, M.V.; et al. SARS-CoV-2 BA.2.86 enters lung cells and evades neutralizing antibodies with high efficiency. Cell 2024, 187, 596–608.e17. [Google Scholar] [CrossRef]
- Wannigama, D.L.; Amarasiri, M.; Phattharapornjaroen, P.; Hurst, C.; Modchang, C.; Chadsuthi, S.; Anupong, S.; Miyanaga, K.; Cui, L.; Fernandez, S.; et al. Increased faecal shedding in SARS-CoV-2 variants BA.2.86 and JN.1. Lancet Infect. Dis. 2024, 24, e348–e350. [Google Scholar] [CrossRef]
- Bouhaddou, M.; Reuschl, A.K.; Polacco, B.J.; Thorne, L.G.; Ummadi, M.R.; Ye, C.; Rosales, R.; Pelin, A.; Batra, J.; Jang, G.M.; et al. SARS-CoV-2 variants evolve convergent strategies to remodel the host response. Cell 2023, 186, 4597–4614.e26. [Google Scholar] [CrossRef]
- Poran, A.; Harjanto, D.; Malloy, M.; Arieta, C.M.; Rothenberg, D.A.; Lenkala, D.; van Buuren, M.M.; Addona, T.A.; Rooney, M.S.; Srinivasan, L.; et al. Sequence-based prediction of SARS-CoV-2 vaccine targets using a mass spectrometry-based bioinformatics predictor identifies immunogenic T cell epitopes. Genome Med. 2020, 12, 70. [Google Scholar] [CrossRef]
- Hurst, K.R.; Koetzner, C.A.; Masters, P.S. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J. Virol. 2013, 87, 9159–9172. [Google Scholar] [CrossRef]
- Almazán, F.; Galán, C.; Enjuanes, L. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 2004, 78, 12683–12688. [Google Scholar] [CrossRef] [PubMed]
- Zúñiga, S.; Sola, I.; Moreno, J.L.; Sabella, P.; Plana-Durán, J.; Enjuanes, L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology 2007, 357, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Han, Y.; Zhou, R.; Zhou, Y.; Lei, J. Structural insights into ribonucleoprotein dissociation by nucleocapsid protein interacting with non-structural protein 3 in SARS-CoV-2. Commun. Biol. 2023, 6, 193. [Google Scholar] [CrossRef] [PubMed]
- Cubuk, J.; Alston, J.J.; Incicco, J.J.; Singh, S.; Stuchell-Brereton, M.D.; Ward, M.D.; Zimmerman, M.I.; Vithani, N.; Griffith, D.; Wagoner, J.A.; et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021, 12, 1936. [Google Scholar] [CrossRef] [PubMed]
- Hurst-Hess, K.R.; Kuo, L.; Masters, P.S. Dissection of amino-terminal functional domains of murine coronavirus nonstructural protein 3. J. Virol. 2015, 89, 6033–6047. [Google Scholar] [CrossRef] [PubMed]
- Minkoff, J.M.; tenOever, B. Innate immune evasion strategies of SARS-CoV-2. Nature reviews. Microbiology 2023, 21, 178–194. [Google Scholar] [CrossRef]
- Liu, H.; Bai, Y.; Zhang, X.; Gao, T.; Liu, Y.; Li, E.; Wang, X.; Cao, Z.; Zhu, L.; Dong, Q.; et al. SARS-CoV-2 N Protein Antagonizes Stress Granule Assembly and IFN Production by Interacting with G3BPs to Facilitate Viral Replication. J. Virol. 2022, 96, e0041222. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Johnson, B.A.; Meliopoulos, V.A.; Ju, X.; Zhang, P.; Hughes, M.P.; Wu, J.; Koreski, K.P.; Clary, J.E.; Chang, T.-C.; et al. Interaction between host G3BP and viral nucleocapsid protein regulates SARS-CoV-2 replication and pathogenicity. Cell Rep. 2024, 43, 113965. [Google Scholar] [CrossRef]
- Carlson, C.R.; Asfaha, J.B.; Ghent, C.M.; Howard, C.J.; Hartooni, N.; Safari, M.; Frankel, A.D.; Morgan, D.O. Phosphoregulation of Phase Separation by the SARS-CoV-2 N Protein Suggests a Biophysical Basis for its Dual Functions. Mol. Cell 2020, 80, 1092–1103.e4. [Google Scholar] [CrossRef]
- Wu, C.H.; Chen, P.J.; Yeh, S.H. Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe 2014, 16, 462–472. [Google Scholar] [CrossRef]
- LeBlanc, K.; Lynch, J.; Layne, C.; Vendramelli, R.; Sloan, A.; Tailor, N.; Deschambault, Y.; Zhang, F.; Kobasa, D.; Safronetz, D.; et al. The Nucleocapsid Proteins of SARS-CoV-2 and Its Close Relative Bat Coronavirus RaTG13 Are Capable of Inhibiting PKR- and RNase L-Mediated Antiviral Pathways. Microbiol. Spectr. 2023, 11, e0099423. [Google Scholar] [CrossRef] [PubMed]
- Keane, S.C.; Giedroc, D.P. Solution structure of mouse hepatitis virus (MHV) nsp3a and determinants of the interaction with MHV nucleocapsid (N) protein. J. Virol. 2013, 87, 3502–3515. [Google Scholar] [CrossRef] [PubMed]
- Hurst, K.R.; Ye, R.; Goebel, S.J.; Jayaraman, P.; Masters, P.S. An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J. Virol. 2010, 84, 10276–10288. [Google Scholar] [CrossRef]
- Yaron, T.M.; Heaton, B.E.; Levy, T.M.; Johnson, J.L.; Jordan, T.X.; Cohen, B.M.; Kerelsky, A.; Lin, T.Y.; Liberatore, K.M.; Bulaon, D.K.; et al. Host protein kinases required for SARS-CoV-2 nucleocapsid phosphorylation and viral replication. Sci. Signal. 2022, 15, eabm0808. [Google Scholar] [CrossRef] [PubMed]
- Barona-Gómez, F.; Delaye, L.; Díaz-Valenzuela, E.; Plisson, F.; Cruz-Pérez, A.; Díaz-Sánchez, M.; García-Sepúlveda, C.A.; Sanchez-Flores, A.; Pérez-Abreu, R.; Valencia-Valdespino, F.J.; et al. Phylogenomics and population genomics of SARS-CoV-2 in Mexico during the pre-vaccination stage reveals variants of interest B.1.1.28.4 and B.1.1.222 or B.1.1.519 and the nucleocapsid mutation S194L associated with symptoms. Microb. Genom. 2021, 7, 000684. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.; Puvar, A.; Kumar, D.; Ansari, A.; Pandya, M.; Raval, J.; Patel, Z.; Trivedi, P.; Gandhi, M.; Pandya, L.; et al. Genomic Variations in SARS-CoV-2 Genomes From Gujarat: Underlying Role of Variants in Disease Epidemiology. Front. Genet. 2021, 12, 586569. [Google Scholar] [CrossRef]
- Adly, A.; Bi, M.; Carlson, C.; Syed, A.M.; Ciling, A.; Doudna, J.; Cheng, Y.; Morgan, D.O. Assembly of SARS-CoV-2 ribonucleosomes by truncated N* variant of the nucleocapsid protein. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cheng, Z.; Wang, F.; Chang, J.; Zhao, Q.; Zhou, H.; Liu, C.; Ruan, J.; Duan, G.; Gao, S. A Negative Feedback Model to Explain Regulation of SARS-CoV-2 Replication and Transcription. Front. Genet. 2021, 12, 641445. [Google Scholar] [CrossRef]
- Bouhaddou, M.; Reuschl, A.-K.; Polacco, B.J.; Thorne, L.G.; Ummadi, M.R.; Ye, C.; Rosales, R.; Pelin, A.; Batra, J.; Jang, G.M.; et al. Global landscape of the host response to SARS-CoV-2 variants reveals viral evolutionary trajectories. bioRxiv 2022. [Google Scholar] [CrossRef]
- Mourier, T.; Shuaib, M.; Hala, S.; Mfarrej, S.; Alofi, F.; Naeem, R.; Alsomali, A.; Jorgensen, D.; Subudhi, A.K.; Ben Rached, F.; et al. SARS-CoV-2 genomes from Saudi Arabia implicate nucleocapsid mutations in host response and increased viral load. Nat. Commun. 2022, 13, 601. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Xing, N.; Meng, K.; Fu, B.; Xue, W.; Dong, P.; Tang, W.; Xiao, Y.; Liu, G.; Luo, H.; et al. Nucleocapsid mutations R203K/G204R increase the infectivity, fitness, and virulence of SARS-CoV-2. Cell Host Microbe 2021, 29, 1788–1801.e6. [Google Scholar] [CrossRef]
- Bozidis, P.; Tsaousi, E.T.; Kostoulas, C.; Sakaloglou, P.; Gouni, A.; Koumpouli, D.; Sakkas, H.; Georgiou, I.; Gartzonika, K. Unusual N Gene Dropout and Ct Value Shift in Commercial Multiplex PCR Assays Caused by Mutated SARS-CoV-2 Strain. Diagnostics 2022, 12, 973. [Google Scholar] [CrossRef]
- Harankhedkar, S.; Chatterjee, G.; Rajpal, S.; Khan, A.; Srivastava, T.; Mirgh, S.; Gokarn, A.; Punatar, S.; Shetty, N.; Joshi, A.; et al. N Gene Target Failure (NGTF) for detection of Omicron: A way out for the ‘stealth’ too? medRxiv 2022. [Google Scholar] [CrossRef]
- Malone, B.; Urakova, N.; Snijder, E.J.; Campbell, E.A. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nature reviews. Mol. Cell Biol. 2022, 23, 21–39. [Google Scholar] [CrossRef]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Pizzato, M.; Baraldi, C.; Boscato Sopetto, G.; Finozzi, D.; Gentile, C.; Gentile, M.D.; Marconi, R.; Paladino, D.; Raoss, A.; Riedmiller, I.; et al. SARS-CoV-2 and the Host Cell: A Tale of Interactions. Front. Virol. 2022, 1, 815388. [Google Scholar] [CrossRef]
- Fehr, A.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. In Coronaviruses: Methods and Protocols; Maier, H.J., Bickerton, E., Eds.; Methods in Molecular Biology; Humana: New York, NY, USA, 2015; Volume 1282. [Google Scholar]
- Patchett, S.; Lv, Z.; Rut, W.; Békés, M.; Drag, M.; Olsen, S.K.; Huang, T.T. A molecular sensor determines the ubiquitin substrate specificity of SARS-CoV-2 papain-like protease. Cell Rep. 2021, 36, 109754. [Google Scholar] [CrossRef]
- Wolff, G.; Limpens, R.; Zevenhoven-Dobbe, J.C.; Laugks, U.; Zheng, S.; de Jong, A.W.M.; Koning, R.I.; Agard, D.A.; Grünewald, K.; Koster, A.J.; et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 2020, 369, 1395–1398. [Google Scholar] [CrossRef] [PubMed]
- Snijder, E.J.; Limpens, R.; de Wilde, A.H.; de Jong, A.W.M.; Zevenhoven-Dobbe, J.C.; Maier, H.J.; Faas, F.; Koster, A.J.; Bárcena, M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020, 18, e3000715. [Google Scholar] [CrossRef]
- Zimmermann, L.; Zhao, X.; Makroczyova, J.; Wachsmuth-Melm, M.; Prasad, V.; Hensel, Z.; Bartenschlager, R.; Chlanda, P. SARS-CoV-2 nsp3 and nsp4 are minimal constituents of a pore spanning replication organelle. Nat. Commun. 2023, 14, 7894. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.; Ulasli, M.; Schepers, H.; Mauthe, M.; V’Kovski, P.; Kriegenburg, F.; Thiel, V.; de Haan, C.A.M.; Reggiori, F. Nucleocapsid Protein Recruitment to Replication-Transcription Complexes Plays a Crucial Role in Coronaviral Life Cycle. J. Virol. 2020, 94, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Bessa, L.M.; Guseva, S.; Camacho-Zarco, A.R.; Salvi, N.; Maurin, D.; Perez, L.M.; Botova, M.; Malki, A.; Nanao, M.; Jensen, M.R.; et al. The intrinsically disordered SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a. Sci. Adv. 2022, 8, eabm4034. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Xue, B.; Schnicker, N.J.; Wong, L.R.; Meyerholz, D.K.; Perlman, S. Nsp3-N interactions are critical for SARS-CoV-2 fitness and virulence. Proc. Natl. Acad. Sci. USA 2023, 120, e2305674120. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Ma-Lauer, Y.; Han, Y.; Thoms, M.; Buschauer, R.; Jores, J.; Thiel, V.; Beckmann, R.; Deng, W.; Leonhardt, H.; et al. The SARS-unique domain (SUD) of SARS-CoV and SARS-CoV-2 interacts with human Paip1 to enhance viral RNA translation. EMBO J. 2021, 40, e102277. [Google Scholar] [CrossRef] [PubMed]
- Zandi, M.; Shafaati, M.; Kalantar-Neyestanaki, D.; Pourghadamyari, H.; Fani, M.; Soltani, S.; Kaleji, H.; Abbasi, S. The role of SARS-CoV-2 accessory proteins in immune evasion. Biomed. Pharmacother. 2022, 156, 113889. [Google Scholar] [CrossRef] [PubMed]
- Kehrer, T.; Cupic, A.; Ye, C.; Yildiz, S.; Bouhaddou, M.; Crossland, N.A.; Barrall, E.A.; Cohen, P.; Tseng, A.; Çağatay, T.; et al. Impact of SARS-CoV-2 ORF6 and its variant polymorphisms on host responses and viral pathogenesis. Cell Host Microbe 2023, 31, 1668–1684.e12. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; Gueli, F.; Gruber, C.; Giombini, E.; Maggi, F. Analysis of SARS-CoV-2 sublineages reported from local cases in mainland China (September 2022-January 2023) and surveillance of outbound passengers. J. Clin. Virol. Plus 2023, 3, 100153. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Buhr, H.d.; Praest, P.; Evers, A.; Brak-Boer, I.; Grinsven, M.v.; Longo, Y.; Vries, L.d.; Nijenhuis, W.; Kapitein, L.C.; et al. The SARS-CoV-2 accessory factor ORF7a downregulates MHC class I surface expression. bioRxiv 2022. [Google Scholar] [CrossRef]
- Timilsina, U.; Duffy, S.; Plianchaisuk, A.; Ito, J.; Sato, K.; Stavrou, S. SARS-CoV-2 ORF7a mutation found in BF.5 and BF.7 sublineages impacts its functions. Int. J. Mol. Sci. 2024, 25, 2351. [Google Scholar] [CrossRef]
- Kriplani, N.; Clohisey, S.; Fonseca, S.; Fletcher, S.; Lee, H.-M.; Ashworth, J.; Kurian, D.; Lycett, S.J.; Tait-Burkard, C.; Baillie, J.K.; et al. Secreted SARS-CoV-2 ORF8 modulates the cytokine expression profile of human macrophages. bioRxiv 2021. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Y.; Li, Y.; Huang, F.; Luo, B.; Yuan, Y.; Xia, B.; Ma, X.; Yang, T.; Yu, F.; et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc. Natl. Acad. Sci. USA 2021, 118, e2024202118. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, M.; Lucas, C.; Monteiro, V.S.; Iwasaki, A. Enhanced inhibition of MHC-I expression by SARS-CoV-2 Omicron subvariants. Proc. Natl. Acad. Sci. USA 2023, 120, e2221652120. [Google Scholar] [CrossRef] [PubMed]
- Kee, J.; Thudium, S.; Renner, D.M.; Glastad, K.; Palozola, K.; Zhang, Z.; Li, Y.; Lan, Y.; Cesare, J.; Poleshko, A.; et al. SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry. Nature 2022, 610, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lu, Z.; Yang, X.; Zhou, Y.; Gao, J.; Zhang, S.; Huang, S.; Cai, J.; Yu, J.; Zhao, W.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 ORF8 Protein Inhibits Type I Interferon Production by Targeting HSP90B1 Signaling. Front. Cell. Infect. Microbiol. 2022, 12, 899546. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, W.; Wang, T.; Wang, J.; Jiang, Y.; Wang, X.; Qiu, Z.; Feng, N.; Sun, W.; Li, C.; et al. SARS-CoV-2 ORF8 Protein Induces Endoplasmic Reticulum Stress-like Responses and Facilitates Virus Replication by Triggering Calnexin: An Unbiased Study. J. Virol. 2023, 97, e0001123. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Fu, B.; Yin, S.; Li, Z.; Liu, H.; Zhang, H.; Xing, N.; Wang, Y.; Xue, W.; Xiong, Y.; et al. ORF8 contributes to cytokine storm during SARS-CoV-2 infection by activating IL-17 pathway. iScience 2021, 24, 102293. [Google Scholar] [CrossRef] [PubMed]
- Hisner, R.; Gueli, F.; Peacock, T. Repeated Loss of ORF8 Expression in Circulating SARS-CoV-2 Lineages. Available online: https://virological.org/t/repeated-loss-of-orf8-expression-in-circulating-sars-cov-2-lineages/931 (accessed on 1 August 2024).
- Tamura, T.; Irie, T.; Deguchi, S.; Yajima, H.; Tsuda, M.; Nasser, H.; Mizuma, K.; Plianchaisuk, A.; Suzuki, S.; Uriu, K.; et al. Virological characteristics of the SARS-CoV-2 XBB.1.5 variant. bioRxiv 2023. [Google Scholar] [CrossRef]
- Thorne, L.G.; Bouhaddou, M.; Reuschl, A.K.; Zuliani-Alvarez, L.; Polacco, B.; Pelin, A.; Batra, J.; Whelan, M.V.X.; Hosmillo, M.; Fossati, A.; et al. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature 2022, 602, 487–495. [Google Scholar] [CrossRef]
- Parker, M.D.; Stewart, H.; Shehata, O.M.; Lindsey, B.B.; Shah, D.R.; Hsu, S.; Keeley, A.J.; Partridge, D.G.; Leary, S.; Cope, A.; et al. Altered subgenomic RNA abundance provides unique insight into SARS-CoV-2 B.1.1.7/Alpha variant infections. Commun. Biol. 2022, 5, 666. [Google Scholar] [CrossRef]
- McBride, D.S.; Garushyants, S.K.; Franks, J.; Magee, A.F.; Overend, S.H.; Huey, D.; Williams, A.M.; Faith, S.A.; Kandeil, A.; Trifkovic, S.; et al. Accelerated evolution of SARS-CoV-2 in free-ranging white-tailed deer. Nat. Commun. 2023, 14, 5105. [Google Scholar] [CrossRef]
- Stukalov, A.; Girault, V.; Grass, V.; Karayel, O.; Bergant, V.; Urban, C.; Haas, D.A.; Huang, Y.; Oubraham, L.; Wang, A.; et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature 2021, 594, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Kemp, S.A.; Collier, D.A.; Datir, R.P.; Ferreira, I.A.T.M.; Gayed, S.; Jahun, A.; Hosmillo, M.; Rees-Spear, C.; Mlcochova, P.; Lumb, I.U.; et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021, 592, 277–282. [Google Scholar] [CrossRef]
- Wilkinson, S.A.J.; Richter, A.; Casey, A.; Osman, H.; Mirza, J.D.; Stockton, J.; Quick, J.; Ratcliffe, L.; Sparks, N.; Cumley, N.; et al. Recurrent SARS-CoV-2 mutations in immunodeficient patients. Virus Evol. 2022, 8, veac050. [Google Scholar] [CrossRef]
- Lin, X.; Sha, Z.; Trimpert, J.; Kunec, D.; Jiang, C.; Xiong, Y.; Xu, B.; Zhu, Z.; Xue, W.; Wu, H. The NSP4 T492I mutation increases SARS-CoV-2 infectivity by altering non-structural protein cleavage. Cell Host Microbe 2023, 31, 1170–1184.e7. [Google Scholar] [CrossRef]
- Willett, J.D.S.; Gravel, A.; Dubuc, I.; Gudimard, L.; dos Santos Pereira Andrade, A.C.; Lacasse, É.; Fortin, P.; Liu, J.-L.; Cervantes, J.A.; Galvez, J.H.; et al. SARS-CoV-2 rapidly evolves lineage-specific phenotypic differences when passaged repeatedly in immune-naïve mice. Commun. Biol. 2024, 7, 191. [Google Scholar] [CrossRef] [PubMed]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20. [Google Scholar] [CrossRef]
- Dadonaite, B.; Brown, J.; McMahon, T.E.; Farrell, A.G.; Asarnow, D.; Stewart, C.; Logue, J.; Murrell, B.; Chu, H.Y.; Veesler, D.; et al. Full-spike deep mutational scanning helps predict the evolutionary success of SARS-CoV-2 clades. bioRxiv 2023. [Google Scholar] [CrossRef]
- Zahradník, J.; Marciano, S.; Shemesh, M.; Zoler, E.; Harari, D.; Chiaravalli, J.; Meyer, B.; Rudich, Y.; Li, C.; Marton, I.; et al. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat. Microbiol. 2021, 6, 1188–1198. [Google Scholar] [CrossRef]
- Bloom, J.D.; Neher, R.A. Fitness effects of mutations to SARS-CoV-2 proteins. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Mears, H.V.; Young, G.R.; Sanderson, T.; Harvey, R.; Crawford, M.; Snell, D.M.; Fowler, A.S.; Hussain, S.; Nicod, J.; Peacock, T.P.; et al. Emergence of new subgenomic mRNAs in SARS-CoV-2. bioRxiv 2022. [Google Scholar] [CrossRef]
- Bräutigam, K.; Reinhard, S.; Wartenberg, M.; Forster, S.; Greif, K.; Granai, M.; Bösmüller, H.; Klingel, K.; Schürch, C.M. Comprehensive analysis of SARS-CoV-2 receptor proteins in human respiratory tissues identifies alveolar macrophages as potential virus entry site. Histopathology 2023, 82, 846–859. [Google Scholar] [CrossRef] [PubMed]
- McBroome, J.; Schneider, A.d.B.; Roemer, C.; Wolfinger, M.T.; Hinrichs, A.S.; O’Toole, A.N.; Ruis, C.; Turakhia, Y.; Rambaut, A.; Corbett-Detig, R. Automated Agnostic Designation of Pathogen Lineages. bioRxiv 2023. [Google Scholar] [CrossRef]
- Smith, K.; Ye, C.; Turakhia, Y. Tracking and curating putative SARS-CoV-2 recombinants with RIVET. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D. A Web Tool to Estimate Baseline Anti-Spike Monoclonal Antibody Efficacy Based on Regional Genomic Surveillance. Viruses 2023, 15, 1048. [Google Scholar] [CrossRef]
- Pung, R.; Kong, X.P.; Cui, L.; Chae, S.-R.; Chen, M.I.C.; Lee, V.J.; Ho, Z.J.M. Severity of SARS-CoV-2 Omicron XBB subvariants in Singapore. Lancet Reg. Health-West. Pac. 2023, 37, 100849. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Focosi, D.; Spezia, P.G.; Maggi, F. Subsequent Waves of Convergent Evolution in SARS-CoV-2 Genes and Proteins. Vaccines 2024, 12, 887. https://doi.org/10.3390/vaccines12080887
Focosi D, Spezia PG, Maggi F. Subsequent Waves of Convergent Evolution in SARS-CoV-2 Genes and Proteins. Vaccines. 2024; 12(8):887. https://doi.org/10.3390/vaccines12080887
Chicago/Turabian StyleFocosi, Daniele, Pietro Giorgio Spezia, and Fabrizio Maggi. 2024. "Subsequent Waves of Convergent Evolution in SARS-CoV-2 Genes and Proteins" Vaccines 12, no. 8: 887. https://doi.org/10.3390/vaccines12080887





