Molecular Design of Encapsulin Protein Nanoparticles to Display Rotavirus Antigens for Enhancing Immunogenicity
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid DNA (pDNA)
2.2. Production and Purification of Recombinant Protein pDNAs
2.3. Production of ENC-Based Nanoparticles (NPs)
2.3.1. Cell Lysis and Initial Preparation
2.3.2. Purification of ENC-Based NPs
2.3.3. Final Purification and Quality Control
2.4. Characterization of ENC-Based NPs with TEM
2.5. DLS Analysis of ENC-Based NPs
2.6. Mouse Immunizations with ENC-Based NPs
2.7. Detection of Binding Antibodies by Enzyme-Linked Immunosorbent Assay
2.8. Measurement of Neutralization Activity in Serum against Rotavirus
2.9. Statistical Analyses
3. Results
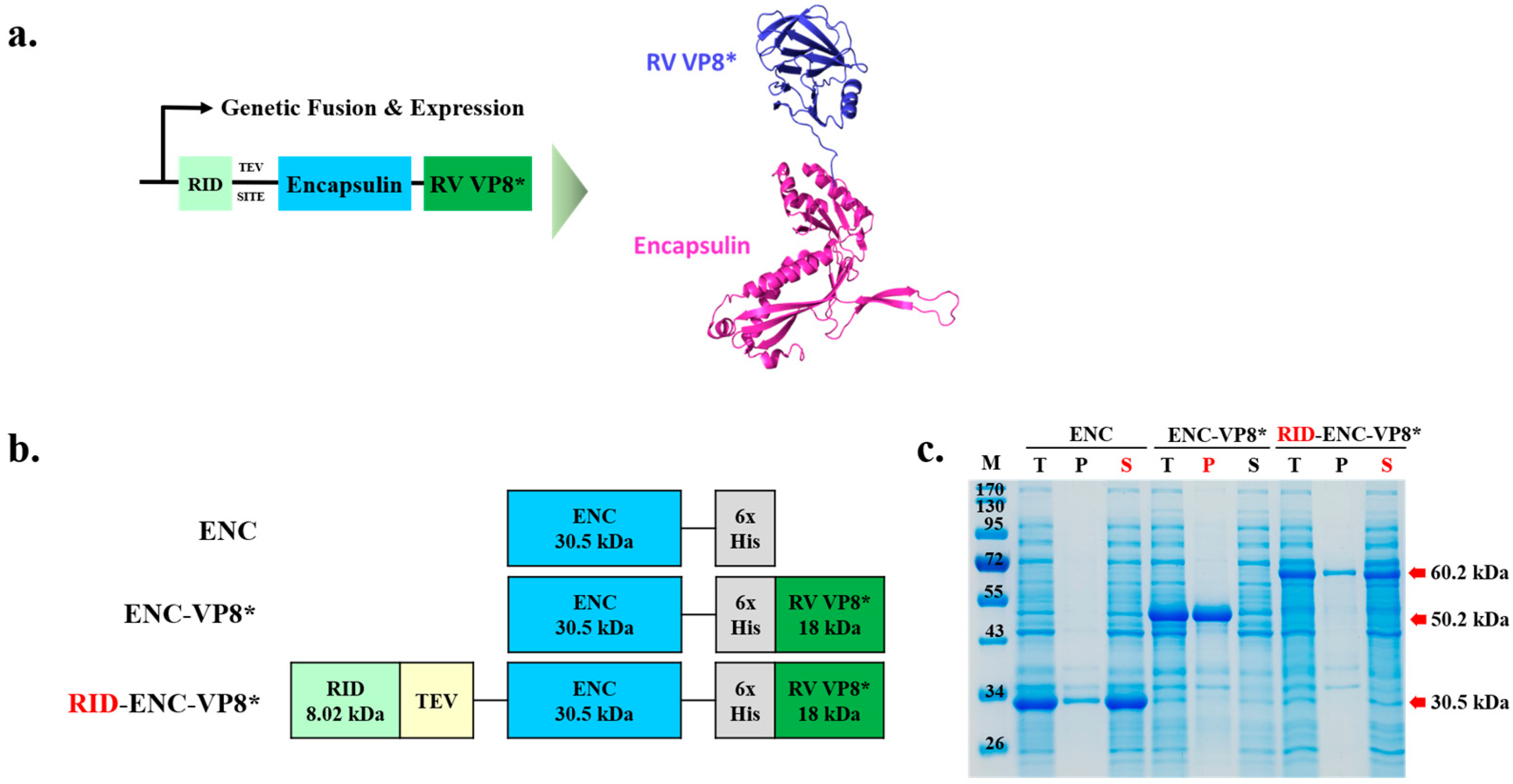
3.1. Construction of Expression Vectors for the Soluble Expression of ENC Rotavirus VP8* Fusion Protein in E. coli
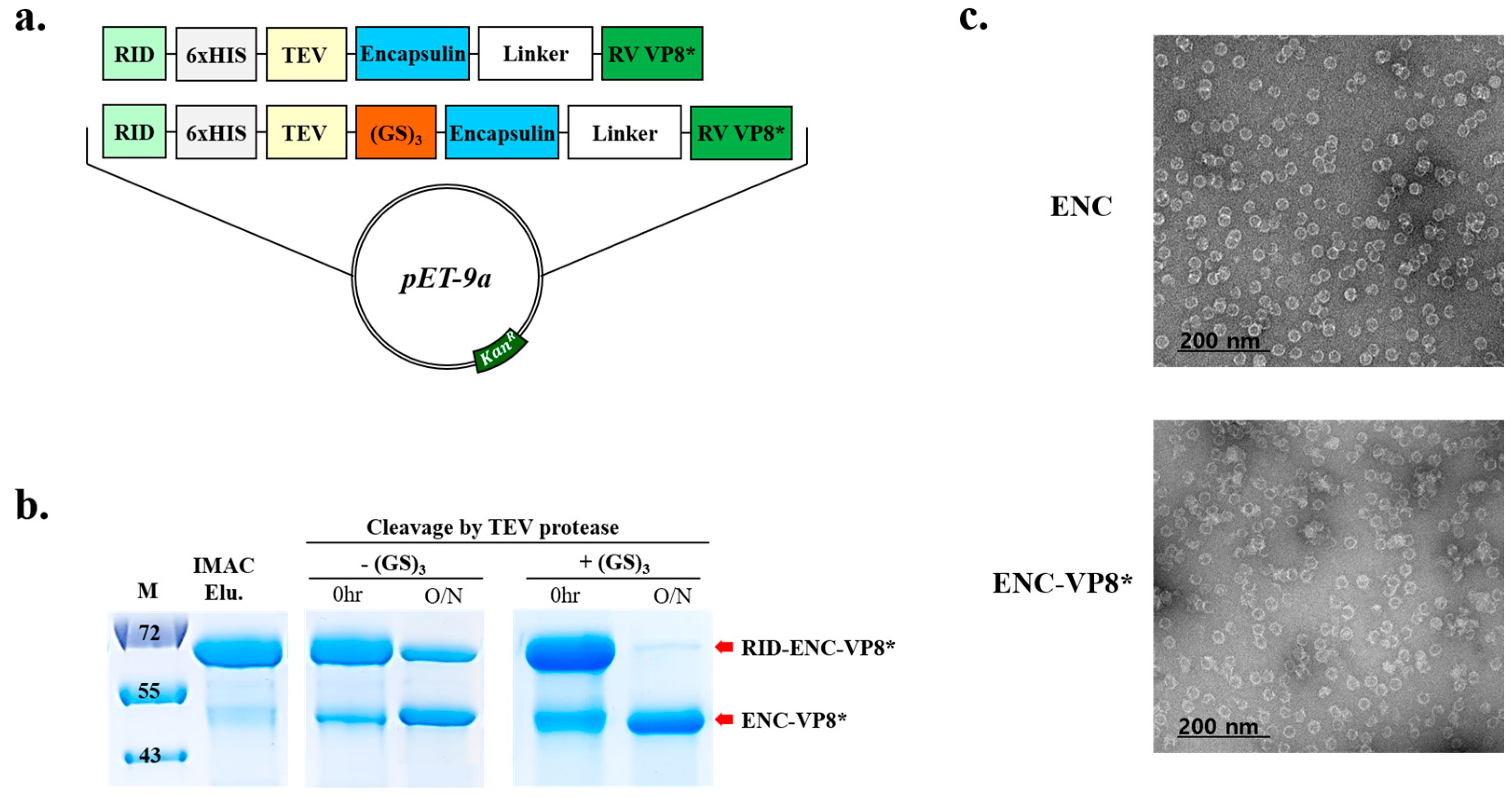
3.2. Enhanced Efficiency of TEV Cleavage and Assembly of ENC-VP8* via the Introduction of (GS)3 Linker
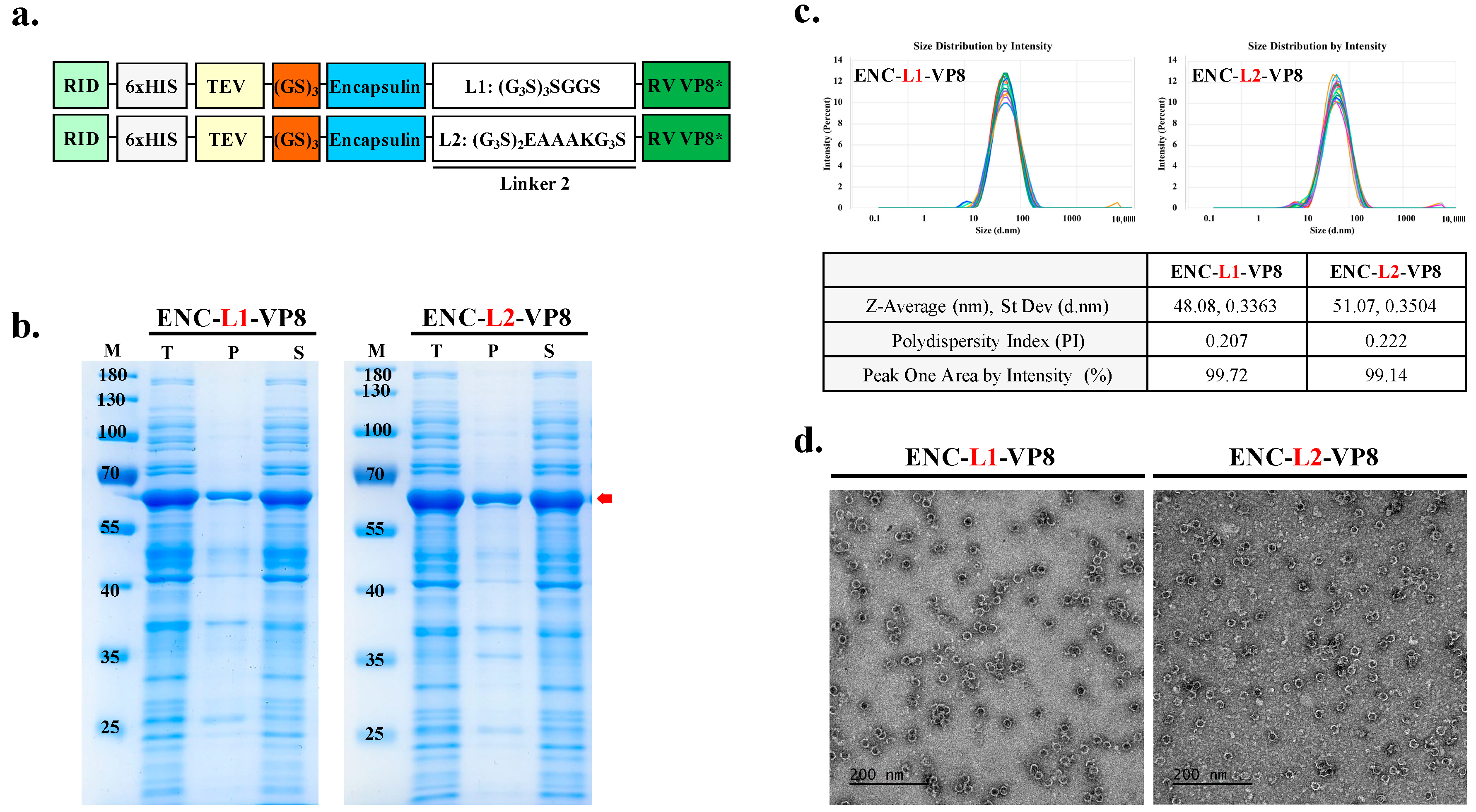
3.3. Role of L2 in Enhancing Solubility and High-Yield Expression of ENC-VP8* NPs
3.4. Simple Two-Step Purification Process Enables the Production of ENC-VP8* NPs in a Stable and Efficient Manner
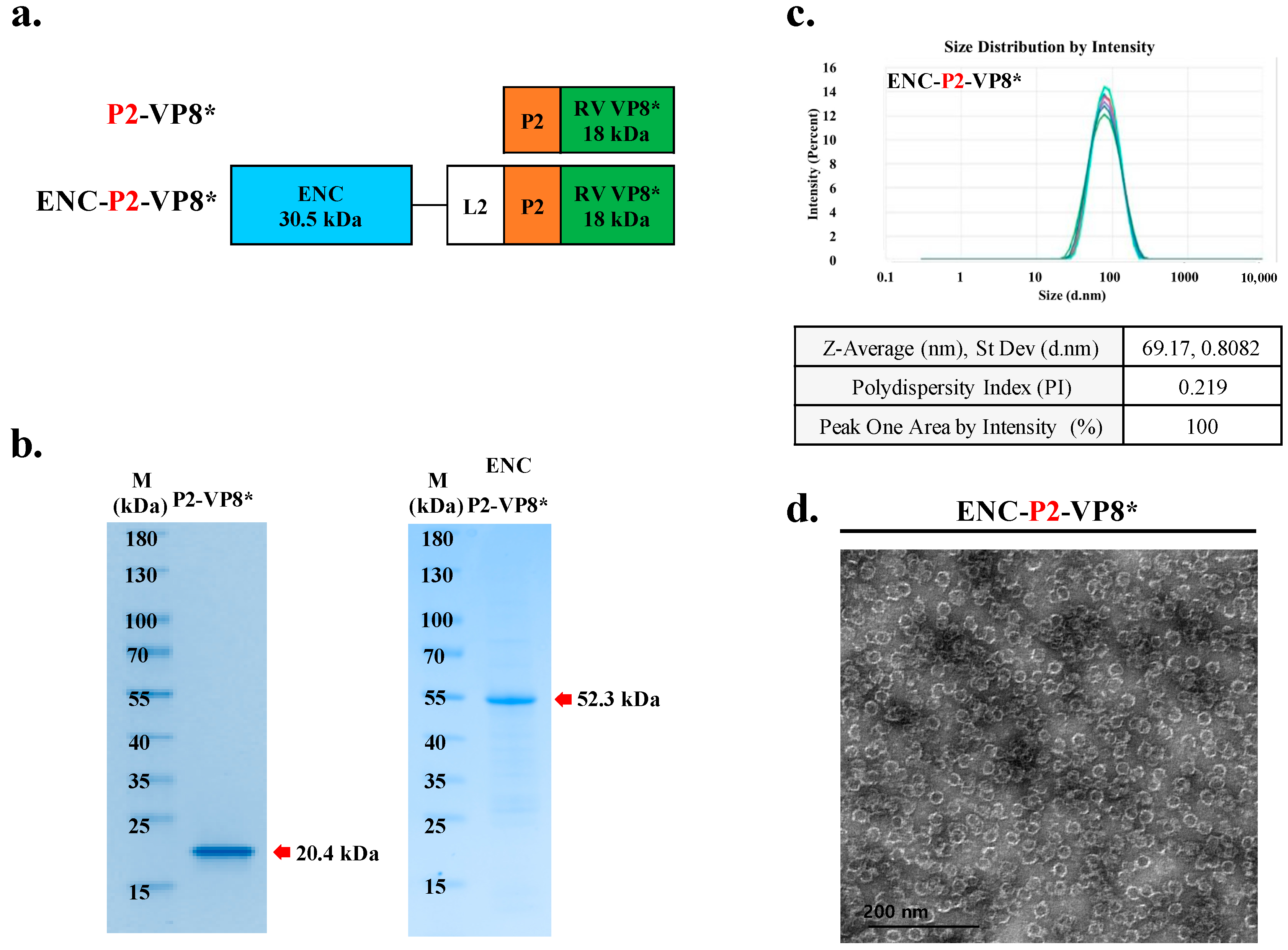
3.5. Preparation of ENC-P2-VP8* and P2-VP8* for the Comparison Study
3.6. NP-Based ENC-P2-VP8* Enhances Binding and Neutralizing Antibody Responses Compared to the Subunit P2-VP8* in Mice
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burnett, E.; Jonesteller, C.L.; Tate, J.E.; Yen, C.; Parashar, U.D. Global Impact of Rotavirus Vaccination on Childhood Hospitalizations and Mortality from Diarrhea. J. Infect. Dis. 2017, 215, 1666–1672. [Google Scholar] [CrossRef] [PubMed]
- Angel, J.; Franco, M.A.; Greenberg, H.B. Rotavirus Vaccines: Recent Developments and Future Considerations. Nat. Rev. Microbiol. 2007, 5, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Vetter, V.; Gardner, R.C.; Debrus, S.; Benninghoff, B.; Pereira, P. Established and New Rotavirus Vaccines: A Comprehensive Review for Healthcare Professionals. Hum. Vaccin. Immunother. 2022, 18, 1870395. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, L.; Yu, H.; Lv, P.; Lei, Z.; Zeng, Y.; Liu, G.; Cheng, T. Ferritin Nanocage-Based Antigen Delivery Nanoplatforms: Epitope Engineering for Peptide Vaccine Design. Biomater. Sci. 2019, 7, 1794–1800. [Google Scholar] [CrossRef]
- Negahdaripour, M.; Golkar, N.; Hajighahramani, N.; Kianpour, S.; Nezafat, N.; Ghasemi, Y. Harnessing Self-Assembled Peptide Nanoparticles in Epitope Vaccine Design. Biotechnol. Adv. 2017, 35, 575–596. [Google Scholar] [CrossRef]
- Rodrigues, M.Q.; Alves, P.M.; Roldão, A. Functionalizing Ferritin Nanoparticles for Vaccine Development. Pharmaceutics 2021, 13, 1621. [Google Scholar] [CrossRef]
- Bellido, D.; Craig, P.O.; Mozgovoj, M.V.; Gonzalez, D.D.; Wigdorovitz, A.; Goldbaum, F.A.; Dus Santos, M.J. Brucella spp. Lumazine Synthase as a Bovine Rotavirus Antigen Delivery System. Vaccine 2009, 27, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.M.; Allende-Ballestero, C.; Cornelissen, J.J.L.M.; Castón, J.R. Nanotechnological Applications Based on Bacterial Encapsulins. Nanomaterials 2021, 11, 1467. [Google Scholar] [CrossRef]
- Ross, J.; McIver, Z.; Lambert, T.; Piergentili, C.; Bird, J.E.; Gallagher, K.J.; Cruickshank, F.L.; James, P.; Zarazúa-Arvizu, E.; Horsfall, L.E.; et al. Pore Dynamics and Asymmetric Cargo Loading in an Encapsulin Nanocompartment. Sci. Adv. 2022, 8, eabj4461. [Google Scholar] [CrossRef]
- Hwang, B.J.; Jang, Y.; Kwon, S.B.; Yu, J.E.; Lim, J.; Roh, Y.H.; Seong, B.L. RNA-Assisted Self-Assembly of Monomeric Antigens into Virus-like Particles as a Recombinant Vaccine Platform. Biomaterials 2021, 269, 120650. [Google Scholar] [CrossRef]
- Yang, S.W.; Jang, Y.H.; Kwon, S.B.; Lee, Y.J.; Chae, W.; Byun, Y.H.; Kim, P.; Park, C.; Lee, Y.J.; Kang Kim, C.; et al. Harnessing an RNA-Mediated Chaperone for the Assembly of Influenza Hemagglutinin in an Immunologically Relevant Conformation. FASEB J. 2018, 32, 2658–2675. [Google Scholar] [CrossRef] [PubMed]
- Francin, M.; Kaminska, M.; Kerjan, P.; Mirande, M. The N-Terminal Domain of Mammalian Lysyl-TRNA Synthetase Is a Functional TRNA-Binding Domain. J. Biol. Chem. 2002, 277, 1762–1769. [Google Scholar] [CrossRef] [PubMed]
- Agou, F.; Waller, J.P.; Yang, Y.; Guittet, E.; Gesquière, J.C. Polyanion-Induced α-Helical Structure of a Synthetic 23-Residue Peptide Representing the Lysine-Rich Segment of the N-Terminal Extension of Yeast Cytoplasmic Aspartyl-tRNA Synthetase. Biochemistry 1995, 34, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.B.; Yu, J.E.; Park, C.; Lee, J.; Seong, B.L. Nucleic Acid-Dependent Structural Transition of the Intrinsically Disordered n-Terminal Appended Domain of Human Lysyl-Trna Synthetase. Int. J. Mol. Sci. 2018, 19, 3016. [Google Scholar] [CrossRef]
- Kim, Y.S.; Son, A.; Kim, J.; Kwon, S.B.; Kim, M.H.; Kim, P.; Kim, J.; Byun, Y.H.; Sung, J.; Lee, J.; et al. Chaperna-Mediated Assembly of Ferritin-Based Middle East Respiratory Syndrome-Coronavirus Nanoparticles. Front. Immunol. 2018, 9, 1093. [Google Scholar] [CrossRef]
- Arai, R.; Ueda, H.; Kitayama, A.; Kamiya, N.; Nagamune, T. Design of the Linkers Which Effectively Separate Domains of a Bifunctional Fusion. Protein 2001, 14, 529–532. [Google Scholar] [CrossRef]
- Li, G.; Huang, Z.; Zhang, C.; Dong, B.J.; Guo, R.H.; Yue, H.W.; Yan, L.T.; Xing, X.H. Construction of a Linker Library with Widely Controllable Flexibility for Fusion Protein Design. Appl. Microbiol. Biotechnol. 2016, 100, 215–225. [Google Scholar] [CrossRef]
- Wen, X.; Cao, D.; Jones, R.W.; Li, J.; Szu, S.; Hoshino, Y. Construction and Characterization of Human Rotavirus Recombinant VP8* Subunit Parenteral Vaccine Candidates. Vaccine 2012, 30, 6121–6126. [Google Scholar] [CrossRef]
- Fix, A.D.; Harro, C.; McNeal, M.; Dally, L.; Flores, J.; Robertson, G.; Boslego, J.W.; Cryz, S. Safety and Immunogenicity of a Parenterally Administered Rotavirus VP8 Subunit Vaccine in Healthy Adults. Vaccine 2015, 33, 3766–3772. [Google Scholar] [CrossRef]
- Kim, J.M.; Choi, H.S.; Seong, B.L. The Folding Competence of HIV-1 Tat Mediated by Interaction with TAR RNA. RNA Biol. 2017, 14, 926–937. [Google Scholar] [CrossRef]
- Zinelli, R.; Soni, S.; Cornelissen, J.J.L.M.; Michel-Souzy, S.; Nijhuis, C.A. Charge Transport across Proteins inside Proteins: Tunneling across Encapsulin Protein Cages and the Effect of Cargo Proteins. Biomolecules 2023, 13, 174. [Google Scholar] [CrossRef] [PubMed]
- Putri, R.M.; Fredy, J.W.; Cornelissen, J.J.L.M.; Koay, M.S.T.; Katsonis, N. Labelling Bacterial Nanocages with Photo-Switchable Fluorophores. ChemPhysChem 2016, 17, 1815–1818. [Google Scholar] [CrossRef] [PubMed]
- Blondel, A.; Nageotte, R.; Bedouelle, H. Destabilizing Interactions between the Partners of a Bifunctional Fusion Protein. Protein Eng. 1996, 9, 231–238. [Google Scholar] [CrossRef][Green Version]
- Shih, Y.-P. Self-Cleavage of Fusion Protein in Vivo Using TEV Protease to Yield Native Protein. Protein Sci. 2005, 14, 936–941. [Google Scholar] [CrossRef]
- Chen, X.; Zaro, J.L.; Shen, W.C. Fusion Protein Linkers: Property, Design and Functionality. Adv. Drug Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef]
- Pati, R.; Shevtsov, M.; Sonawane, A. Nanoparticle Vaccines against Infectious Diseases. Front. Immunol. 2018, 9, 2224. [Google Scholar] [CrossRef]
- Chen, H.; Chen, Z.; Wu, B.; Ullah, J.; Zhang, T.; Jia, J.; Wang, H.; Tan, T. Influences of Various Peptide Linkers on the Thermotoga Maritima MSB8 Nitrilase Displayed on the Spore Surface of Bacillus Subtilis. J. Mol. Microbiol. Biotechnol. 2017, 27, 64–71. [Google Scholar] [CrossRef]
- Mohan, T.; Sharma, C.; Bhat, A.A.; Rao, D.N. Modulation of HIV Peptide Antigen Specific Cellular Immune Response by Synthetic α- and β-Defensin Peptides. Vaccine 2013, 31, 1707–1716. [Google Scholar] [CrossRef] [PubMed]
- Nezafat, N.; Ghasemi, Y.; Javadi, G.; Khoshnoud, M.J.; Omidinia, E. A Novel Multi-Epitope Peptide Vaccine against Cancer: An in Silico Approach. J. Theor. Biol. 2014, 349, 121–134. [Google Scholar] [CrossRef]
- Jones, J.A.; Giessen, T.W. Advances in Encapsulin Nanocompartment Biology and Engineering. Biotechnol. Bioeng. 2021, 118, 491–505. [Google Scholar] [CrossRef]
- Cassidy-Amstutz, C.; Oltrogge, L.; Going, C.C.; Lee, A.; Teng, P.; Quintanilla, D.; East-Seletsky, A.; Williams, E.R.; Savage, D.F. Identification of a Minimal Peptide Tag for in Vivo and in Vitro Loading of Encapsulin. Biochemistry 2016, 55, 3461–3468. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.H.; Cai, H.; Steinmetz, N.F. Viral Nanoparticles for Drug Delivery, Imaging, Immunotherapy, and Theranostic Applications. Adv. Drug Deliv. Rev. 2020, 156, 214–235. [Google Scholar] [CrossRef] [PubMed]
- Sutter, M.; Boehringer, D.; Gutmann, S.; Günther, S.; Prangishvili, D.; Loessner, M.J.; Stetter, K.O.; Weber-Ban, E.; Ban, N. Structural Basis of Enzyme Encapsulation into a Bacterial Nanocompartment. Nat. Struct. Mol. Biol. 2008, 15, 939–947. [Google Scholar] [CrossRef]
- Putri, R.M.; Allende-Ballestero, C.; Luque, D.; Klem, R.; Rousou, K.A.; Liu, A.; Traulsen, C.H.H.; Rurup, W.F.; Koay, M.S.T.; Castón, J.R.; et al. Structural Characterization of Native and Modified Encapsulins as Nanoplatforms for in Vitro Catalysis and Cellular Uptake. ACS Nano 2017, 11, 12796–12804. [Google Scholar] [CrossRef]
- Lakatos, K.; McAdams, D.; White, J.A.; Chen, D. Formulation and Preclinical Studies with a Trivalent Rotavirus P2-VP8 Subunit Vaccine. Hum. Vaccin. Immunother. 2020, 16, 1957–1968. [Google Scholar] [CrossRef]
- HogenEsch, H. Mechanism of Immunopotentiation and Safety of Aluminum Adjuvants. Front. Immunol. 2012, 3, 406. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Wei, X.; Ran, X.; Ni, H.; Cao, S.; Zhang, Y. Immunogenicity of Porcine P[6], P[7]-Specific VP8 Rotavirus Subunit Vaccines with a Tetanus Toxoid Universal T Cell Epitope. Vaccine 2015, 33, 4533–4539. [Google Scholar] [CrossRef]
- Moore, J.P.; Klasse, P.J. SARS-CoV-2 Vaccines: “Warp Speed” Needs Mind Melds Not Warped Minds. J. Virol. 2020, 5, 268. [Google Scholar]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like Particles: Preparation, Immunogenicity and Their Roles as Nanovaccines and Drug Nanocarriers. J. Nanobiotechnol. 2021, 19, 59. [Google Scholar] [CrossRef]
- Van Tilbeurgh, M.; Lemdani, K.; Beignon, A.S.; Chapon, C.; Tchitchek, N.; Cheraitia, L.; Lopez, E.M.; Pascal, Q.; Le Grand, R.; Maisonnasse, P.; et al. Predictive Markers of Immunogenicity and Efficacy for Human Vaccines. Vaccines 2021, 9, 579. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Zha, L.; Cabral-Miranda, G.; Bachmann, M.F. Major Findings and Recent Advances in Virus–like Particle (VLP)-Based Vaccines. Semin. Immunol. 2017, 34, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Curley, S.M.; Putnam, D. Biological Nanoparticles in Vaccine Development. Front. Bioeng. Biotechnol. 2022, 10, 867119. [Google Scholar] [CrossRef]
- Zepeda-Cervantes, J.; Ramírez-Jarquín, J.O.; Vaca, L. Interaction Between Virus-Like Particles (VLPs) and Pattern Recognition Receptors (PRRs) From Dendritic Cells (DCs): Toward Better Engineering of VLPs. Front. Immunol. 2020, 11, 1100. [Google Scholar] [CrossRef] [PubMed]
- Hathout, R.M.; Metwally, A.A.; Woodman, T.J.; Hardy, J.G. Prediction of Drug Loading in the Gelatin Matrix Using Computational Methods. ACS Omega 2020, 5, 1549–1556. [Google Scholar] [CrossRef]
- Liu, S.; Hu, M.; Liu, X.; Liu, X.; Chen, T.; Zhu, Y.; Liang, T.; Xiao, S.; Li, P.; Ma, X. Nanoparticles and Antiviral Vaccines. Vaccines 2024, 12, 30. [Google Scholar] [CrossRef]
- Diaz, D.; Care, A.; Sunna, A. Bioengineering Strategies for Protein-Based Nanoparticles. Genes 2018, 9, 370. [Google Scholar] [CrossRef] [PubMed]
- Schiller, J.; Lowy, D. Explanations for the High Potency of HPV Prophylactic Vaccines. Vaccine 2018, 36, 4768–4773. [Google Scholar] [CrossRef]
- Schiller, J.T.; Castellsagué, X.; Garland, S.M. A Review of Clinical Trials of Human Papillomavirus Prophylactic Vaccines. Vaccine 2012, 30, F123–F138. [Google Scholar] [CrossRef] [PubMed]
- De Vincenzo, R.; Conte, C.; Ricci, C.; Scambia, G.; Capelli, G. Long-Term Efficacy and Safety of Human Papillomavirus Vaccination. Int. J. Womens Health 2014, 6, 999–1010. [Google Scholar] [CrossRef]
- Fougeroux, C.; Goksøyr, L.; Idorn, M.; Soroka, V.; Myeni, S.K.; Dagil, R.; Janitzek, C.M.; Søgaard, M.; Aves, K.L.; Horsted, E.W.; et al. Capsid-like Particles Decorated with the SARS-CoV-2 Receptor-Binding Domain Elicit Strong Virus Neutralization Activity. Nat. Commun. 2021, 12, 324. [Google Scholar] [CrossRef]
- Jain, S.; Pillai, J. Bacterial Membrane Vesicles as Novel Nanosystems for Drug Delivery. Int. J. Nanomed. 2017, 12, 6329–6341. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, B.; Ou, L.; Qiu, Q.; Wang, L.; Bylund, T.; Kong, W.P.; Shi, W.; Tsybovsky, Y.; Wu, L.; et al. Extraordinary Titer and Broad Anti-SARS-CoV-2 Neutralization Induced by Stabilized RBD Nanoparticles from Strain BA.5. Vaccines 2024, 12, 37. [Google Scholar] [CrossRef]
- Yurkova, M.S.; Sharapova, O.A.; Zenin, V.A.; Fedorov, A.N. Versatile Format of Minichaperone-Based Protein Fusion System. Sci. Rep. 2019, 9, 15063. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Wen, K.; Cao, D.; Li, G.; Jones, R.W.; Li, J.; Szu, S.; Hoshino, Y.; Yuan, L. Inclusion of a Universal Tetanus Toxoid CD4+ T Cell Epitope P2 Significantly Enhanced the Immunogenicity of Recombinant Rotavirus ΔVP8* Subunit Parenteral Vaccines. Vaccine 2014, 32, 4420–4427. [Google Scholar] [CrossRef] [PubMed]
- Groome, M.J.; Koen, A.; Fix, A.; Page, N.; Jose, L.; Madhi, S.A.; McNeal, M.; Dally, L.; Cho, I.; Power, M.; et al. Safety and Immunogenicity of a Parenteral P2-VP8-P[8] Subunit Rotavirus Vaccine in Toddlers and Infants in South Africa: A Randomised, Double-Blind, Placebo-Controlled Trial. Lancet Infect. Dis. 2017, 17, 843–853. [Google Scholar] [CrossRef]
- Groome, M.J.; Fairlie, L.; Morrison, J.; Fix, A.; Koen, A.; Masenya, M.; Jose, L.; Madhi, S.A.; Page, N.; McNeal, M.; et al. Safety and Immunogenicity of a Parenteral Trivalent P2-VP8 Subunit Rotavirus Vaccine: A Multisite, Randomised, Double-Blind, Placebo-Controlled Trial. Lancet Infect. Dis. 2020, 20, 851–863. [Google Scholar] [CrossRef]
- McAdams, D.; Estrada, M.; Holland, D.; Singh, J.; Sawant, N.; Hickey, J.M.; Kumar, P.; Plikaytis, B.; Joshi, S.B.; Volkin, D.B.; et al. Concordance of in Vitro and in Vivo Measures of Non-Replicating Rotavirus Vaccine Potency. Vaccine 2022, 40, 5069–5078. [Google Scholar] [CrossRef]
- Roier, S.; Mangala Prasad, V.; McNeal, M.M.; Lee, K.K.; Petsch, B.; Rauch, S. mRNA-Based VP8* Nanoparticle Vaccines against Rotavirus Are Highly Immunogenic in Rodents. NPJ Vaccines 2023, 8, 190. [Google Scholar] [CrossRef]
- Xia, M.; Huang, P.; Jiang, X.; Tan, M. A Nanoparticle-Based Trivalent Vaccine Targeting the Glycan Binding Vp8* Domains of Rotaviruses. Viruses 2021, 13, 72. [Google Scholar] [CrossRef]
- Ramesh, A.; Mao, J.; Lei, S.; Twitchell, E.; Shiraz, A.; Jiang, X.; Tan, M.; Yuan, L. Parenterally Administered P24-VP8* Nanoparticle Vaccine Conferred Strong Protection against Rotavirus Diarrhea and Virus Shedding in Gnotobiotic Pigs. Vaccines 2019, 7, 177. [Google Scholar] [CrossRef]
- Xia, M.; Huang, P.; Jiang, X.; Tan, M. Immune Response and Protective Efficacy of the S Particle Presented Rotavirus VP8* Vaccine in Mice. Vaccine 2019, 37, 4103–4110. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Feng, J.; Li, J.; Li, J.; Wang, Z.; Khalique, A.; Yang, M.; Ni, X.; Zeng, D.; Zhang, D.; et al. Surface Display of Antigen Protein VP8* of Porcine Rotavirus on Bacillus Subtilis Spores Using COtB as a Fusion Partner. Molecules 2019, 24, 3793. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Huang, P.; Sun, C.; Han, L.; Vago, F.S.; Li, K.; Zhong, W.; Jiang, W.; Klassen, J.S.; Jiang, X.; et al. Bioengineered Norovirus S60 Nanoparticles as a Multifunctional Vaccine Platform. ACS Nano 2018, 12, 10665–10682. [Google Scholar] [CrossRef] [PubMed]
- Giessen, T.W. Encapsulins: Microbial Nanocompartments with Applications in Biomedicine, Nanobiotechnology and Materials Science. Curr. Opin. Chem. Biol. 2016, 34, 1–10. [Google Scholar] [CrossRef]
- Choi, B.; Moon, H.; Hong, S.J.; Shin, C.; Do, Y.; Ryu, S.; Kang, S. Effective Delivery of Antigen-Encapsulin Nanoparticle Fusions to Dendritic Cells Leads to Antigen-Specific Cytotoxic T Cell Activation and Tumor Rejection. ACS Nano 2016, 10, 7339–7350. [Google Scholar] [CrossRef]
- Son, S.; Song, W.J. Programming Interchangeable and Reversible Heterooligomeric Protein Self-Assembly Using a Bifunctional Ligand. Chem. Sci. 2024, 15, 2975–2983. [Google Scholar] [CrossRef]
- Khaleeq, S.; Sengupta, N.; Kumar, S.; Patel, U.R.; Rajmani, R.S.; Reddy, P.; Pandey, S.; Singh, R.; Dutta, S.; Ringe, R.P.; et al. Neutralizing Efficacy of Encapsulin Nanoparticles against SARS-CoV2 Variants of Concern. Viruses 2023, 15, 346. [Google Scholar] [CrossRef]
- Rennie, C.; Sives, C.; Boyton, I.; Diaz, D.; Gorrie, C.; Vittorio, O.; Collins-Praino, L.; Care, A. In Vivo Behavior of Systemically Administered Encapsulin Protein Nanocages and Implications for Their Use in Targeted Drug Delivery. Adv. Ther. 2024, 7, 2300360. [Google Scholar] [CrossRef]
- Nyblade, C.; Hensley, C.; Parreño, V.; Zhou, P.; Frazier, M.; Frazier, A.; Ramesh, A.; Lei, S.; Degiuseppe, J.I.; Tan, M.; et al. A New Gnotobiotic Pig Model of P[6] Human Rotavirus Infection and Disease for Preclinical Evaluation of Rotavirus Vaccines. Viruses 2022, 14, 2803. [Google Scholar] [CrossRef]
- Lamontagne, F.; Khatri, V.; St-Louis, P.; Bourgault, S.; Archambault, D. Vaccination Strategies Based on Bacterial Self-Assembling Proteins as Antigen Delivery Nanoscaffolds. Vaccines 2022, 10, 1920. [Google Scholar] [CrossRef]
- Blutt, S.E.; Conner, M.E. Rotavirus: To the Gut and Beyond! Curr. Opin. Gastroenterol. 2007, 23, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Blutt, S.E.; Matson, D.O.; Crawford, S.E.; Staat, M.A.; Azimi, P.; Bennett, B.L.; Piedra, P.A.; Conner, M.E. Rotavirus Antigenemia in Children Is Associated with Viremia. PLoS Med. 2007, 4, e121. [Google Scholar] [CrossRef] [PubMed]
- Whitton, J.L.; Slifka, M.K.; Liu, F.; Nussbaum, A.K.; Whitmire, J.K. The Regulation and Maturation of Antiviral Immune Responses. Adv. Virus Res. 2004, 63, 181. [Google Scholar] [CrossRef] [PubMed]
- Piersma, S.J. Tissue-Specific Features of Innate Lymphoid Cells in Antiviral Defense. Cell Mol. Immunol. 2024, 21, 1036–1050. [Google Scholar] [CrossRef]
- Xue, M.; Yu, L.; Che, Y.; Lin, H.; Zeng, Y.; Fang, M.; Li, T.; Ge, S.; Xia, N. Characterization and Protective Efficacy in an Animal Model of a Novel Truncated Rotavirus VP8 Subunit Parenteral Vaccine Candidate. Vaccine 2015, 33, 2606–2613. [Google Scholar] [CrossRef]


| Antigen | Dose | ENC-P2-VP8* Mid | ENC-P2-VP8* Low | P2-VP8* High | P2-VP8* Mid | P2-VP8* Low |
|---|---|---|---|---|---|---|
| ENC-P2-VP8* high | 1st | ns (p = 0.6905) | ** (p = 0.0079) | ** (p = 0.0079) | ** (p = 0.0079) | ** (p = 0.0079) |
| 2nd | ns (p = 0.4206) | * (p = 0.0317) | ** (p = 0.0079) | ** (p = 0.0079) | ** (p = 0.0079) | |
| 3rd | ns (p = 0.4206) | ns (p = 0.2222) | * (p = 0.0159) | * (p = 0.0159) | ** (p = 0.0079) | |
| ENC-P2-VP8* mid | 1st | - | * (p = 0.0476) | ns (p = 0.1032) | * (p = 0.0476) | * (p = 0.0476) |
| 2nd | - | * (p = 0.0159) | ** (p = 0.0079) | ** (p = 0.0079) | ** (p = 0.0079) | |
| 3rd | - | ns (p = 0.0952) | ** (p = 0.0079) | ** (p = 0.0079) | ** (p = 0.0079) | |
| ENC-P2-VP8* low | 1st | - | - | ns (p = 0.4444) | ns (p > 0.9999) | ns (p > 0.9999) |
| 2nd | - | - | * (p = 0.0159) | * (p = 0.0159) | ** (p = 0.0079) | |
| 3rd | - | - | ns (p = 0.0556) | ns (p = 0.0556) | ** (p = 0.0079) | |
| P2-VP8* high | 1st | - | - | - | ns (p = 0.4444) | ns (p = 0.4444) |
| 2nd | - | - | - | ns (p = 0.1508) | ** (p = 0.0079) | |
| 3rd | - | - | - | ns (p = 0.2222) | ** (p = 0.0079) | |
| P2-VP8* mid | 1st | - | - | - | - | ns (p > 0.9999) |
| 2nd | - | - | - | - | ** (p = 0.0079) | |
| 3rd | - | - | - | - | ** (p = 0.0079) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, H.-G.; Jeong, S.; Kang, M.-J.; Hong, I.; Park, Y.-S.; Ko, E.; Kim, J.-O.; Choi, D.-Y. Molecular Design of Encapsulin Protein Nanoparticles to Display Rotavirus Antigens for Enhancing Immunogenicity. Vaccines 2024, 12, 1020. https://doi.org/10.3390/vaccines12091020
Jung H-G, Jeong S, Kang M-J, Hong I, Park Y-S, Ko E, Kim J-O, Choi D-Y. Molecular Design of Encapsulin Protein Nanoparticles to Display Rotavirus Antigens for Enhancing Immunogenicity. Vaccines. 2024; 12(9):1020. https://doi.org/10.3390/vaccines12091020
Chicago/Turabian StyleJung, Hyun-Gyo, Seonghun Jeong, Min-Ji Kang, Ingi Hong, Young-Shin Park, Eunbyeol Ko, Jae-Ouk Kim, and Deog-Young Choi. 2024. "Molecular Design of Encapsulin Protein Nanoparticles to Display Rotavirus Antigens for Enhancing Immunogenicity" Vaccines 12, no. 9: 1020. https://doi.org/10.3390/vaccines12091020
APA StyleJung, H.-G., Jeong, S., Kang, M.-J., Hong, I., Park, Y.-S., Ko, E., Kim, J.-O., & Choi, D.-Y. (2024). Molecular Design of Encapsulin Protein Nanoparticles to Display Rotavirus Antigens for Enhancing Immunogenicity. Vaccines, 12(9), 1020. https://doi.org/10.3390/vaccines12091020





